Abstract
Nonsense-mediated mRNA decay (NMD) represents a key mechanism to control the expression of wild-type and aberrant mRNAs. Phosphorylation of the protein UPF1 in the context of translation termination contributes to committing mRNAs to NMD. We report that translation termination is inhibited by UPF1 and stimulated by cytoplasmic poly(A)-binding protein (PABPC1). UPF1 binds to eRF1 and to the GTPase domain of eRF3 both in its GTP- and GDP-bound states. Importantly, mutation studies show that UPF1 can interact with the exon junction complex (EJC) alternatively through either UPF2 or UPF3b to become phosphorylated and to activate NMD. On this basis, we discuss an integrated model where UPF1 halts translation termination and is phosphorylated by SMG1 if the termination-promoting interaction of PABPC1 with eRF3 cannot readily occur. The EJC, with UPF2 or UPF3b as a cofactor, interferes with physiological termination through UPF1. This model integrates previously competing models of NMD and suggests a mechanistic basis for alternative NMD pathways.
Keywords: exon junction complex, NMD, release factors, translation termination, UPF1
Introduction
Nonsense-mediated mRNA decay (NMD) represents one of the core mechanisms that help ensure fidelity of gene expression. NMD has been highly conserved during evolution, and identifies and limits the expression of transcripts with premature termination codons (Hentze and Kulozik, 1999; Maquat, 2004; Behm-Ansmant et al, 2007b; Chang et al, 2007; Isken and Maquat, 2007). Such transcripts encode C-terminally truncated proteins that may be non-functional or could exert an effect in a dominant-negative manner. The biological and medical significance of NMD is highlighted by an increasing number of hereditary and acquired genetic diseases that are modified by the activity of this mRNA surveillance pathway (Holbrook et al, 2004; Neu-Yilik and Kulozik, 2004; Tarpey et al, 2007). Furthermore, NMD also has an important function in the development of novel treatment strategies for diseases that are caused by premature termination codons (Durand et al, 2007; Welch et al, 2007; Linde et al, 2007a, 2007b).
Mechanistically, the mammalian NMD machinery distinguishes a normal stop codon from a premature termination codon in a splicing- and translation-dependent manner (Carter et al, 1996; Thermann et al, 1998; Le Hir et al, 2000; Danckwardt et al, 2002), although in a few cases NMD may also exert an effect independently from splicing (Buhler et al, 2006). Splicing deposits a protein complex 20–24 nt upstream of the splice junction (Kataoka et al, 2000; Le Hir et al, 2000; Lykke-Andersen et al, 2001), which is referred to as the exon junction complex (EJC). The core of the EJC consists of the proteins eIF4AIII, BTZ, Y14 and MAGOH, which is thought to recruit additional components, including UPF3b and UPF2 (Kataoka et al, 2000; Le Hir et al, 2000; Lykke-Andersen et al, 2000, 2001; Gehring et al, 2003, 2005; Chan et al, 2004; Palacios et al, 2004; Bono et al, 2006). In case of premature translation termination, UPF2/UPF3b interact with UPF1 and the release factors eRF1 and eRF3 at the A-site of the terminating ribosome (Kashima et al, 2006), promoting phosphorylation of UPF1 by the PI3-kinase-like SMG1 kinase. This in turn recruits SMG5, SMG6 and SMG7, leading to degradation of the mRNA (Yamashita et al, 2001; Ohnishi et al, 2003; Grimson et al, 2004; Unterholzner and Izaurralde, 2004; Glavan et al, 2006).
This linear model of mammalian NMD is likely oversimplified because examples have been described where NMD can be UPF2 or UPF3b independent, or even occur independently of splicing (Gehring et al, 2005; Buhler et al, 2006; Chan et al, 2007). Furthermore, inactivating UPF3b mutations in humans do not abolish NMD, suggesting the existence of alternative pathways (Tarpey et al, 2007).
In yeast, physiological translation termination requires a permissive mRNP environment. A termination codon is recognized by the NMD machinery, if it is part of an inappropriate mRNP, that has been termed ‘faux 3′-UTR'. Poly(A)-binding protein (Pab1p) appears to represent an important component of a termination-permissive 3′ mRNP because the expression of NMD substrate mRNAs can be rescued by tethering Pab1p downstream of a premature stop codon (Amrani et al, 2004). Tethered cytoplasmic poly(A)-binding protein (PABPC1) has been shown to suppress NMD also in Drosophila cells (Behm-Ansmant et al, 2007a).
In this study, we analyse the interplay between the mammalian release factors eRF1 and eRF3, the NMD proteins UPF1, UPF2, UPF3b, the EJC and PABPC1. On the basis of our results and the published literature, we discuss an integrated model for mammalian NMD. As its novel features, it postulates that UPF1 inhibits the efficiency of translation termination, and that the functionally important phosphorylation of UPF1 is stimulated alternatively by its interaction with UPF2 or UPF3b. This model thus offers a biochemical basis for recently described alternative NMD pathways and for the redundancy of UPF3b in patients with inactivating mutations.
Results
PABPC1 inhibits NMD in a position-dependent manner
In yeast, efficient termination depends on the interaction between the terminating ribosome and a specific 3′ mRNP including cytoplasmic Pab1p. Prematurely terminating ribosomes that encounter an improper, ‘faux', 3′ mRNP, are not released efficiently, and the mRNA is targeted to the NMD pathway. ‘Faux' 3′-UTRs can be ‘repaired' by tethering Pab1p sufficiently close to the translation termination codon (Amrani et al, 2004).
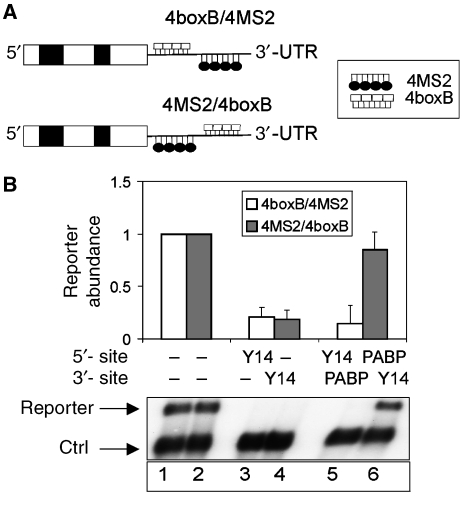
We first analysed the role of PABPC1 and its interplay with the EJC in human NMD. We designed a set of reporter constructs with 4MS2 and 4boxB-binding sites in their 3′-UTRs (Figure 1A), which allows to tether simultaneously two different, λN- and MS2-tagged proteins to the mRNA in a position-defined manner. Importantly, tethering of the EJC proteins Y14, MAGOH and RNPS1 solidly downmodulates reporter mRNA expression in a predictable manner that is independent of the tag and the tethering site within the 3′-UTR (Figure 1B and Supplementary Figure 1). Furthermore, positioning of the multimerizing MS2 coat protein between the termination codon and a tagged EJC protein does not suppress NMD, indicating that the ability of a tethered EJC protein to trigger NMD is not suppressed by mere steric hindrance from a protein tethered between it and the stop codon (Figure 1B, lane 4). We next tested if positioning of PABPC1 between the tethered EJC protein and the termination codon would antagonize NMD. Simultaneous transfection of MS2–PABPC1 and λNV5–Y14 together with the 4MS2/4boxB reporter resulted in a three- to four-fold higher abundance of the reporter mRNA than the same construct following co-transfection of λNV5–Y14 and MS2 without PABPC1 (Figure 1B, compare lanes 4 and 6). This effect is position dependent because PABPC1 tethering 3′ instead of 5′ of the EJC protein does not affect reporter mRNA abundance (Figure 1B, lane 5). Similar results were obtained when tethered RNPS1 or MAGOH was used instead of tethered Y14 (Supplementary Figure 1). These data show that PABPC1 can overcome the effect of tethered EJC proteins when positioned closer to the termination codon.
Figure 1.
Antagonistic relationship of PABPC1 and the EJC in stop codon definition. (A) Schematic representation of the 4boxB/4MS2 and 4MS2/4boxB reporters. White and black boxes represent exons and introns of the β-globin gene, respectively. BoxB and MS2 sites are shown as white boxes or black loops, respectively. (B) Tethering of PABPC1 and the EJC protein Y14. HeLa cells were transfected with the indicated 4boxB/4MS2 or 4MS2/4boxB constructs and the plasmids expressing the MS2–PABPC1 and λNV5–Y14 fusion proteins as indicated. Total RNA samples were analysed by northern blotting. The levels of the 4boxB/4MS2 and the 4MS2/4boxB reporter mRNAs were normalized to the control β-globin reporter (ctrl). For each reporter, mRNA levels of the reporter without any tethered factor (MS2 protein and λN peptide alone) were set as 1.0 (lanes 1 and 2). Values were calculated from three independent experiments, error bars represent standard deviations.
Both the yeast and mammalian poly(A)-binding proteins interact with eRF3 (Hoshino et al, 1999; Cosson et al, 2002a, 2002b). In yeast, this interaction stimulates translation termination and the release of the nascent polypeptide from the ribosome. It is noteworthy that the domain of eRF3 that is directly bound by PABPC1 corresponds to that bound by UPF1 (see below). These data suggest a possible functional inter-relationship between these three proteins in NMD, which we explored by further analyses in mammalian cells.
UPF1 and PABPC1 modulate the efficiency of translation termination
To test the role of mammalian UPF1 in translation termination, we used a readthrough assay in a dual luciferase reporter (Grentzmann et al, 1998; Howard et al, 2001). This reporter is composed of two in-frame open reading frames (ORFs) encoding Renilla and firefly luciferase, respectively, which are interrupted by a stop codon (Figure 2A). The 5′ Renilla luciferase ORF is constitutively translated and serves as an internal standard, against which the expression of the 3′ firefly luciferase ORF is used as a quantitative measure of readthrough by the elongating ribosome. The ratio between firefly luciferase and Renilla luciferase activities thus reflects the efficiency of translation termination. The design of this reporter ensures that potentially nonspecific effects on RNA synthesis, RNA stability or translation are internally controlled.
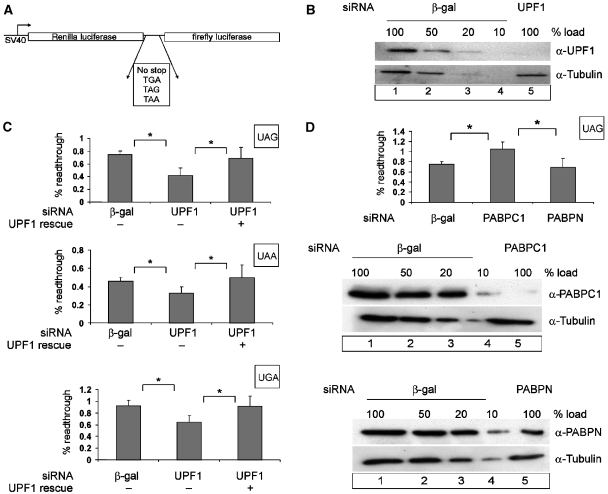
Figure 2.
Effect of UPF1 and PABPC1 on translational readthrough and termination. (A) Schematic representation of the dual luciferase reporters used for measuring translation termination readthrough. (B, C) UPF1 knockdown decreases the level of readthrough at all three stop codons. Protein lysates from HeLa cells that were transfected with siRNAs against β-galactosidase (β-gal) (B, lanes 1–4) or UPF1 (B, lane 5) were immunoblotted with an anti-UPF1 antibody. A dilution series corresponding to 50, 20 and 10% of the protein amount that was used in lane 1 (20 μg) was loaded to assess the efficiency of UPF1 depletion. Reprobing with a tubulin-specific antibody was performed as a loading control. At 48 h after siRNA depletion, HeLa cells were transfected with the dual luciferase construct (p2luc-stop). The percentage of readthrough at the three different stop codons is shown (C). The results obtained with the construct without a stop codon were set as 100% readthrough. To complement the UPF1 depletion, cells were transfected with 0.2 μg of siRNA-resistant FLAG-UPF1R. The values were calculated from six independent depletion experiments, error bars represent standard deviations. *Statistical significance at P<0.05. (D) PABPC1 knockdown increases the level of readthrough at a UAG stop codon. Depletions of PABPC1 and nuclear PABP were achieved by siRNA treatment as described in (B). HeLa cells were transfected with the dual luciferase construct p2luc-TAG or with the construct without an interposed stop codon p2luc-if 48 hours after siRNA treatment. The percentage of readthrough at the UAG stop codon was calculated as described in (C). *Statistical significance at P<0.05.
The ratio of Renilla/firefly luciferase activity in HeLa cells transfected with the reporter lacking an intervening stop codon and hence encoding a fusion protein was defined as 100% readthrough. Cells co-transfected with reporters bearing any of the three stop codons and a β-galactosidase siRNA display readthrough activities of 0.45±0.07% for UAA, 0.75±0.05% for UAG and 0.95±0.06% for UGA (Figure 2C). In cells that were co-transfected with the same reporters but with UPF1-specific siRNA and in which UPF1 levels were depleted to <10% (Figure 2B), readthrough decreased 1.5- to 2-fold (P<0.05) (Figure 2C). The specificity of this effect was controlled by re-establishing termination efficiency with UPF1 constructs that carry a functionally silent mutation of the sequences that were targeted in the endogenous transcript by the siRNA (Gehring et al, 2005; Figure 2C).
Because of the role of the poly(A)-binding protein in translation termination (Cosson et al, 2002b), and because of the interference of this protein with NMD in yeast (Amrani et al, 2004), Drosophila cells (Behm-Ansmant et al, 2007a) and in our functional tethering assays (Figure 1B), we next tested the effect of PABPC1 depletion on translation termination in our reporter system. Interestingly, PABPC1 depletion significantly increases readthrough 1.5-fold (±0.12), in contrast to the depletion of the control, nuclear PABP (PABPN), that lacks the interaction domain with eRF3 (Figure 2D). These data indicate an antagonistic relationship between UPF1 and PABPC1, where UPF1 interferes with and PABPC1 stimulates the efficiency of translation termination. Potentially, the antagonistic effects of these proteins in NMD may relate to their respective roles in translation termination.
Determinants of the interactions between UPF1, eRF1, eRF3 and PABPC1
A role of UPF1 in termination-coupled NMD has been suggested by the identification of the SURF complex consisting of UPF1, its kinase SMG1 and the release factors eRF1 and eRF3 (Kashima et al, 2006; Supplementary Figure 2). Our results described above also implicate PABPC1, and we next characterized the biochemical interactions between these proteins.
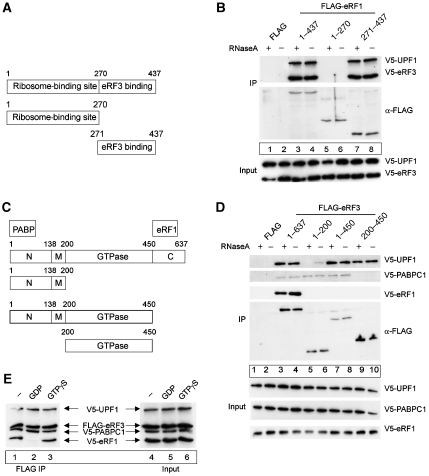
Human eRF1 is composed of two functionally distinct domains (Figure 3A) (Frolova et al, 1994, 2000). The N-terminal part mediates binding to the ribosome and is fully competent as a release factor in translation termination. The C-terminal third of the polypeptide is responsible for binding eRF3 and protein phosphatase 2A (PP2A) (Andjelkovic et al, 1996; Merkulova et al, 1999; Frolova et al, 2000; Song et al, 2000). To identify the region of eRF1 that binds UPF1, we co-transfected HeLa cells with plasmids expressing V5-tagged UPF1 and FLAG-tagged full-length eRF1, or the eRF1 C- and N-terminal domains, respectively. As a positive control, V5-eRF3 was also co-transfected. Following immunoprecipitation of eRF1 with anti-FLAG antibodies, the interaction with UPF1 and eRF3 was analysed by western blotting with anti-V5 antibodies. As predicted, full-length eRF1 and its C-terminal domain (eRF1 271–437) efficiently bind eRF3 (Figure 3B, lanes 3 and 4 and 7 and 8). Of note, UPF1 binds to the C-terminal part of eRF1 (lanes 7 and 8), whereas the N-terminal domain extending from residues 1–270 does not detectably bind UPF1 (lanes 5 and 6). These results indicate that eRF1 either binds UPF1 and eRF3 with the same domain or interacts with UPF1 through eRF3, which in both cases suggests an intricate relationship of these three proteins in translation termination (Figure 7).
Figure 3.
Definition of biochemical interactions between UPF1, release factors and PABPC1. (A, B) UPF1 and eRF3 interact with the C-terminal part of eRF1. (A) Schematic representation of the domain structure of human release factor eRF1. The ribosome- and the eRF3-binding domain are depicted. (B) Co-immunoprecipitation experiments with eRF1 fragments. HeLa cells were co-transfected with an empty FLAG plasmid (lanes 1 and 2), with FLAG-eRF1 (lanes 3 and 4) or the indicated FLAG-tagged fragments of eRF1 (lanes 5–8) together with a plasmid for V5-UPF1 and V5-eRF3. Immunoprecipitations were carried out in the presence or absence of RNaseA. V5-tagged proteins were detected by immunoblotting with an anti-V5 antibody. Lysate (5%) used for the immunoprecipitations was loaded in the input lanes. (C, D) Interaction of eRF3 with eRF1, UPF1 and PABPC1. (C) Schematic representation of the domain structure of the human release factor eRF3. N-terminal (N; PABPC1 binding), middle (M), GTPase- and C-terminal (C; eRF1 binding) parts of eRF3 are indicated. (D) Co-immunoprecipitation experiments with eRF3 fragments. HeLa cells were transfected with an empty FLAG plasmid (lanes 1 and 2), with FLAG-eRF3 (lanes 3 and 4) or the indicated FLAG-tagged fragments of eRF3 (lanes 5–10) together with a plasmid for V5-UPF1, V5-PABPC1 and V5-eRF1. Immunoprecipitations were carried out as described in (B). V5-tagged proteins were detected by immunoblotting with an anti-V5 antibody. Lysate (5%) used for the immunoprecipitations was loaded in the input lanes. (E) UPF1 interacts with eRF3 in the presence of GDP or GTP. Co-immunoprecipitation experiments with eRF3 in the presence of GTP or GDP. HeLa cells were transfected with the plasmid FLAG-eRF3 (lanes 1–6) and with plasmids V5-UPF1, V5-PABPC1 and V5-eRF1, respectively. Immunoprecipitation was carried out in the presence of RNaseA and the guanine nucleotides (1 μM) as indicated (lanes 2 and 3). V5-tagged proteins were detected by immunoblotting with an anti-V5 antibody as described above. Lysate (2.5%) used for the immunoprecipitations was loaded in the input lanes (lanes 4–6).
As a next step, we characterized which of the functional domains of human eRF3 participate in this interaction. Human eRF3a (referred to as eRF3 throughout the text) consists of three domains (Figure 3C), an N-terminal domain extending from residues 1–138, which interacts with PABPC1 (Cosson et al, 2002b), a positively charged middle domain extending from residues 139–200 and a large C-terminal GTPase domain extending from residues 201–637, which has been shown to be essential for translation termination (Alkalaeva et al, 2006). This GTPase domain can be further subdivided into two parts with an N-terminal portion containing four GTP-binding motifs (G1–G4; 200–450) and a C-terminal part that interacts with eRF1 (Stansfield et al, 1995; Zhouravleva et al, 1995; Frolova et al, 1996, 1998; Jakobsen et al, 2001). Accordingly, we co-transfected different FLAG-tagged eRF3 variants with V5-UPF1, V5-eRF1 and V5-PABPC1 and immunoprecipitated on FLAG antibody beads in the presence or absence of RNAse A (Figure 3D). PABPC1 and eRF1 show efficient binding to the expected N- and C-terminal domains of eRF3, respectively, serving as positive controls for the reaction (lanes 3–8 and lanes 3 and 4). UPF1 binds to the GTPase domain of eRF3 (Figure 3D, lanes 9 and 10) both in the presence and absence of RNA, whereas it shows weak binding to the N terminus of eRF3 in the absence of RNaseA (Figure 3D, compare lanes 5 and 6).
Considering that UPF1 binds to the GTPase domain of eRF3, we next analysed if the interaction is influenced by whether eRF3 is in the GTP- or the GDP-bound states. eRF3 stimulates termination in a GTP-dependent manner, and eRF1 together with the ribosome exerts an effect as a composite GTPase-activating protein towards eRF3 (Frolova et al, 1996). The eRF1:eRF3:GTP:Mg2+ complex binds to the pre-termination ribosome and, following hydrolysis of GTP, induces efficient peptide release from the ribosome (Alkalaeva et al, 2006; Hauryliuk et al, 2006; Mitkevich et al, 2006). We performed immunoprecipitation analysis between FLAG-eRF3 and V5-UPF1 either in the presence of GTPγS, a non-hydrolysable GTP analogue, or in the presence of GDP. The binding of eRF1 to eRF3 served as a positive control because eRF1 binds eRF3 under non-equilibrium conditions such as an immunoprecipitation only in the presence of GTP, that is, before peptide release (Pisareva et al, 2006). We also monitored PABPC1 binding to eRF3 because PABPC1 interacts with the N-terminal domain of eRF3 and not with the GTPase domain as eRF1 and UPF1 do (see Figure 3D). As expected, the eRF1:eRF3 interaction occurs in the presence of GTP (Figure 3E, lanes 2 and 3), whereas UPF1 binds to both eRF3:GDP and a complex containing eRF3:GTP (such as eRF1:eRF3:GTP (Figure 3E, lanes 2 and 3). These data indicate that the interaction of UPF1 with the release factors can occur prior to GTP hydrolysis and peptide release, and that it can be maintained post-terminationally (Figure 7).
eRF3 binds to the cysteine–histidine-rich domain of UPF1
We next analysed which domain of UPF1 interacts with eRF3. UPF1 is a multifunctional protein that has RNA binding, ATPase and RNA helicase properties and is structurally organized into at least two domains. One of these domains is a cysteine- and histidine-rich region (Cys–His-rich region, CHR, amino acids 123–213) near its N-terminus, which includes three zinc-finger motifs (Kadlec et al, 2006). The other domain, referred to as the core domain, includes six conserved helicase motifs that are common to members of the helicase I superfamily (Altamura et al, 1992; Czaplinski et al, 1995; Weng et al, 1996a). This two-domain structure is flanked by amino-acid sequences, which are important for SMG1-dependent phosphorylation (Yamashita et al, 2001, 2005; Ohnishi et al, 2003).
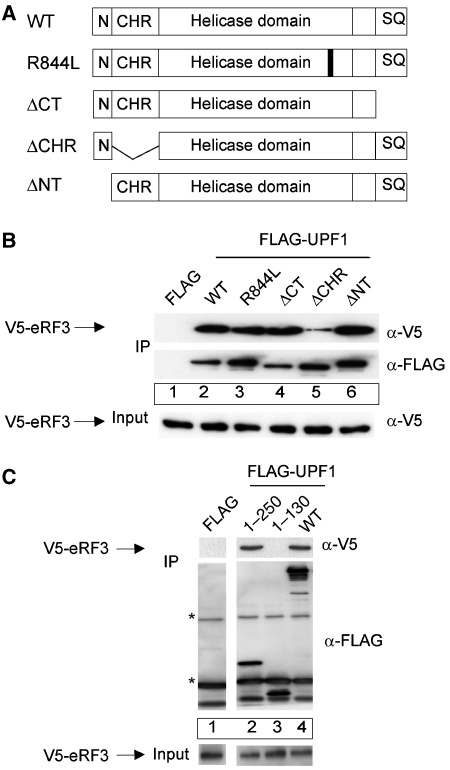
We mapped the regions of UPF1 that are important for the interaction with eRF3 by immunoprecipitation analyses with FLAG-UPF1 mutants (Figure 4A). These mutants included the R844L dominant-negative that inactivates the helicase domain (Sun et al, 1998), a C-terminal deletion mutant of residues 1074–1118 (ΔCT), an N-terminal deletion mutant of residues 1–40 (ΔNT) and a mutant with a deletion of residues 130–250 including the Cys–His-rich region (ΔCHR). The ΔCHR mutant displays reduced binding to eRF3 compared with wild type (Figure 4B, compare lanes 2 and 5), whereas the other deletion mutants and the R844L mutant show similar binding as the WT protein (Figure 4B, compare lane 2 with lanes 3, 4 and 6). We confirmed that the UPF1 CHR domain represents the bona fide eRF3-binding site by immunoprecipitation analysis with V5-tagged eRF3 and FLAG-tagged UPF1 fragments (Figure 4C). The UPF1 fragment containing residues 1–250 including the CHR (residues 123–213) shows a similar interaction with eRF3 as UPF1 wild type does (Figure 4C, compare lanes 2 and 4), whereas the UPF1 fragment including residues 1–130, thus excluding the CHR, does not display eRF3 binding (Figure 4C, lane 3). The protein–protein contact between UPF1 and eRF3 is thus established by binding between the CHR of UPF1 and the C-terminal GTPase domain of eRF3 (Figure 7).
Figure 4.
eRF3 binds to the cysteine–histidine-rich region (CHR) of UPF1. (A) Schematic representation of the domain structure of human UPF1. Abbreviations used: WT—wild-type UPF1; R844L—dominant-negative point mutant of the helicase domain; ΔCT—mutant lacking the C-terminal amino acids 1074–1118; ΔCHR—mutant lacking amino acids 130–250 including the CHR; ΔNT—mutant lacking the N-terminal amino acids 1–40 (N); SQ—serine/threonine-glutamine-rich ((S/T)-Q-rich) motifs. (B) Co-immunoprecipitation experiments with UPF1 mutants. HeLa cells were transfected with an empty FLAG plasmid (lane 1), with wild-type FLAG-UPF1 (lane 2), the point mutant FLAG-R844L (lane 3) or the indicated FLAG-tagged fragments of UPF1 (lanes 4–6) together with a V5-eRF3 plasmid. Immunoprecipitations were carried out as described in Figure 3B in the presence of RNaseA. V5-eRF3 was detected by immunoblotting with an anti-V5 antibody. The input lane shows 4% of the lysate used per assay. (C) Co-immunoprecipitation experiments with the N-terminal fragment of UPF1. HeLa cells were transfected with an empty FLAG plasmid (lane 1), with FLAG-UPF1 (lane 4) or the indicated FLAG-tagged fragments of UPF1 (lanes 2 and 3) together with a V5-eRF3 plasmid. Immunoprecipitation and the detection of V5-eRF3 were carried out as described in (B). *Immunoglobulin heavy and light chains. Lysate (5%) used for the immunoprecipitations was loaded in the input lane.
Phosphorylation of UPF1 and NMD can be alternatively supported by UPF2 or UPF3b
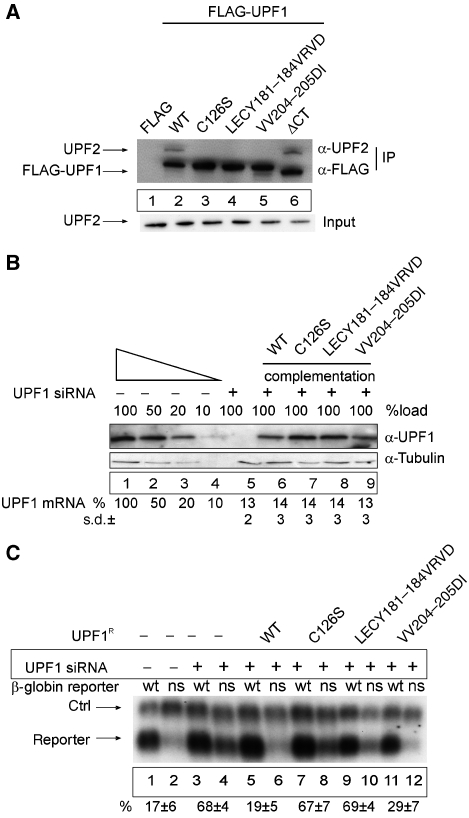
The CHR of UPF1, which we have shown to be critical for eRF3 binding, has previously been demonstrated to be important also for the interaction of UPF1 with UPF2 (Applequist et al, 1997; Kadlec et al, 2006) and to modulate translation termination and stop codon readthrough in yeast (Weng et al, 1996b, 2001). This domain thus represents a critical region of UPF1 that establishes its contact with both the translation termination apparatus and the EJC. We analysed this domain of UPF1 in more detail by mutagenesis screening of conserved residues in this region. From a pool of UPF1 mutants, we selected three variants that completely abolished the interaction with UPF2 (Figure 5A). The affected residues (C126, LECY181–184 and VV204–205) are located in those parts of the protein that have been shown in structural analyses to directly contact UPF2 (Kadlec et al, 2006) and to be critical for bridging UPF1 to the EJC (Kashima et al, 2006).
Figure 5.
An UPF1 mutant that triggers NMD but fails to interact with UPF2. (A) Mutational analysis of the UPF2-binding site within the UPF1 CHR region. HeLa cells were transfected with empty FLAG plasmid (lane 1), wild-type FLAG-UPF1 (lane 2) or with the indicated FLAG-tagged mutants of UPF1 (lanes 3–6). Cell extract preparation, immunoprecipitation procedure and immunoblotting were carried out as described in Figure 3B in the presence of RNaseA. The UPF2 protein was detected by immunoblotting with an anti-UPF2 antibody. Lysate (5%) used for the immunoprecipitations was loaded in the input lane. (B, C) Functional analysis of UPF1 CHR mutants. (B) Functional complementation of UPF1 depletion. Immunoblotting analysis of lysates obtained from UPF1-depleted and UPF1-complemented cells was performed as described in Figure 2B and as described previously (Gehring et al, 2005). For the complementation of UPF1-depleted cells, cells were transfected with 0.2 μg FLAG-UPF1R. UPF1 mRNA levels were determined by qRT–PCR using primers that are specific for the endogenous mRNA and do not amplify the plasmid-derived UPF1 mRNA. The values were calculated from four independent experiments with standard deviations (±s.d.). (C) Northern blot analysis of RNA isolated from HeLa cells transfected with siRNA against β-galactosidase (lanes 1–2) or UPF1 (lanes 3–12). At 48 h after siRNA transfections, the cells were co-transfected with plasmids for the transfection efficiency control (ctrl), the NMD reporters (reporter; wt—wild-type β-globin, ns—NS39 β-globin mutant) and plasmids expressing the depicted FLAG-UPF1R variants. The values were calculated from three independent experiments. Error bars represent s.d.
We functionally tested the NMD activity of these mutants in HeLa cells that had been treated with siRNA against endogenous UPF1 (or β-galactosidase siRNA as a negative control). UPF1 RNAi was efficient and depleted the endogenous protein to <10% (Figure 5B, lanes 1–5). Subsequently, these cells were co-transfected with a β-globin NMD reporter carrying either the wild-type sequence or a nonsense mutation at position 39 (NS39), and with one of the UPF1 variants that were engineered to be resistant to the UPF1-specific siRNA (UPF1R; Figure 5B, lanes 6–9). The NS39 reporter is expressed at a level of 17±6% of wild-type mRNA in cells treated with β-galactosidase-specific siRNA (Figure 5C, lanes 1 and 2) Upon depletion of UPF1, the expression of the NS39 mRNA increases four-fold (Figure 5C, lanes 3 and 4). As predicted, co-transfected WT-UPF1R completely rescues NMD of NS39 (Figure 5C, lanes 5 and 6). The UPF1R mutants C126S and LECY181–184VRVD fail to rescue NMD activity (Figure 5C, lanes 7–10), which is consistent with the important role of UPF1:UPF2 binding in NMD (Kashima et al, 2006). Surprisingly, the UPF1R mutant VV204–205DI does rescue NMD activity (Figure 5C, lanes 11 and 12), although similar to the other two mutants it was confirmed not to detectably interact with UPF2 (Figure 5A, lanes 3–5). This result shows that NMD can be triggered even by an UPF1 mutant that does not display UPF2-binding capacity and hence that other features of UPF1 may suffice for the NMD activity of this mutant.
We next explored which feature of UPF1 could complement the UPF2-binding incompetent mutant. The phosphorylation of UPF1 by SMG1 is a key event of the NMD pathway (Page et al, 1999; Pal et al, 2001; Ohnishi et al, 2003; Kashima et al, 2006). Therefore, we tested the capacity of the UPF1 mutants to be phosphorylated. We transfected HeLa cells with FLAG-tagged mutants of UPF1, and immunoprecipitated these mutants in the presence of phosphatase inhibitors. The immunoprecipitates were eluted and aliquots immunoblotted either with a phospho-specific UPF1 antibody (Yamashita et al, 2001), or a FLAG antibody that was used as an internal control for protein loading (Figure 6B). As expected, the C126S, the LECY181–184VRVD and the ΔCT mutants display strongly reduced phosphorylation when compared with WT-UPF1 (Figure 6B, compare lanes 3–5 and 1). Importantly, the VV204–205DI mutant is phosphorylated almost normally (Figure 6B, lane 2), which indicates that UPF2 is not absolutely required for UPF1 phosphorylation and NMD activity.
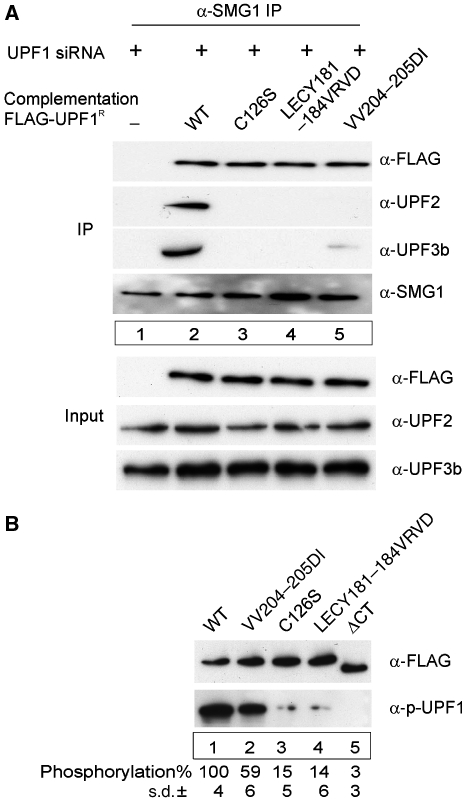
Figure 6.
SMG1-dependent phosphorylation of UPF1 correlates with binding of UPF2 or UPF3b. (A) SMG1 precipitates UPF1 mutants. Complementation of the UPF1 depletion with the indicated Flag-UPF1R variants was carried out as described in Figure 5B. An anti-SMG1 antibody was used to immunoprecipitate SMG1-complexes. Anti-FLAG, anti-UPF2, anti-UPF3b and anti-SMG1 antibodies were used to visualize the respective proteins in the immunoprecipitates. Lysate (10%) used for the immunoprecipitations was loaded in the input lanes. (B) Mutations in the Cys–His-rich region of UPF1 affect its phosphorylation. HeLa cells were transfected with plasmids expressing the indicated FLAG-UPF1 mutants. After 48 h, cells were lysed and UPF1 mutants were immunoprecipitated with anti-FLAG agarose beads in the presence of phosphatase inhibitors. Immunoprecipitates were extensively washed, bound proteins were eluted and equal amounts of the eluted proteins were separated by SDS–PAGE. Immunoblotting was performed with an anti-phospho-UPF1 antibody (α-p-UPF1) or a FLAG antibody. The bands were quantified using an Odyssey Infrared Imaging System (LI-COR Biosciences).
Because SMG1 is the UPF1 kinase and is contained within the functionally critical SURF complex (Kashima et al, 2006), we next tested whether the UPF1 mutations affect SMG1 binding. We treated cells with UPF1-specific siRNAs and complemented with FLAG-tagged UPF1R mutants in a similar way as described in Figure 5B. We then immunoprecipitated endogenous SMG1 with specific antibodies and assayed for cofactor binding with FLAG-, UPF2- and UPF3b-specific antibodies (Figure 6A). SMG1 antibodies efficiently pull down both wild-type and mutant UPF1 (anti-FLAG, Figure 6A, lanes 2–5). Further immunoblotting analyses of the precipitates demonstrate that the WT-UPF1:SMG1 complex efficiently precipitates both UPF2 and UPF3b (Figure 6A, lane 2), whereas the SMG1 complex with the UPF1 mutants C126S and LECY181–184VRVD interacts with neither UPF2 nor UPF3b (Figure 6A, lanes 3–4). Of note, the SMG1:UPF1-VV204–205DI complex fails to bind UPF2, but interacts with UPF3b albeit somewhat less efficiently than WT-UPF1 (Figure 6A, lane 5). These results confirm that phosphorylation of UPF1 is a critical step of the NMD pathway. Significantly, they support the notion that UPF1 phosphorylation and NMD can be mediated by an interaction with UPF2 or with UPF3b. These data thus also provide a biochemical and mechanistic correlate of previously proposed alternative NMD pathways (Gehring et al, 2005; Chan et al, 2007), and for the NMD activity of patients with inactivating UPF3b mutations (Tarpey et al, 2007).
Discussion
Nonsense-mediated mRNA decay is triggered by translation termination at premature stop codons, which induces the assembly of a 3′ mRNP that leads to the rapid degradation of the transcript. Upf1 proteins have a central function in NMD in all organisms that have been studied. Yeast and human Upf1 proteins are recruited to prematurely terminating ribosomes through interactions with the release factors (Czaplinski et al, 1998; Kashima et al, 2006), and the NMD activity of UPF1 is regulated by phosphorylation and dephosphorylation (Page et al, 1999; Chiu et al, 2003; Ohnishi et al, 2003; Fukuhara et al, 2005).
In yeast, translation termination efficiency differs at natural and premature stop codons; whereas termination at natural stop codons is efficient and quick, termination at premature stop codons is inefficient and slow (Amrani et al, 2004, 2006). This inefficiency of termination at a premature termination codon has been explained by the failure to assemble an appropriate 3′ mRNP and instead the assembly of an inappropriate 3′ mRNP termed faux 3′-UTR. Poly(A)-binding protein appears to be an important architectural component of an appropriate 3′ mRNP because repositioning of the protein to sufficient proximity to a premature termination codon redefines it to being normal, leading to efficient translation termination and NMD suppression in Saccharomyces cerevisiae (Amrani et al, 2004) and in Drosophila melanogaster cells (Behm-Ansmant et al, 2007a, 2007b). In contrast to yeast and fly cells, an EJC downstream of a stop codon provides critical positional information for mammalian NMD to discriminate a premature termination codon from a natural stop codon (Carter et al, 1996; Thermann et al, 1998; Le Hir et al, 2000). The EJC is thought to interact with the mRNP recruited to the vicinity of the termination codon, referred to as SURF complex and to induce phosphorylation of UPF1 (Kashima et al, 2006).
Our results indicate that PABPC1 stimulates (Figure 2D) and UPF1 inhibits (Figure 2C) efficient translation termination in human cells. Furthermore, PABPC1 inhibits (Figure 1B and Supplementary Figure 1) and UPF1 stimulates NMD (Applequist et al, 1997). These results correlate with in vitro toeprint experiments in yeast extracts, which show that intermediates of termination complexes are sufficiently stable to be detectable only in the presence of a premature termination codon and of Upf1p (Amrani et al, 2004). Although depletion of Upf1p in yeast has been shown to cause nonsense suppression in vivo and amino-acid substitutions within the highly conserved CHR domain of Upf1p have previously been shown to affect nonsense suppression and NMD (Weng et al, 1996a, 1996b), other data have challenged this view (Bidou et al, 2000; Harger and Dinman, 2004), and the effect of Upf1p on translation termination may be context dependent (Keeling et al, 2004). Significantly, human UPF1 is not only homologous to yeast Upf1p but also to another yeast helicase with a CHR domain named Mtt1p. Similar to human UPF1, Mtt1p binds to the N-terminus and to the GTPase domains of eRF3 and can inhibit translation termination (Czaplinski et al, 2000). In the course of evolution, human UPF1 may thus have acquired characteristics of both yeast Upf1p and Mtt1p.
In analogy to the faux 3′-UTR model in yeast, we propose that the juxtaposition of PABPC1 and a stop codon can overcome the NMD-stimulating effect of a 3′ EJC (Figure 1). However, the NMD-inducing effect of the EJC–UPF1 interaction is dominant over the effect of PABPC1, if the EJC is placed closer to the stop codon than PABPC1 (Figure 1B). This dominance is maintained even when PABPC1 is located in the immediate 3′ neighbourhood of the EJC, showing that absolute linear distance of PABPC1 to the stop codon per se is not a sufficient determinant of a proper 3′ mRNP. These results provide a basis to suggest that human NMD has evolutionarily retained components of the NMD machinery of lower eukaryotes.
The biochemical correlate of the functionally antagonistic effect of UPF1 on translation termination is reflected by eRF1 binding to UPF1 and to eRF3 with the same domain (Figure 3B). These data thus suggest that the presence of UPF1 may interfere with functionally effective eRF1:eRF3 binding and, consequently, efficient translation termination; this hypothesis remains to be tested. Furthermore, UPF1 binds to the GTPase domain of eRF3, which has previously been implicated in termination-coupled processes and is considered to function as a molecular switch connecting GTP hydrolysis to constitutive mRNA decay and to NMD through shortening of poly(A) tails (Hosoda et al, 2003; Kobayashi et al, 2004). Finally, the data reported here demonstrate that the ability of Upf1 proteins to interact with both the pre-termination eRF1:eRF3:GTP complex and the post-termination eRF3:GDP-containing complex, is conserved between human UPF1 and yeast Upf1p (Kobayashi et al, 2004). We further suggest that the inhibitory effect of UPF1 on termination may result in a delay of the termination process with inefficient disassembly of the terminating ribosome and release of the nascent peptide. Thus, in case of premature termination there is sufficient time for the EJC to interact with UPF1, induce UPF1 phosphorylation by SMG1 and rapid RNA degradation. By contrast, in case of normal termination and without an EJC being present, PABPC1 interacts with the termination complex, which stimulates termination efficiency and results in the dissociation of the release factors and UPF1 from the mRNA, thus maintaining its stability (Figure 7).
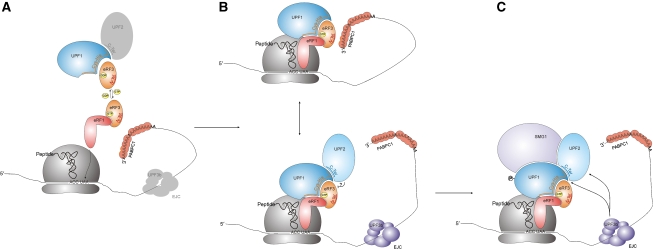
Figure 7.
An integrated model for mammalian NMD pathways. Schematic diagram of the structural and the functional interaction networks between release factors, UPF proteins, PABPC1, SMG1 and the EJC. Note that the diagram is not time-resolved, and hence some depicted interactions may not occur simultaneously and/or be competitive. (A) The Cys–His-rich domain of UPF1 (Cys–His) binds to the C terminus of UPF2 (see also Kadlec et al, 2006) and to the GTPase domain of eRF3. In the presence of GTP (reflecting the pre-termination state), the C-terminal end of eRF1 binds to the C-terminal end of eRF3, whereas this interaction does not occur in the presence of GDP (reflecting the post-termination state). The C terminus of eRF1 may directly bind to UPF1 or this interaction may be bridged by eRF3. (B) The C terminus of PABPC1 binds to the N terminus of eRF3 (N-Ter). During translation termination, UPF1 monitors the correct position of the termination codon. An interaction between PABPC1 and eRF3 ‘certifies' a normal termination event (top panel), whereas a downstream EJC is indicating premature termination (bottom panel). (?)—a possible competition between UPF2 and eRF3 for UPF1 binding. (C) In case of premature termination, the EJC triggers the SMG1-dependent UPF1 phosphorylation through UPF2 or UPF3b, which leads to NMD.
Our data also shed new light on the question of how the EJC can interact with UPF1 to become phosphorylated and to trigger NMD. A number of previous studies indicated that the EJC consisting of the proteins eIF4AIII, RNPS1, BTZ, Y14 and MAGOH interacts with UPF3b and UPF2, which in turn bind to and induce phosphorylation of UPF1 (Lykke-Andersen et al, 2000, 2001; Kim et al, 2001; Gehring et al, 2003; Kashima et al, 2006; Kunz et al, 2006). Such a simple linear model of NMD has been challenged by the finding of both UPF2- and UPF3b-independent NMD and by the identification of patients with inactivating UPF3b mutations and normal NMD function (Gehring et al, 2005; Chan et al, 2007; Tarpey et al, 2007). Our data now identify a mutant of UPF1 (VV204–205DI) that can be phosphorylated (Figure 6B) and is NMD active in spite of its inability to interact detectably with UPF2 (Figure 5A–C). Interestingly, the VV204–205 motif is not conserved in Upf1 proteins of some organisms such as Encephalitozoon cuniculi, Entamoeba histolytica and Giardia lamblia. Significantly, the genomes of these organisms do not contain UPF2 homologous genes, suggesting that Upf1 proteins in these organisms function in an UPF2-independent manner (Kadlec et al, 2006). Importantly, the VV204–205DI mutant is capable to interact with UPF3b (Figure 6A) establishing an UPF2-independent interaction of UPF1 with the EJC.
These data thus provide direct evidence that SMG1-dependent phosphorylation of UPF1 can be stimulated independently by UPF2 or UPF3b, and unveil a potential biochemical correlate for previously identified alternative NMD pathways that can function independently of either UPF2 or UPF3b. Finally, our integrated NMD model (Figure 7) could also help explain cases of splicing-independent mammalian NMD (Buhler et al, 2006), if PABPC1-stimulated efficient termination is blocked and/or UPF1 phosphorylation triggered by other features of the mRNA or factors binding to it.
Materials and methods
Plasmid constructs
β-Globin 4boxB/4MS2 and 4MS2/4boxB were constructed by introducing sequences for the 4MS2 or 4boxB into the 3′-UTR of 4boxB-β-globin or 4MS2-β-globin, respectively. pCI-neo-MS2, pCI-neo-λNV5, pCI-FLAG, pCI-V5, wt300+e3, 4boxB- and 4MS2-β-globin, and expression vectors for UPF1, Y14, MAGOH, RNPS1 and UPF2 were described previously (Danckwardt et al, 2002; Gehring et al, 2003, 2005; Schell et al, 2003). Plasmid pET15b-eRF3a expressing 6His-eRF3a was kind gifts of LY Frolova (Jakobsen et al, 2001). eRF1 and PABPC1 cDNAs were PCR-amplified from HeLa cDNA, eRF3a was amplified from pET15b-eRF3a and used for subcloning to pCI-neo-FLAG, pCI-neo-V5, pCI-neo-MS2 and pCI-neo-λNV5. Deletion mutants of PABPC1, eRF3, eRF1 and UPF1 were created using appropriate PCR primer pairs (Supplementary Table I). Site-specific mutants of UPF1 were generated by site-directed mutagenesis (GeneTailor, Invitrogen). The plasmid p2luc-if was a generous gift of JF Atkins (Howard et al, 2001). The plasmids p2luc-TAA, p2luc-TAG and p2luc-TGA were produced by site-directed mutagenesis of p2luc-if plasmid.
Cell culture, plasmid and siRNA transfections
HeLa cells were grown and transfected with DNA or siRNA as described (Gehring et al, 2003, 2005). For tethering experiments, 2 μg of 4MS2/4boxB or 4boxB/4MS2 reporter, 1 μg of control WT globin, 0.2 μg of GFP reporter and 1 μg of respective pCI-neo-MS2 or pCI-neo-λNV5 reporter were used. For transfection of β-globin WT or NS39, we used 1 μg of pCI-neo-WT or pCI-neo-NS39 reporter, 0.5 μg of the control plasmid wt300+e3, and 0.1 μg of the GFP-expressing plasmid. Transfections for IPs were carried out with 2 μg of the FLAG expression vector and 0.5–3 μg of V5 expression plasmids together with 0.5 μg of GFP vector.
siRNA transfection and UPF1-complementation assays were carried out as described (Gehring et al, 2005).
Dual luciferase assay
HeLa cells transfected with 2 μg p2luc-TAA, p2luc-TAG or p2luc-TGA were collected 48 h after transfection, lysed with passive lysis buffer (Promega) and luciferase activity was determined using the Dual Luciferase reporter assay (Promega) as described in Howard et al (2001).
RNA extraction and analysis
Total cytoplasmic RNA was analysed by northern blotting and qRT–PCR as described (Gehring et al, 2003). Quantification of signals was carried out in FLA-3000 fluorescent analyser. Percentages and standard deviations were calculated as described in Kunz et al (2006).
Protein extraction, western blotting and immunoprecipitations
Complexes of FLAG-tagged proteins were immunoprecipitated as described (Gehring et al, 2003). Unless otherwise stated, cells were lysed for IP reactions in lysis buffer (20 mM Tris–HCl, pH 7.2, 150 mM KCl, 2 mM MgCl2, 0.5% NP-40 and protease cocktail inhibitor ‘complete' (Roche Diagnostics)). The lysis buffer for immunoprecipitation with eRFs was supplemented with 10 mM MgCl2.
Anti-SMG1 (Cell Signalling) antibodies were used for immunoprecipitations using Protein A/G Plus-Agarose (Santa Cruz Biotechnology) according to the manufacturer's recommendations. The 14 000 g supernatants (S14), used as cytosolic fractions, were pre-cleared by incubation with the Protein A/G Plus-Agarose beads pre-blocked with BSA. Subsequently, samples were incubated overnight with mouse anti-SMG1 at 4°C. The pre-blocked beads were then added to the samples for 2–3 h at 4°C. Afterwards, beads were washed three times with washing buffer (20 mM Tris–HCl, pH 7.2, 200 mM KCl, 2 mM MgCl2). The immunoprecipitates were eluted with 1 × SDS–PAGE loading buffer.
For immunoblotting analysis following siRNA depletion, 20 μg of cytoplasmic lysates was subjected to SDS–PAGE (Gehring et al, 2005).
For quantification of UPF1 phosphorylation levels, FLAG-UPF1 mutants were immunoprecipitated as described above. The lysis buffer was supplemented with a phosphatase inhibitor cocktail (‘PhosSTOP'; Roche Diagnostics). An anti-phospho-UPF1 antibody was used for detection of phosphorylated UPF1 (Kashima et al, 2006), FLAG antibodies were used as a control for protein loading. Infrared IRDye-labeled secondary antibodies were used for the detection of bands using an Odyssey Infrared Imaging System (LI-COR Biosciences).
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Information
Acknowledgments
We are grateful to L Kisselev and L Frolova for antibodies against eRF3 and for the plasmids pET15b-eRF3a/pET15b-eRFb; E Wahle for antibodies against PABPN; C Thoma and P Hundsdoerfer for PABPC1-siRNA and α-PABPC1 antibodies; J Atkins for the plasmid p2Luc-if; I Kashima, A Yamashita and S Ohno for the α-phospho-UPF1 antibodies; J Lykke-Andersen for α-UPF2 antibodies. We thank S Danckwardt, G Neu-Yilik, M Viegas, S Breit and the team of the MMPU for helpful discussions. This study was financially supported by a FEBS Long-Term Fellowship to Pavel Ivanov, by the Young Investigator Award of the Medical Faculty of Heidelberg to Joachim Kunz and by grants from the Deutsche Forschungsgemeinschaft to Andreas Kulozik and Matthias Hentze.
References
- Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV (2006) In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell 125: 1125–1136 [DOI] [PubMed] [Google Scholar]
- Altamura N, Groudinsky O, Dujardin G, Slonimski PP (1992) NAM7 nuclear gene encodes a novel member of a family of helicases with a Zn-ligand motif and is involved in mitochondrial functions in Saccharomyces cerevisiae. J Mol Biol 224: 575–587 [DOI] [PubMed] [Google Scholar]
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A (2004) A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432: 112–118 [DOI] [PubMed] [Google Scholar]
- Amrani N, Dong S, He F, Ganesan R, Ghosh S, Kervestin S, Li C, Mangus DA, Spatrick P, Jacobson A (2006) Aberrant termination triggers nonsense-mediated mRNA decay. Biochem Soc Trans 34: 39–42 [DOI] [PubMed] [Google Scholar]
- Andjelkovic N, Zolnierowicz S, Van Hoof C, Goris J, Hemmings BA (1996) The catalytic subunit of protein phosphatase 2A associates with the translation termination factor eRF1. EMBO J 15: 7156–7167 [PMC free article] [PubMed] [Google Scholar]
- Applequist SE, Selg M, Raman C, Jack HM (1997) Cloning and characterization of HUPF1, a human homolog of the Saccharomyces cerevisiae nonsense mRNA-reducing UPF1 protein. Nucleic Acids Res 25: 814–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Gatfield D, Rehwinkel J, Hilgers V, Izaurralde E (2007a) A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO J 26: 1591–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Kashima I, Rehwinkel J, Sauliere J, Wittkopp N, Izaurralde E (2007b) mRNA quality control: an ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett 581: 2845–2853 [DOI] [PubMed] [Google Scholar]
- Bidou L, Stahl G, Hatin I, Namy O, Rousset JP, Farabaugh PJ (2000) Nonsense-mediated decay mutants do not affect programmed -1 frameshifting. RNA 6: 952–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono F, Ebert J, Lorentzen E, Conti E (2006) The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell 126: 713–725 [DOI] [PubMed] [Google Scholar]
- Buhler M, Steiner S, Mohn F, Paillusson A, Muhlemann O (2006) EJC-independent degradation of nonsense immunoglobulin-mu mRNA depends on 3′ UTR length. Nat Struct Mol Biol 13: 462–464 [DOI] [PubMed] [Google Scholar]
- Carter MS, Li S, Wilkinson MF (1996) A splicing-dependent regulatory mechanism that detects translation signals. EMBO J 15: 5965–5975 [PMC free article] [PubMed] [Google Scholar]
- Chan CC, Dostie J, Diem MD, Feng W, Mann M, Rappsilber J, Dreyfuss G (2004) eIF4A3 is a novel component of the exon junction complex. RNA 10: 200–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan WK, Huang L, Gudikote JP, Chang YF, Imam JS, MacLean JA II, Wilkinson MF (2007) An alternative branch of the nonsense-mediated decay pathway. EMBO J 26: 1820–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YF, Imam JS, Wilkinson MF (2007) The nonsense-mediated decay RNA surveillance pathway. Ann Rev Biochem 76: 51–74 [DOI] [PubMed] [Google Scholar]
- Chiu SY, Serin G, Ohara O, Maquat LE (2003) Characterization of human Smg5/7a: a protein with similarities to Caenorhabditis elegans SMG5 and SMG7 that functions in the dephosphorylation of Upf1. RNA 9: 77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson B, Berkova N, Couturier A, Chabelskaya S, Philippe M, Zhouravleva G (2002a) Poly(A)-binding protein and eRF3 are associated in vivo in human and Xenopus cells. Biol Cell 94: 205–216 [DOI] [PubMed] [Google Scholar]
- Cosson B, Couturier A, Chabelskaya S, Kiktev D, Inge-Vechtomov S, Philippe M, Zhouravleva G (2002b) Poly(A)-binding protein acts in translation termination via eukaryotic release factor 3 interaction and does not influence [PSI(+)] propagation. Mol Cell Biol 22: 3301–3315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K, Majlesi N, Banerjee T, Peltz SW (2000) Mtt1 is a Upf1-like helicase that interacts with the translation termination factors and whose overexpression can modulate termination efficiency. RNA 6: 730–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K, Ruiz-Echevarria MJ, Paushkin SV, Han X, Weng Y, Perlick HA, Dietz HC, Ter-Avanesyan MD, Peltz SW (1998) The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev 12: 1665–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K, Weng Y, Hagan KW, Peltz SW (1995) Purification and characterization of the Upf1 protein: a factor involved in translation and mRNA degradation. RNA 1: 610–623 [PMC free article] [PubMed] [Google Scholar]
- Danckwardt S, Neu-Yilik G, Thermann R, Frede U, Hentze MW, Kulozik AE (2002) Abnormally spliced beta-globin mRNAs: a single point mutation generates transcripts sensitive and insensitive to nonsense-mediated mRNA decay. Blood 99: 1811–1816 [DOI] [PubMed] [Google Scholar]
- Durand S, Cougot N, Mahuteau-Betzer F, Nguyen CH, Grierson DS, Bertrand E, Tazi J, Lejeune F (2007) Inhibition of nonsense-mediated mRNA decay (NMD) by a new chemical molecule reveals the dynamic of NMD factors in P-bodies. J Cell Biol 178: 1145–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova L, Le Goff X, Rasmussen HH, Cheperegin S, Drugeon G, Kress M, Arman I, Haenni AL, Celis JE, Philippe M, Justesen J, Kisselev L (1994) A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature 372: 701–703 [DOI] [PubMed] [Google Scholar]
- Frolova L, Le Goff X, Zhouravleva G, Davydova E, Philippe M, Kisselev L (1996) Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA 2: 334–341 [PMC free article] [PubMed] [Google Scholar]
- Frolova LY, Merkulova TI, Kisselev LL (2000) Translation termination in eukaryotes: polypeptide release factor eRF1 is composed of functionally and structurally distinct domains. RNA 6: 381–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova LY, Simonsen JL, Merkulova TI, Litvinov DY, Martensen PM, Rechinsky VO, Camonis JH, Kisselev LL, Justesen J (1998) Functional expression of eukaryotic polypeptide chain release factors 1 and 3 by means of baculovirus/insect cells and complex formation between the factors. Eur J Biochem 256: 36–44 [DOI] [PubMed] [Google Scholar]
- Fukuhara N, Ebert J, Unterholzner L, Lindner D, Izaurralde E, Conti E (2005) SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol Cell 17: 537–547 [DOI] [PubMed] [Google Scholar]
- Gehring NH, Kunz JB, Neu-Yilik G, Breit S, Viegas MH, Hentze MW, Kulozik AE (2005) Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol Cell 20: 65–75 [DOI] [PubMed] [Google Scholar]
- Gehring NH, Neu-Yilik G, Schell T, Hentze MW, Kulozik AE (2003) Y14 and hUpf3b form an NMD-activating complex. Mol Cell 11: 939–949 [DOI] [PubMed] [Google Scholar]
- Glavan F, Behm-Ansmant I, Izaurralde E, Conti E (2006) Structures of the PIN domains of SMG6 and SMG5 reveal a nuclease within the mRNA surveillance complex. EMBO J 25: 5117–5125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grentzmann G, Ingram JA, Kelly PJ, Gesteland RF, Atkins JF (1998) A dual-luciferase reporter system for studying recoding signals. RNA 4: 479–486 [PMC free article] [PubMed] [Google Scholar]
- Grimson A, O'Connor S, Newman CL, Anderson P (2004) SMG-1 is a phosphatidylinositol kinase-related protein kinase required for nonsense-mediated mRNA decay in Caenorhabditis elegans. Mol Cell Biol 24: 7483–7490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harger JW, Dinman JD (2004) Evidence against a direct role for the Upf proteins in frameshifting or nonsense codon readthrough. RNA 10: 1721–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauryliuk V, Zavialov A, Kisselev L, Ehrenberg M (2006) Class-1 release factor eRF1 promotes GTP binding by class-2 release factor eRF3. Biochimie 88: 747–757 [DOI] [PubMed] [Google Scholar]
- Hentze MW, Kulozik AE (1999) A perfect message: RNA surveillance and nonsense-mediated decay. Cell 96: 307–310 [DOI] [PubMed] [Google Scholar]
- Holbrook JA, Neu-Yilik G, Hentze MW, Kulozik AE (2004) Nonsense-mediated decay approaches the clinic. Nat Genet 36: 801–808 [DOI] [PubMed] [Google Scholar]
- Hoshino S, Imai M, Kobayashi T, Uchida N, Katada T (1999) The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-poly(A) tail of mRNA. Direct association of eRF3/GSPT with polyadenylate-binding protein. J Biol Chem 274: 16677–16680 [DOI] [PubMed] [Google Scholar]
- Hosoda N, Kobayashi T, Uchida N, Funakoshi Y, Kikuchi Y, Hoshino S, Katada T (2003) Translation termination factor eRF3 mediates mRNA decay through the regulation of deadenylation. J Biol Chem 278: 38287–38291 [DOI] [PubMed] [Google Scholar]
- Howard MT, Shirts BH, Zhou J, Carlson CL, Matsufuji S, Gesteland RF, Weeks RS, Atkins JF (2001) Cell culture analysis of the regulatory frameshift event required for the expression of mammalian antizymes. Genes Cells 6: 931–941 [DOI] [PubMed] [Google Scholar]
- Isken O, Maquat LE (2007) Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev 21: 1833–1856 [DOI] [PubMed] [Google Scholar]
- Jakobsen CG, Segaard TM, Jean-Jean O, Frolova L, Justesen J (2001) Identification of a novel termination release factor eRF3b expressing the eRF3 activity in vitro and in vivo. Mol Biol (Mosk) 35: 672–681 [PubMed] [Google Scholar]
- Kadlec J, Guilligay D, Ravelli RB, Cusack S (2006) Crystal structure of the UPF2-interacting domain of nonsense-mediated mRNA decay factor UPF1. RNA 12: 1817–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S (2006) Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev 20: 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka N, Yong J, Kim VN, Velazquez F, Perkinson RA, Wang F, Dreyfuss G (2000) Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol Cell 6: 673–682 [DOI] [PubMed] [Google Scholar]
- Keeling KM, Lanier J, Du M, Salas-Marco J, Gao L, Kaenjak-Angeletti A, Bedwell DM (2004) Leaky termination at premature stop codons antagonizes nonsense-mediated mRNA decay in S. cerevisiae. RNA 10: 691–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Kataoka N, Dreyfuss G (2001) Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon–exon junction complex. Science 293: 1832–1836 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Funakoshi Y, Hoshino S, Katada T (2004) The GTP-binding release factor eRF3 as a key mediator coupling translation termination to mRNA decay. J Biol Chem 279: 45693–45700 [DOI] [PubMed] [Google Scholar]
- Kunz JB, Neu-Yilik G, Hentze MW, Kulozik AE, Gehring NH (2006) Functions of hUpf3a and hUpf3b in nonsense-mediated mRNA decay and translation. RNA 12: 1015–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ (2000) The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J 19: 6860–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linde L, Boelz S, Neu-Yilik G, Kulozik AE, Kerem B (2007a) The efficiency of nonsense-mediated mRNA decay is an inherent character and varies among different cells. Eur J Hum Genet 15: 1156–1162 [DOI] [PubMed] [Google Scholar]
- Linde L, Boelz S, Nissim-Rafinia M, Oren YS, Wilschanski M, Yaacov Y, Virgilis D, Neu-Yilik G, Kulozik AE, Kerem E, Kerem B (2007b) Nonsense-mediated mRNA decay affects nonsense transcript levels and governs response of cystic fibrosis patients to gentamicin. J Clin Invest 117: 683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J, Shu MD, Steitz JA (2000) Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 103: 1121–1131 [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J, Shu MD, Steitz JA (2001) Communication of the position of exon–exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science 293: 1836–1839 [DOI] [PubMed] [Google Scholar]
- Maquat LE (2004) Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol 5: 89–99 [DOI] [PubMed] [Google Scholar]
- Merkulova TI, Frolova LY, Lazar M, Camonis J, Kisselev LL (1999) C-terminal domains of human translation termination factors eRF1 and eRF3 mediate their in vivo interaction. FEBS Lett 443: 41–47 [DOI] [PubMed] [Google Scholar]
- Mitkevich VA, Kononenko AV, Petrushanko IY, Yanvarev DV, Makarov AA, Kisselev LL (2006) Termination of translation in eukaryotes is mediated by the quaternary eRF1*eRF3*GTP*Mg2+ complex. The biological roles of eRF3 and prokaryotic RF3 are profoundly distinct. Nucleic Acids Res 34: 3947–3954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu-Yilik G, Kulozik AE (2004) mRNA metabolism and hereditary disorders: a tale of surveillance and escape. Klin Padiatr 216: 304–314 [DOI] [PubMed] [Google Scholar]
- Ohnishi T, Yamashita A, Kashima I, Schell T, Anders KR, Grimson A, Hachiya T, Hentze MW, Anderson P, Ohno S (2003) Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol Cell 12: 1187–1200 [DOI] [PubMed] [Google Scholar]
- Page MF, Carr B, Anders KR, Grimson A, Anderson P (1999) SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol Cell Biol 19: 5943–5951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal M, Ishigaki Y, Nagy E, Maquat LE (2001) Evidence that phosphorylation of human Upfl protein varies with intracellular location and is mediated by a wortmannin-sensitive and rapamycin-sensitive PI 3-kinase-related kinase signaling pathway. RNA 7: 5–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios IM, Gatfield D, St Johnston D, Izaurralde E (2004) An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature 427: 753–757 [DOI] [PubMed] [Google Scholar]
- Pisareva VP, Pisarev AV, Hellen CU, Rodnina MV, Pestova TV (2006) Kinetic analysis of interaction of eukaryotic release factor 3 with guanine nucleotides. J Biol Chem 281: 40224–40235 [DOI] [PubMed] [Google Scholar]
- Schell T, Kocher T, Wilm M, Seraphin B, Kulozik AE, Hentze MW (2003) Complexes between the nonsense-mediated mRNA decay pathway factor human upf1 (up-frameshift protein 1) and essential nonsense-mediated mRNA decay factors in HeLa cells. Biochem J 373: 775–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Mugnier P, Das AK, Webb HM, Evans DR, Tuite MF, Hemmings BA, Barford D (2000) The crystal structure of human eukaryotic release factor eRF1––mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell 100: 311–321 [DOI] [PubMed] [Google Scholar]
- Stansfield I, Jones KM, Kushnirov VV, Dagkesamanskaya AR, Poznyakovski AI, Paushkin SV, Nierras CR, Cox BS, Ter-Avanesyan MD, Tuite MF (1995) The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J 14: 4365–4373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Perlick HA, Dietz HC, Maquat LE (1998) A mutated human homologue to yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proc Natl Acad Sci USA 95: 10009–10014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarpey PS, Lucy Raymond F, Nguyen LS, Rodriguez J, Hackett A, Vandeleur L, Smith R, Shoubridge C, Edkins S, Stevens C, O'Meara S, Tofts C, Barthorpe S, Buck G, Cole J, Halliday K, Hills K, Jones D, Mironenko T, Perry J et al. (2007) Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat Genet 39: 1127–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thermann R, Neu-Yilik G, Deters A, Frede U, Wehr K, Hagemeier C, Hentze MW, Kulozik AE (1998) Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J 17: 3484–3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner L, Izaurralde E (2004) SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol Cell 16: 587–596 [DOI] [PubMed] [Google Scholar]
- Wang W, Czaplinski K, Rao Y, Peltz SW (2001) The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J 20: 880–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch EM, Barton ER, Zhuo J, Tomizawa Y, Friesen WJ, Trifillis P, Paushkin S, Patel M, Trotta CR, Hwang S, Wilde RG, Karp G, Takasugi J, Chen G, Jones S, Ren H, Moon YC, Corson D, Turpoff AA, Campbell JA et al. (2007) PTC124 targets genetic disorders caused by nonsense mutations. Nature 447: 87–91 [DOI] [PubMed] [Google Scholar]
- Weng Y, Czaplinski K, Peltz SW (1996a) Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol Cell Biol 16: 5477–5490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y, Czaplinski K, Peltz SW (1996b) Identification and characterization of mutations in the UPF1 gene that affect nonsense suppression and the formation of the Upf protein complex but not mRNA turnover. Mol Cell Biol 16: 5491–5506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Kashima I, Ohno S (2005) The role of SMG-1 in nonsense-mediated mRNA decay. Biochim Biophys Acta 1754: 305–315 [DOI] [PubMed] [Google Scholar]
- Yamashita A, Ohnishi T, Kashima I, Taya Y, Ohno S (2001) Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev 15: 2215–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhouravleva G, Frolova L, Le Goff X, Le Guellec R, Inge-Vechtomov S, Kisselev L, Philippe M (1995) Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J 14: 4065–4072 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Information