Abstract
The yeast telomerase holoenzyme, which adds telomeric repeats at the chromosome ends, is composed of the TLC1 RNA and the associated proteins Est1, Est2 and Est3. To study the biogenesis of telomerase in endogenous conditions, we performed fluorescent in situ hybridization on the native TLC1 RNA. We found that the telomerase RNA colocalizes with telomeres in G1- to S-phase cells. Strains lacking any one of the Est proteins accumulate TLC1 RNA in their cytoplasm, indicating that a critical stage of telomerase biogenesis could take place outside of the nucleus. We were able to demonstrate that endogenous TLC1 RNA shuttles between the nucleus and the cytoplasm, in association with the Crm1p exportin and the nuclear importins Mtr10p–Kap122p. Furthermore, nuclear retention of the TLC1 RNA is impaired in the absence of yKu70p, Tel1p or the MRX complex, which recruit telomerase to telomeres. Altogether, our results reveal that the nucleo-cytoplasmic trafficking of the TLC1 RNA is an important step in telomere homeostasis, and link telomerase biogenesis to its recruitment to telomeres.
Keywords: Crm1p, FISH, nucleo-cytoplasmic shuttling, telomerase, TLC1 RNA
Introduction
Telomeres are essential chromosomal substructures constituted of DNA repeats and associated proteins. They ensure stability of chromosome ends by protecting them from non-homologous end-joining (NHEJ) and degradation. The overall length of telomeric repeat arrays is determined genetically and, in most eukaryotic cells, maintained by a ribonucleoprotein complex (RNP) called the telomerase (Kelleher et al, 2002; Hug and Lingner, 2006). The RNA moiety of telomerase is used as a template via reverse transcription to allow addition of DNA repeats to the telomere ends (Greider and Blackburn, 1985, 1989; Morin, 1989). In yeast, the telomerase complex is composed of the RNA molecule TLC1, which is used as a scaffold for the binding of a number of proteins, including three Est proteins (for ever shorter telomere) (Lundblad and Szostak, 1989; Singer and Gottschling, 1994; Zappulla and Cech, 2004). One of these, Est2p, constitutes the catalytic subunit of the holoenzyme (Counter et al, 1997; Lingner et al, 1997), whereas Est1p and Est3p are involved in regulating the activity of the telomerase (Qi and Zakian, 2000; Evans and Lundblad, 2002; Taggart et al, 2002). The yKu70/80 heterodimer complex also associates with the TLC1 RNA, and is directly implicated in the recruitment of telomerase to the telomeres (Stellwagen et al, 2003; Fisher et al, 2004).
Several factors are known to regulate the activity and recruitment of telomerase at the telomeres. Among these, the yeast ATM homologue Tel1p is a central actor. Besides its role in the DNA damage checkpoint, Te1lp also regulates the association of telomerase with telomeres (Goudsouzian et al, 2006). In cells lacking TEL1, telomeres are short but stable (Lustig and Petes, 1986; Ritchie et al, 1999). Recently, Tel1p has been shown to be bound preferentially to short telomeres (Hector et al, 2007; Sabourin et al, 2007), allowing the recruitment of telomerase and promoting telomere elongation. Another key actor in the regulation of telomerase association with telomeres is the three-component MRX complex, consisting of Mre11p, Rad50p and Xrs2p. The MRX complex is required for homologous recombination and NHEJ during double-strand break repair of chromosomes (D'Amours and Jackson, 2002). Moreover, cells lacking any component of MRX have short telomeres, as do cells lacking Tel1p (Boulton and Jackson, 1998), and the telomeric overhangs in these cells are altered (Larrivee et al, 2004). Deletion of MRE11 reduces the association of Est1p and Est2p with telomeres (Goudsouzian et al, 2006), and there is evidence that both Tel1p and MRX act on telomeres by the same pathway (Ritchie and Petes, 2000).
Although the TLC1 RNA is a large polymerase II transcript with both poly(A)+ and poly(A)− forms detectable in yeast cells (Chapon et al, 1997; Ferrezuelo et al, 2002), it shares many characteristics with snRNAs. Hence, TLC1 has a 2,2,7-trimethylguanosine (TMG) cap structure and is known to bind Sm proteins (Chapon et al, 1997; Seto et al, 1999). Thus, although most of the individual components that constitute the telomerase holoenzyme have been identified, the assembly of the RNP and its regulation in vivo are still poorly understood. Recent studies have reported on the trafficking of telomerase RNA in yeast (Ferrezuelo et al, 2002; Teixeira et al, 2002) and mammalian (Jady et al, 2006; Tomlinson et al, 2006) cells, but the biological significance of this phenomenon remains unclear.
Here, we show that besides their role in telomerase activity, Est1–3 and yKu proteins act independently to promote the nuclear localization of this holoenzyme. Indeed, in the absence of any single Est protein or yKu70p, the TLC1 RNA accumulates in the cytoplasm. Yeast cells bearing a deletion of TEL1, MRE11 or XRS2, which are defective in the recruitment of telomerase to telomeres, also show reduced nuclear accumulation of TLC1 RNA. We defined the nucleo-cytoplasmic shuttling pathway involved in this process and found that it requires hypermethylation of the 5′ cap of TLC1 RNA in the nucleolus, followed by nuclear export of this RNA by the Crm1p pathway, and Mtr10p–Kap122p for its nuclear import. Our study reveals a subcellular trafficking of telomerase that is essential for its biogenesis and its association with the telomeres.
Results
The TLC1 RNA colocalizes with telomeres in G1 cells
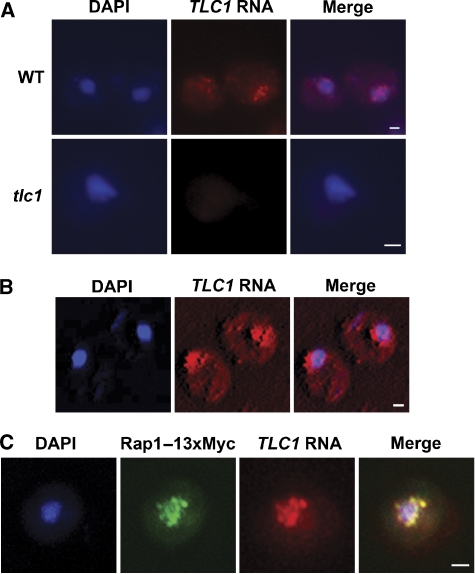
To directly visualize in situ the endogenous form of the TLC1 RNA, a set of five TLC1-specific oligonucleotide probes coupled to Cy3 fluorophores were designed and fluorescent in situ hybridization (FISH) experiments were performed on a wild-type (WT) yeast strain. As shown in Figure 1A, the signal observed with the use of these probes clustered in the form of nuclear foci. To assess the specificity of these probes, the same experiment was performed in a tlc1 strain (Figure 1A) and this resulted in little or no fluorescent signal, confirming that the probes were specific for the TLC1 RNA. Single plane projection of a three-dimensional reconstruction of yeast cells hybridized with the TLC1 probes showed that the TLC1 RNA clustered into 8–10 foci inside the cell nucleus (Figure 1B), which is consistent with a normal telomere clustering in yeast (Gotta et al, 1996; Laroche et al, 2000). To determine if the observed localization of TLC1 RNA foci was associated with nuclear telomere clustering, the detection of TLC1 RNA by FISH was coupled with immunofluorescence (IF) against a 13xMyc-tagged Rap1 protein, a telomere-binding protein commonly used for the detection of telomere position in yeast (Klein et al, 1992). As shown in Figure 1C, the FISH and IF signals overlap extensively and in G1–S phase of the cell cycle, 92% of the TLC1 RNA and Rap1p foci colocalized. These results suggest that TLC1 RNA is present at the telomeres in G1–S, at which time the catalytic subunit Est2p and the yKu complex are also associated with the telomeres (Taggart et al, 2002; Fisher et al, 2004).
Figure 1.
The TLC1 RNA clusters into 8–10 foci inside the cell nucleus that colocalized with telomeres. (A) Detection of the endogenous form of the TLC1 RNA in a WT strain (W303) using FISH assay. FISH against TLC1 RNA in a tlc1 strain was used as a control. DAPI, DNA staining. Scale bar=1 μm. The same intensity levels were set for the WT and tlc1 images. (B) FISH against TLC1 RNA, followed by acquisition of a stack of 100 images reconstructed with the Metamorph program and merged with the DAPI signal. Scale bar=1 μm. (C) Dual FISH against TLC1 RNA and immunolocalization of the Rap1–13xMyc protein. Signals were deconvoluted, merged and colocalization was measured with the Metamorph program. Scale bar=1 μm.
Est1–3 and yKu are essential for the nuclear accumulation of the TLC1 RNA
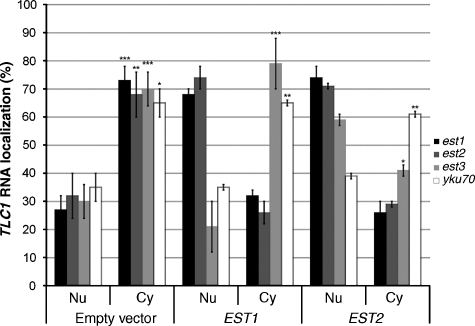
A deletion of any of the EST genes leads to progressive shortening of telomeres followed by growth arrest (Lundblad and Szostak, 1989). To investigate the contribution of these proteins to TLC1 RNA sorting and telomerase holoenzyme biogenesis, the localization of this RNA was determined in strains lacking the EST1–3 genes. As shown in Figure 2A, in strains lacking an EST gene, the probes for the TLC1 RNA detected cytoplasmic foci with little signal associated with the nuclei. The proportions at which a cytoplasmic phenotype was observed are indicated in Figure 3 and reflect the penetrance of the phenotype in a global yeast population. These results show that the absence of any one of the Est proteins affected not only the activity of the telomerase but also the accumulation of the TLC1 RNA in the nucleus. As Est1p and Est2p bind the TLC1 RNA in the absence of each other (Hughes et al, 2000; Teixeira et al, 2002) and both accumulate in the nucleus when overexpressed (Zhou et al, 2000; Teixeira et al, 2002), overexpression of these proteins may promote the nuclear accumulation of the TLC1 RNA by carrying this RNA in the nucleus. Therefore, overexpression of Est1 and Est2 proteins was performed to determine if they could rescue the cytoplasmic accumulation of TLC1 RNA observed in the est1, est2 and est3 strains. As shown in Figure 3, overexpression of either EST1 in an est1 strain or EST2 in an est2 strain efficiently restored the nuclear phenotype of the TLC1 RNA. Unexpectedly, both proteins compensated for the deletion of one another by restoring the nuclear localization of the TLC1 RNA in the various knockout backgrounds (EST2 in an est1 strain or EST1 in an est2 strain). However, only the overexpression of EST2 restored the nuclear localization of TLC1 RNA in an est3 strain (Figure 3). The reason why only Est2p and not Est1p overexpression suppresses the est3 localization phenotype remains unclear.
Figure 2.
Deletion of ESTs or YKU70 resulted in the cytoplasmic accumulation of the TLC1 RNA. (A) FISH against TLC1 RNA in WT (BY4742), est1, est2 or est3 strain. Scale bar=1 μm. (B) FISH against TLC1 RNA in yku70 and a tlc1-Δstemyku strains. Scale bar=1 μm. (C) TLC1 RNA expression level measured by qPCR in the various strains. TLC1 RNA level was normalized against that of U1 snRNA and compared with the RNA levels in the WT strain (BY4742). (D) Level of TMG-capped TLC1 RNA in the various strains measured by immunoprecipitation with an anti-TMG antibody followed by semiquantitative RT–PCR amplification of the TLC1 RNA. Numbers indicate PCR cycles. T (total): RNA from total extract; B (bound): RNA immunoprecipitated with anti-TMG antibody. Anti-TMG immunoprecipitation and RT–PCR amplification (30 cycles) of ACT1 mRNA served as a negative control.
Figure 3.
Overexpression of EST1 or EST2 in ESTs and YKU70 knockout strains and their effect on the nuclear accumulation of the TLC1 RNA. The est1, est2, est3 or yku70 strain was transformed with either the empty vector or a plasmid overexpressing EST1 or EST2. The percentage of cells with TLC1 RNA in the cytoplasm (Cy) or the nucleus (Nu) is shown. The data are from at least three independent experiments, where 100 cells were scored. *P<0.01, **P<0.005 and ***P<0.001. Standard deviations compare mutants versus the WT strain (BY4742).
Deletion of the yKu genes also leads to shorter telomeres and an alteration in the chromosomal DNA end structure, but these cells retain their ability to grow (Porter et al, 1996; Gravel et al, 1998). To determine the contribution of yKu in TLC1 RNA nuclear sorting, FISH experiments were performed on a yku70 strain and demonstrated that the TLC1 RNA was not retained in the cell nuclei in the absence of the yKu70/80p heterodimer (Figure 2B). To confirm this result, a strain expressing endogenous levels of a TLC1 RNA deleted for the yKu70/80p binding stem (tlc1-Δstemyku) was used, and a phenotype similar to a YKU70 deletion was observed (Figure 2B), suggesting that the binding of the TLC1 RNA by the yKu70/80p heterodimer is necessary to ensure nuclear localization of the telomerase holoenzyme. Overexpression of EST1 or EST2 in a yku70 strain could not bypass the requirement of yKu70p for the nuclear localization of TLC1 RNA (Figure 3), suggesting that the ESTs and yKU have different roles in the nuclear localization of TLC1 RNA.
To determine whether the TLC1 RNA mislocalization is linked to a flawed processing or modification of TLC1 RNA level, quantitative PCR (qPCR) experiments were performed to assess expression levels of the TLC1 RNA in the various mutants. Deletion of either EST genes or YKU70 did not significantly modify the expression level of the TLC1 RNA (Figure 2C). To determine the state of maturation of the TLC1 RNA found in the various mutants, immunoprecipitation of TMG-capped RNAs was performed, followed by RT–PCR of the TLC1 RNA. The results in Figure 2D show that deletions of the ESTs and YKU70 genes had little effect on the hypermethylation of the TLC1 cap or on the modification of polyadenylation of this RNA (data not shown).
Association of TLC1 RNA with telomeres maintains its nuclear retention
The previous results show that at least two independent pathways may be involved in the nuclear retention of the TLC1 RNA. One is dependent on the Est proteins, suggesting that proper assembly of the telomerase holoenzyme is required for its accumulation in the nucleus. The other pathway is dependent on the yKu heterodimer, which is involved in the recruitment of telomerase to the telomeres (Stellwagen et al, 2003; Fisher et al, 2004), suggesting that anchoring at the telomeres may be important for the nuclear retention of telomerase. To explore this possibility, mutants that affect the association of telomerase with telomeres were tested. Previous data have shown that the kinase Tel1p and the MRX complex, key mediators of DNA damage and DNA repair pathways, are both required for the recruitment of telomerase components to the telomeres (Goudsouzian et al, 2006). Using FISH, the distribution of TLC1 RNA was determined in a tel1 strain and in strains deleted of MRE11 and XRS2, which encode two components of the MRX complex. As shown in Figure 4A, deletion of these genes resulted in a redistribution of TLC1 RNA from the nucleus to the cytoplasm. Quantification of the phenotypes of these cells shows that the number of cells with TLC1 RNA mostly in the nucleus dropped by two-fold, whereas the number of cells with TLC1 RNA in both nucleus and cytoplasm, or mostly cytoplasmic, increased by over three-fold (Figure 4B). Combined with the results on yKu, these data suggest that nuclear retention of the telomerase holoenzyme requires its anchoring to telomeres.
Figure 4.
Deletion of TEL1, MRE11 or XRS2 decreases the nuclear accumulation of TLC1 RNA. (A) FISH against TLC1 RNA in tel1, mre11 or xrs2 strain. Scale bar=1 μm. (B) Quantification of the TLC1 RNA distribution observed in WT (BY4742), tel1, mre11 or xrs2 strain. A total of 100 cells were counted for each time point, in at least three independent experiments.
Nuclear export of TLC1 RNA occurs by the Crm1p pathway
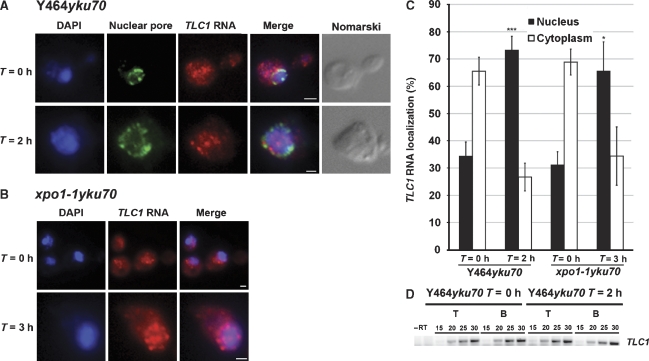
The previous experiments strongly suggest that the TLC1 RNA has a cytoplasmic phase and thus must transit through the nuclear pore complex. To characterize the RNA export machinery required for the nuclear export of TLC1 RNA, the distribution of this RNA was examined in strains harbouring different mutations affecting the Crm1p export pathway, the main export pathway for non-coding RNAs in yeast (Moy and Silver, 2002). Leptomycin B (LMB) was used to inhibit the binding of Crm1p to nuclear export signal-containing proteins in an LMB-sensitive strain (Y464) deleted of its YKU70 gene, as the TLC1 RNA was cytoplasmic in a yku70 background. In this strain, the TLC1 RNA accumulated in the cytoplasm before LMB exposure, but was sequestered in the nucleus following 2 h of LMB treatment (Figure 5A–C), indicating a defect in the export mechanism of TLC1 RNA in the absence of Crm1p function. These results were confirmed using a thermosensitive form of Crm1p (xpo1-1) in a yku70 background (Figure 5B and C), which demonstrates that Crm1p has a central role in the nuclear export of this RNA. Furthermore, we determined that TLC1 RNA was exported independent of the rRNA and mRNA nuclear export machineries by using specific export mutants (Supplementary data, Supplementary Figure S1).
Figure 5.
The TLC1 RNA is exported from the nucleus through the Crm1p-dependent pathway. (A) The LMB-sensitive strain Y464yku70 was treated with 100 ng/ml LMB for 0 or 2 h. IF against the nuclear pore complex was used to have the absolute position of the nuclear membrane. Scale bar=1 μm. (B) The xpo1-1 yku70 strain was grown at 30°C and shifted to 37°C for 3 h. FISH against the TLC1 RNA was realized after 0 or 3 h at restrictive temperature. Scale bar=1 μm. (C) Quantification of the TLC1 RNA distribution observed in the Y464yku70 and xpo1-1 yku70 strains. A total of 100 cells were counted for each time point, in at least three independent experiments. *P<0.01 and ***P<0.001. (D) Level of TMG-capped TLC1 RNA in the Y464yku70 strain after 0 or 2 h of LMB treatment as measured by immunoprecipitation with an anti-TMG antibody followed by semiquantitative RT–PCR amplification of the TLC1 RNA. Numbers indicate PCR cycles. T: RNA from total extract; B: RNA immunoprecipitated with anti-TMG antibody.
Hypermethylation of the 5′ cap of TLC1 RNA requires the methyltransferase Tgs1p and occurs in the nucleolus, before the nuclear export of this RNA
In metazoans, Crm1p is involved in the nuclear export of monomethyl guanosine (m7G)-capped snRNAs, leading to their hypermethylation into a trimethyl guanosine (m3G) cap in the cytoplasm before their re-import in the nucleus as snRNPs (Matera et al, 2007). Although the results presented above may suggest a similar pathway for the TLC1 RNA, such a mechanism is still controversial for snRNA maturation in yeast (Hopper, 2006). To determine if the TLC1 RNA is TMG-capped before or after its nuclear export, this RNA was retained in the nucleus by LMB treatment in the Y464yku70 strain, immunoprecipitated with an anti-TMG antibody and detected by RT–PCR. As shown in Figure 5D, the TLC1 RNA was detected in the pellet after immunoprecipitation, suggesting that the hypermethylation of this RNA occurs before its nuclear export. This result is supported by the observation that the TLC1 RNA was properly capped in all est mutants, which blocks telomerase biogenesis at a cytoplasmic step (Figure 2D).
The modification of the m7G cap of snRNAs and snoRNAs has been shown to depend on the methyltransferase Tgs1p in yeast (Mouaikel et al, 2002). To test the requirement of Tgs1p for the hypermethylation of the 5′ cap of TLC1 RNA, an anti-TMG cap antibody was used to immunoprecipitate the TLC1 RNA in WT and tgs1 strains. The hypermethylated form of TLC1 RNA can be immunoprecipitated normally in a WT strain, whereas the TLC1 RNA can no longer be found in the bound fraction in a tgs1 strain (Figure 6A), indicating that the TLC1 5′ cap is no longer hypermethylated in this mutant. Tgs1p is a nucleolar protein and a previous study has found that the deletion of TGS1 resulted in the nucleolar accumulation of RNA substrates of this methyltransferase (Mouaikel et al, 2002). To determine if this cap methylation defect could have an impact on the localization of the TLC1 RNA, FISH was performed against the TLC1 RNA in a tgs1 strain containing an Nop1–GFP marker to visualize the nucleolus. As shown in Figure 6B, blocking the hypermethylation of TLC1 RNA cap induced a nucleolar accumulation of this transcript, suggesting that this maturation step of the TLC1 RNA occurs in the nucleolus, as for other substrates of the Tgs1p.
Figure 6.
Tgs1p hypermethylates the 5′ cap of TLC1 RNA in the nucleolus. (A) RT–PCR amplification of TLC1 RNA from WT (W303) and tgs1 strains. T: total RNA extract; B: immunoprecipitated TMG-capped TLC1 RNA. The ACT1 mRNA was used as a negative control. (B) Deletion of TGS1 resulted in a nucleolar accumulation of the TLC1 RNA. FISH experiment was performed against TLC1 RNA in a tgs1 strain. The Nop1–GFP was used to visualize the nucleolus. Scale bar=1 μm. Number in panel d indicates the percentage of cells with this phenotype.
The TLC1 RNA shuttles between the nucleus and the cytoplasm, and requires the importins Mtr10p and Kap122p for its nuclear import
We have shown that the TLC1 RNA can be found both in the nucleus and the cytoplasm of yeasts, and that its nuclear export is dependent on Crm1p. However, it is not clear if this RNA truly shuttles between these cellular compartments, or if the nuclear export observed resulted only from a defect in the anchoring of a misassembled telomerase complex at telomeres. Evidence that the TLC1 RNA shuttles comes from the work of Teixeira and colleagues, which used an heterokaryon assay to show that overexpressed TLC1 RNA was able to shuttle between nuclei (Teixeira et al, 2002). To determine if endogenous TLC1 RNA can shuttle between the nucleus and the cytoplasm, an heterokaryon shuttling assay was also performed. A tlc1 strain was mated with a strain containing a WT TLC1 gene and a modified kar1 allele (MS739). This allele allows mating of the two strains but prevents the fusion of their nuclei (Vallen et al, 1992). Heterokaryons were identified by morphologic criteria and the presence of two or more nuclei per cell. Even if these heterokaryons have WT alleles of the ESTs and YKUs genes, they are not like WT cells. If the TLC1 RNA shuttles between the nucleus and the cytoplasm, it should be detected in every nucleus of an heterokaryon. As shown in Figure 7A, the endogenous TLC1 RNA was able to shuttle from a nucleus to another in 47% of the heterokaryons observed. To eliminate the possibility that nonspecific leakage from the nucleus may be responsible for this phenotype, the non-shuttling nuclear protein Loc1p was used as a negative control in the heterokaryon assay (Long et al, 2001). When Loc1p–Myc was transiently expressed in the kar1 strain before mating, it remained in only one nucleus in 100% of the heterokaryons observed (Supplementary Figure 2), suggesting that the nucleus was not leaky during the assay. Altogether, these experiments demonstrate that TLC1 RNA normally shuttles between the nucleus and the cytoplasm.
Figure 7.
The TLC1 RNA shuttles between the nucleus and the cytoplasm. (A) Heterokaryon shuttling assay. Heterokaryons were created by mating a Mata strain deleted of the TLC1 gene with a Matα strain carrying the kar1-1 allele and a WT TLC1 gene (MS739). Upon mating and FISH, the distribution of the TLC1 RNA in the nuclei of heterokaryons was determined. Although 47% of the heterokaryons contained TLC1 RNA in every nucleus, indicating that this RNA was shuttling between the nuclei (a–c), 53% of the heterokaryons still contained TLC1 RNA restricted to one nucleus (e–g). (B) Deletion of KAP122 and MTR10 resulted in the cytoplasmic accumulation of the TLC1 RNA. The TLC1 RNA showed a normal nuclear localization in the msn5 strain. Scale bar=1 μm. (C) Quantification of the TLC1 RNA distribution observed in the various importin alpha and beta mutant strains. Distribution at permissive (30°C) and restrictive (37°C) temperature is shown for the temperature-sensitive mutants pse1-1 and srp1-31. WT strain is BY4742. *P<0.01 and ***P<0.001.
It could be hypothesized that a shuttling TLC1 RNA requires a component of the nuclear import machinery to ensure its normal nuclear localization. Previous data suggest that the importin Mtr10p is required for TLC1 RNA biogenesis and nuclear accumulation (Ferrezuelo et al, 2002). To assess the requirement of importin proteins for the nuclear import of TLC1 RNA, a collection of importin deletion strains (either deletions or, if the gene is essential, conditional alleles) were screened for the localization of the TLC1 RNA. Deletion of most importins did not induce a cytoplasmic accumulation of TLC1 RNA (Figure 7B and C). However, deletion of KAP122 and MTR10 resulted in a significant reduction of the nuclear accumulation of the TLC1 RNA (Figure 7B and C). Mtr10p-lacking cells express low levels of TLC1 RNA (Ferrezuelo et al, 2002), a phenotype that was not observed with other mutants that promote cytoplasmic accumulation of this RNA (Figure 2C). Therefore, Mtr10p may affect TLC1 RNA by a pathway that is independent of nuclear import. Indeed, overexpression of TLC1 in an mtr10 strain restored WT telomere length but did not increase its nuclear accumulation (Ferrezuelo et al, 2002), suggesting the presence of redundant nuclear import pathways for this RNA.
Discussion
Our study provides the first direct visualization in yeast cells of the endogenous telomerase RNA subunit, the TLC1 RNA, of which there are only about 30 copies per cell (Mozdy and Cech, 2006). In G1 phase, TLC1 RNA was found to cluster into 8–10 foci inside the cell nucleus, colocalizing with the bona fide telomeric protein Rap1p. Nuclear accumulation of the TLC1 RNA depends on the proper assembly of the telomerase complex, as deleting any of the telomerase proteins (Est1–3) resulted in a mostly cytoplasmic accumulation of this RNA. Other studies have used FISH to detect the TLC1 RNA in yeast cells (Ferrezuelo et al, 2002; Teixeira et al, 2002) and have provided evidence of a nucleo-cytoplasmic trafficking of the telomerase RNA. However, neither of these detected any defect in TLC1 RNA distribution in ESTs knockout strains. As both studies used overexpressed TLC1 RNA, this may result in the titration of a limiting factor essential for TLC1 RNA nuclear export or for its processing (Chapon et al, 1997), thus promoting the nuclear retention of this RNA.
Our data on the EST genes suggest that part of the biogenesis of the telomerase holoenzyme occurs in the cytoplasm, which implies that the cytoplasmic TLC1 RNA is associated with Est proteins. This is supported by a previous study that has shown that the deletion of EST1 (which resulted in a cytoplasmic TLC1 RNA; Figure 2A) did not affect the formation of the TLC1–Est2p complex in vivo, and vice versa (Hughes et al, 2000). Moreover, a TLC1 RNA deleted of its Est1p-binding domain still maintains its association with Est2p in vivo (Osterhage et al, 2006). In the light of our results, this suggests that a cytoplasmic TLC1 RNA is associated with Est proteins. A cytoplasmic assembly of telomerase could prevent the nuclear accumulation of misassembled telomerase complexes, which could behave like dominant negative enzymes. This may happen if the TLC1 RNA is not properly folded and/or cannot interact with one of the Est proteins. Proper assembly of the telomerase holoenzyme may be monitored in the cytoplasm, and only mature and functional ribonucleocomplexes may be imported into the nucleus.
Another possibility is that only the mature telomerase holoenzyme can be retained in the nucleus by its recruitment at the telomeres. Indeed, our study also shows that the association of telomerase with telomeres is essential for maintaining this holoenzyme in the nucleus. First, disruption of YKU70 or deletion of the stem–loop that binds the yKu70/80 heterodimer in the TLC1 RNA results in a cytoplasmic TLC1 RNA. As yKu is involved in the recruitment of telomerase to telomeres, this implies that tethering the telomerase to telomeres is essential for maintaining it in the nucleus. Without an association with telomeres, the telomerase possibly shuttles in and out of the nucleus by the Crm1p pathway and, at steady state, is mostly cytoplasmic. These data are supported by deletions of TEL1 and of components of the MRX complex, both being involved in the recruitment of telomerase to telomeres, which result in a decreased nuclear accumulation of TLC1 RNA. However, the decreased localization of TLC1 RNA in the nucleus of tel1, mre11 and xrs2 strains is not as pronounced as in a yku70 strain, possibly due to the role of yKu as a more direct mediator of the interaction between telomeres and the TLC1 RNA (Stellwagen et al, 2003).
Based on the above results, a model for the trafficking of the telomerase can be suggested. (1) After being generated by RNA polymerase II, the TLC1 RNA traverses the nucleolus where cap hypermethylation occurs. (2) This RNA is then exported to the cytoplasm by the Crm1p export pathway, where part of the assembly of the telomerase holoenzyme occurs (most likely the association of Est and the yKU proteins). (3) The assembled telomerase is re-imported into the nucleus by the Mtr10p or Kap122p importin pathways. (4) Nuclear retention of telomerase requires its association with the telomeres. Interestingly, the TLC1 RNA remained nuclear in all the phases of the cell cycle in a WT background (data not shown). It would therefore appear that even if Est proteins are essential for TLC1 RNA nuclear localization, variations in Est protein levels during the cell cycle do not disturb TLC1 RNA nuclear accumulation. For instance, Est2p and TLC1 RNA are present at the telomeres in G1 (Figure 1C; Taggart et al., 2002), when Est1p levels are extremely low (Osterhage et al, 2006). As the TLC1 RNA has a long half-life (>1 h; Larose et al, 2007), it is possible that the TLC1 RNA detected at the telomeres in G1 was synthesized during the previous mitosis and remained nuclear when the levels of Est1p decrease in G1. Indeed, our data suggest that it is during the biogenesis of telomerase (i.e., with newly synthesized TLC1 RNA) that all Est proteins are required for TLC1 RNA nuclear localization. This also fits with the observation that synthesis of TLC1 RNA drops in G1 and increases in S phase (Chapon et al, 1997), like Est1p levels, suggesting a coordination between Est proteins and TLC1 RNA for telomerase assembly and nuclear localization.
Finally, our data raise the possibility that in yeast, TMG-capped RNAs such as snRNAs, may also undergo a nucleo-cytoplasmic shuttling. Although there is some evidence of such shuttling (Olson and Siliciano, 2003), it is still controversial. Altogether, this study reveals another level of regulation of telomerase assembly and recruitment to telomeres, and provides a different perspective on how telomere homeostasis is achieved.
Materials and methods
Fixation of yeast cells
Fixation protocol of yeast cells has been described previously (Chartrand et al, 2000). All centrifugation steps were carried out at 4°C when not mentioned. The cells were grown in the appropriate media until they reached early- to mid-log phase (OD600 of 0.2–0.6) and then fixed with freshly prepared 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) for 45 min at room temperature. The cells were harvested by centrifugation for 5 min at 3400 r.p.m. and washed twice with 10 ml of ice-cold 1 × buffer B (1.2 M sorbitol, 0.1 M potassium phosphate (pH 7.5)). The cells were resuspended in 1 ml of 1 × buffer B containing 20 mM vanadylribonucleoside complex (VRC), 28 mM β-mercaptoethanol, 0.06 mg/ml phenylmethylsulphonyl fluoride, 5 μg/ml of pepstatin, leupeptin and aprotinin and 120 U/ml RNA guard (all from Sigma) and transferred to a microtube containing dried oxalyticase produced from Escherichia coli DH5α. The cells were incubated at 30°C for 16–18 min to allow cell wall digestion and then harvested by centrifugation for 4 min at 3400 r.p.m. The cells were washed once with 1 ml of 1 × buffer B and resuspended in 750 μl of 1 × buffer B. A 100 μl portion of the spheroplasts was spotted on microscope coverslips coated with poly-L-lysine. The cells were incubated at 4°C for 30 min to allow the binding on the coverslip, washed once with 1 × buffer B and dehydrated in 70% ethanol in DEPC-treated water. The cells were kept at −20°C until fluorescence microscopy experiments were conducted.
Fluorescence microscopy of fixed yeast cells and signal detection
FISH experiments using oligonucleotide DNA probes have been described previously (Chartrand et al, 2000). Following fixation of the yeasts, the cells were rehydrated with two washes of 2 × SSC for 5 min at room temperature. The coverslips were incubated with 2 × SSC 40% formamide for 5 min at room temperature. In parallel, 10 μl of a mix of TLC1 probes (1 ng/μl) was mixed with 4 μl of a 5 mg/ml solution of 1:1 sonicated salmon sperm/E. coli tRNA (Sigma). The mix was lyophilized in a speed vacuum. The pellet was resuspended in 12 μl of 80% formamide and 10 mM sodium phosphate (pH 7.0). The 12 μl probe solution was heated at 95°C for 5 min and 12 μl of 4 × SSC, 20 mM VRC, 4 μg/μl BSA and 50 U of RNA guard was added. The probe preparation was finally dropped on a parafilm sheet and the coverslips were placed on the drop face-down. Hybridization was then carried out overnight at 37°C. Following hybridization, the cells were washed sequentially twice with 2 × SSC 40% formamide for 15 min at 37°C, once with 2 × SSC 0.1% Triton X-100 and twice with 1 × SSC. The coverslips were incubated in 1 × PBS containing diamidino phenylindole (DAPI) for 2 min and mounted on the microscope slides with mounting medium (86% glycerol, 1 mg p-phenylene diamine, 1 × PBS).
Dual FISH and IF protocols were conducted as described previously (Chartrand et al, 2000). Following the last wash of 1 × SSC of the FISH protocol, the cells were incubated with 1 × PBS 0.1% BSA, 1 × PBS 0.1% BSA 0.1% NP-40 (Sigma) and 1 × PBS 0.1% BSA for 5 min each at room temperature. The primary antibody was diluted to the appropriate concentration (ranging from 1/400 to 1/2500) in 1 × PBS containing 0.1% BSA, 20 mM VRC and 120 U/ml RNA guard. Mab414 αFG Nup (Abcam) was used to detect the position of nuclear pores. 9E10 anti-Myc was purchased from Roche. Hybridization was carried out from 2 h to overnight at 37 and 4°C, respectively. The cells were washed sequentially with 1 × PBS containing 0.1% BSA, 1 × PBS containing 0.1% BSA and 0.1% NP-40 and finally 1 × PBS containing 0.1% BSA for 15 min at room temperature. The secondary antibody was diluted in 1 × PBS containing 0.1% BSA, 20 mM VRC and 120 U/ml RNA guard (usually 1/1000) and incubated at room temperature for 60–90 min in the dark. The cells were washed with 1 × PBS containing 0.1% BSA, 1 × PBS containing 0.1% BSA and 0.1% NP-40 and finally 1 × PBS containing 0.1% BSA. The coverslips were finally incubated for 2 min in 1 × PBS containing DAPI and mounted.
Image acquisition, deconvolution and processing
All the images were acquired using a Nikon Eclipse E800 epifluorescence microscope equipped with a Nikon × 100 DIC H (1.4 NA) lens and with a Photometrics CoolSNAP fx CCD camera. Images were acquired using the Metamorph software and processed with Adobe Photoshop. For the deconvolution experiments, images were imported into the AutoDeblur software and the background was removed by autodetection. Deconvolution was performed using a PSF theoretical algorithm, ranging from 10 to 60 iterations every channel. Deconvoluted channels were then overlaid and the level of colocalization was determined using the Metamorph program. The very low level of the TLC1 RNA does not allow the use of Photoshop scripts to process the pictures and intensity levels were adjusted manually to allow processing.
Heterokaryon-based nucleo-cytoplasmic shuttling assay
The tlc1 and TLC1 kar1-1 strains were grown in YEPD to an OD600 of 0.5. A 25 ml portion of tlc1 strain was mated with 12.5 ml of TLC1 kar1-1 strain in 150 ml of YEPD. After 5 h of mating, the cells were fixed and FISH experiment was performed for the TLC1 RNA. For plate mating, 5 ml of tlc1 strain and 2.5 ml of TLC1 kar1-1 strain were harvested by centrifugation, washed twice with YEPD and resuspended in 300 μl of YEPD. The cells were mixed in a microtube and 200 μl was spotted on a YEPD plate. After adsorption (15 min), the plates were placed at 30°C for 5 h. After this, the cells were recovered with a cell scraper (Fisher), diluted in 50 ml of YEPD and fixed. FISH experiment was carried out as described.
As a negative control, the kar1-1 strain was transformed with pRL134 (pGal-LOC1-6xMyc; kindly provided by Roy Long). Transformants were selected and grown overnight at 30°C in SD media containing 2% raffinose. Cells were diluted at OD600 0.2 in 50 ml of SD media containing 2% galactose. Induction of Loc1p–Myc was carried out at 30°C for 2 h. After induction, expression of Loc1–Myc was repressed by adding glucose to a final concentration of 2% for 2 h at 30°C. kar1-1+pRL134 cells were mated with tlc1 cells and processed as above.
Quantitative RT–PCR analysis
Total RNA was extracted from yeast strains using the yeast RNA miniprep protocol as described previously (Schmitt et al, 1990). A 2 μg portion of total RNA was reverse-transcribed using ACT1 mRNA-, U1 snRNA- and TLC1 RNA-specific primers. Total RNA was resuspended in DEPC-treated water containing 2 pmol of gene-specific primers and 1 μl of 25 mM dNTP to 12 μl. The samples were heated for 5 min at 65°C and chilled on ice rapidly. A 4 μl volume of 5 × RT–PCR buffer (250 mM Tris–HCl (pH 8.3), 250 mM KCl, 20 mM MgCl2, 50 mM DTT), 2 μl of 1 M DTT and 1 μl of RNA guard were added to the reaction. The samples were preheated for 2 min at 42°C and 1 μl (200 U) of Revert-aid M-MuLV-RT was added to start the reaction. The samples were reverse-transcribed for 50 min at 42°C. Subsequently, gene expression level was determined using primer and probe sets from the Universal Probe Library (Roche Scientific; https://www.roche-applied-science.com). PCRs for 384-well plate formats were performed using 2 μl of cDNA samples (50 ng), 5 μl of TaqMan PCR Master Mix (Applied Biosystems, Foster City, CA), 2 μM of each primer and 1 μM of the Universal TaqMan probe in a total volume of 10 μl. The ABI PRISM® 7900HT Sequence Detection System (Applied Biosystems) was used to detect the amplification level programmed to an initial step of 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. All the reactions were run in triplicate and the average values were used for quantification. The ACT1 or snR19 (U1 snRNA) was used as an endogenous control. The relative quantification of target genes was determined using the rCT method. Briefly, the Ct (threshold cycle) values of target genes were normalized to an endogenous control gene (ACT1 or snR19) (rCT=Ct target−Ct ACT1 or snR19) and compared to each other.
Immunoprecipitation of TMG-capped RNA
The total RNA was extracted using the yeast RNA miniprep protocol as described previously (Schmitt et al, 1990). A 60 μg portion of total RNA was resuspended in 150 μl of DEPC-treated water and 2 μl of anti-TMG antibody (Calbiochem) was added. Extracts were incubated on a roller drum at 4°C for 2 h. Following this, 40 μl of protein-A Sepharose beads was added and the immunoprecipitation was incubated at 4°C on a roller drum for 2 h. The extracts were then spun for 4 min at 4500 r.p.m. at 4°C and washed three times for 4 min with washing buffer (25 mM HEPES (pH 7.5), 35 mM potassium chloride, 2 mM MgCl2). A 200 μl volume of elution buffer (50 mM Tris (pH 8), 100 mM NaCl, 20 mM EDTA, 1% SDS) was added and incubated at 65°C for 15 min or at 95°C for 5 min. The immunoprecipitated RNA was extracted in phenol–chloroform and subjected to ethanol precipitation. The pellets were resuspended in 45 μl of DEPC-treated water and 5 μl of DNase I digestion buffer was added, following which 1 μl of DNase I was added and the samples were incubated for 1 h at 37°C to eliminate DNA contamination. The RNA was extracted with 1 volume of phenol–chloroform and ethanol precipitated. The pellet was resuspended in 30 μl of DEPC-treated water and 3 μl was used for RT–PCR. The same protocol was used for the amplification of total RNA without the antibody and bead incubation, and 1.5 μl of RNA was used in RT–PCR. PCR amplification of the reverse-transcribed product was performed for 15, 20, 25 or 30 cycles and analysed on 1.5% agarose gel.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary data
Acknowledgments
We thank the laboratories of P Silver, P-E Gleizes, E Hurt, R Long, V Lundblad and J Vogel for providing yeast strains and plasmids. This work was supported by a grant from the Natural Sciences and Engineering Research Council of Canada to PC and grant MOP74438 from the Canadian Institutes for Health Research to RJW. PC was supported by fellowships from the Fond de Recherche sur la Nature et les Technologies du Québec and the Fond de Recherche en Santé du Québec.
References
- Boulton SJ, Jackson SP (1998) Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J 17: 1819–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapon C, Cech TR, Zaug AJ (1997) Polyadenylation of telomerase RNA in budding yeast. RNA 3: 1337–1351 [PMC free article] [PubMed] [Google Scholar]
- Chartrand P, Bertrand E, Singer RH, Long RM (2000) Sensitive and high-resolution detection of RNA in situ. Methods Enzymol 318: 493–506 [DOI] [PubMed] [Google Scholar]
- Counter CM, Meyerson M, Eaton EN, Weinberg RA (1997) The catalytic subunit of yeast telomerase. Proc Natl Acad Sci USA 94: 9202–9207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amours D, Jackson SP (2002) The MRE11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat Rev Mol Cell Biol 3: 317–327 [DOI] [PubMed] [Google Scholar]
- Evans SK, Lundblad V (2002) The Est1 subunit of Saccharomyces cerevisiae telomerase makes multiple contributions to telomere length maintenance. Genetics 162: 1101–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrezuelo F, Steiner B, Aldea M, Futcher B (2002) Biogenesis of yeast telomerase depends on the importin Mtr10. Mol Cell Biol 22: 6046–6055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher TS, Taggart AKP, Zakian VA (2004) Cell cycle-dependent regulation of yeast telomerase by Ku. Nat Struct Mol Biol 11: 1198–1205 [DOI] [PubMed] [Google Scholar]
- Gotta M, Laroche T, Formenton A, Maillet L, Scherthan H, Gasser SM (1996) The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J Cell Biol 134: 1349–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudsouzian LK, Tuzon CT, Zakian VA (2006) S.cerevisiae Tel1p and Mre11p are required for normal levels of Est1p and Est2p telomere association. Mol Cell 24: 603–610 [DOI] [PubMed] [Google Scholar]
- Gravel S, Larrivée M, Labrecque P, Wellinger RJ (1998) Yeast Ku as a regulator of chromosomal DNA end structure. Science 280: 741–744 [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH (1985) Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43: 405–413 [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH (1989) A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337: 331–337 [DOI] [PubMed] [Google Scholar]
- Hector RE, Shtofman RL, Ray A, Chen B-R, Nyun T, Berkner KL, Runge KW (2007) Tel1p preferentially associates with short telomeres to stimulate their elongation. Mol Cell 27: 851–858 [DOI] [PubMed] [Google Scholar]
- Hopper AK (2006) Cellular dynamics of small RNAs. Crit Rev Biochem Mol Biol 41: 3–19 [DOI] [PubMed] [Google Scholar]
- Hug N, Lingner J (2006) Telomere length homeostasis. Chromosoma 115: 413–425 [DOI] [PubMed] [Google Scholar]
- Hughes TR, Evans SK, Weilbaecher RG, Lundblad V (2000) The Est3 protein is a subunit of yeast telomerase. Curr Biol 10: 809–812 [DOI] [PubMed] [Google Scholar]
- Jady BE, Richard P, Bertrand E, Kiss T (2006) Cell cycle-dependent recruitment of telomerase RNA and cajal bodies to human telomeres. Mol Biol Cell 17: 944–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher C, Teixeira MT, Forstemann K, Lingner J (2002) Telomerase: biochemical considerations for enzyme and substrate. Trends Biochem Sci 27: 572–579 [DOI] [PubMed] [Google Scholar]
- Klein F, Laroche T, Cardenas ME, Hofmann JF, Schweizer D, Gasser SM (1992) Localization of RAP1 and topoisomerase II in nuclei and meiotic chromosomes of yeast. J Cell Biol 117: 935–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroche T, Martin SG, Tsai-Pflugfelder M, Gasser SM (2000) The dynamics of yeast telomeres and silencing proteins through the cell cycle. J Struct Biol 129: 159–174 [DOI] [PubMed] [Google Scholar]
- Larose S, Laterreur N, Ghazal G, Gagnon J, Wellinger RJ, Elela SA (2007) RNAse III-dependent regulation of yeast telomerase. J Biol Chem 282: 4373–4381 [DOI] [PubMed] [Google Scholar]
- Larrivee M, LeBel C, Wellinger RJ (2004) The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev 18: 1391–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR (1997) Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276: 561–567 [DOI] [PubMed] [Google Scholar]
- Long RM, Gu W, Meng X, Gonsalvez G, Singer RH, Chartrand P (2001) An exclusively nuclear RNA-binding protein affects asymmetric localization of ASH1 mRNA and Ash1p in yeast. J Cell Biol 153: 307–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V, Szostak JW (1989) A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57: 633–643 [DOI] [PubMed] [Google Scholar]
- Lustig AJ, Petes TD (1986) Identification of yeast mutants with altered telomere structure. Proc Natl Acad Sci USA 83: 1398–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Terns RM, Terns MP (2007) Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol 8: 209–220 [DOI] [PubMed] [Google Scholar]
- Morin GB (1989) The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell 59: 521–529 [DOI] [PubMed] [Google Scholar]
- Mouaikel J, Verheggen C, Bertrand E, Tazi J, Bordonne R (2002) Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol Cell 9: 891–901 [DOI] [PubMed] [Google Scholar]
- Moy TI, Silver PA (2002) Requirements for the nuclear export of the small ribosomal subunit. J Cell Sci 115: 2985–2995 [DOI] [PubMed] [Google Scholar]
- Mozdy AD, Cech TR (2006) Low abundance of telomerase in yeast: implications for telomerase haploinsufficiency. RNA 12: 1721–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson BL, Siliciano PG (2003) A diverse set of nuclear RNAs transfer between nuclei of yeast heterokaryons. Yeast (Chichester, England) 20: 893–903 [DOI] [PubMed] [Google Scholar]
- Osterhage JL, Talley JM, Friedman KL (2006) Proteasome-dependent degradation of Est1p regulates the cell cycle-restricted assembly of telomerase in Saccharomyces cerevisiae. Nat Struct Mol Biol 13: 720–728 [DOI] [PubMed] [Google Scholar]
- Porter SE, Greenwell PW, Ritchie KB, Petes TD (1996) The DNA-binding protein Hdf1p (a putative Ku homologue) is required for maintaining normal telomere length in Saccharomyces cerevisiae. Nucleic Acids Res 24: 582–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H, Zakian VA (2000) The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated Est1 protein. Genes Dev 14: 1777–1788 [PMC free article] [PubMed] [Google Scholar]
- Ritchie KB, Mallory JC, Petes TD (1999) Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol Cell Biol 19: 6065–6075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie KB, Petes TD (2000) The Mre11p/Rad50p/Xrs2p COMPLEX and the Tel1p function in a single pathway for telomere maintenance in yeast. Genetics 155: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabourin M, Tuzon CT, Zakian VA (2007) Telomerase and Tel1p preferentially associate with short telomeres in S.cerevisiae. Mol Cell 27: 550–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt ME, Brown TA, Trumpower BL (1990) A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res 18: 3091–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto AG, Zaug AJ, Sobel SG, Wolin SL, Cech TR (1999) Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature 401: 177–180 [DOI] [PubMed] [Google Scholar]
- Singer MS, Gottschling DE (1994) TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266: 404–409 [DOI] [PubMed] [Google Scholar]
- Stellwagen AE, Haimberger ZW, Veatch JR, Gottschling DE (2003) Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev 17: 2384–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taggart AKP, Teng S-C, Zakian VA (2002) Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science 297: 1023–1026 [DOI] [PubMed] [Google Scholar]
- Teixeira TM, Förstemann K, Gasser SM, Lingner J (2002) Intracellular trafficking of yeast telomerase components. EMBO Rep 3: 652–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson RL, Ziegler TD, Supakorndej T, Terns RM, Terns MP (2006) Cell cycle-regulated trafficking of human telomerase to telomeres. Mol Biol Cell 17: 955–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallen EA, Hiller MA, Scherson TY, Rose MD (1992) Separate domains of KAR1 mediate distinct functions in mitosis and nuclear fusion. J Cell Biol 117: 1277–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappulla DC, Cech TR (2004) Yeast telomerase RNA: a flexible scaffold for protein subunits. Proc Natl Acad Sci USA 101: 10024–10029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Hidaka K, Futcher B (2000) The Est1 subunit of yeast telomerase binds the Tlc1 telomerase RNA. Mol Cell Biol 20: 1947–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary data