Abstract
In the developing nervous system, building a functional neuronal network relies on coordinating the formation, specification and survival to diverse neuronal and glial cell subtypes. The establishment of neuronal connections further depends on sequential neuron–neuron and neuron–glia interactions that regulate cell-migration patterns and axon guidance. The visual system of Drosophila has a highly regular, retinotopic organization into reiterated interconnected synaptic circuits. It is therefore an excellent invertebrate model to investigate basic cellular strategies and molecular determinants regulating the different developmental processes that lead to network formation. Studies in the visual system have provided important insights into the mechanisms by which photoreceptor axons connect with their synaptic partners within the optic lobe. In this review, we highlight that this system is also well suited for uncovering general principles that underlie glial cell biology. We describe the glial cell subtypes in the visual system and discuss recent findings about their development and migration. Finally, we outline the pivotal roles of glial cells in mediating neural circuit assembly, boundary formation, neural proliferation and survival, as well as synaptic function.
Keywords: Optic lobe, photoreceptor axons, axon guidance, migration, neurogenesis
INTRODUCTION
Recent studies have made it increasingly clear that to understand the mechanisms directing the formation of complex neuronal connections, it is important to focus on both neurons and glial cells. About a century ago, Ramòn y Cajal described his appreciation of the delicate, complex architecture of the insect compound eye and its associated processing centers in his autobiography (Ramòn y Cajal, 1937). Since then, the Drosophila visual system has emerged as an excellent model to investigate the mechanisms underlying the development and function of the nervous system (reviewed in Clandinin and Zipursky, 2002; Choe and Clandinin, 2005; Mast et al., 2006; Ting and Lee, 2007).
The visual system of the adult fly consists of the compound eye and the optic lobe, which is subdivided into three ganglia, the lamina, medulla and lobula complex. Within the retina, photoreceptor cells (R-cells) are organized into ∼800 regular subunits, called ommatidia. Each subunit contains eight R-cells (R1–R8): R1–R6 axons innervate the lamina, whereas R7 and R8 axons pass through the lamina and terminate in two of ten distinct neuropil layers in the medulla (Fig. 1). Projection pattern formation is initiated during the mid-3rd instar larval stage, when R-cell growth cones extend from the eye disc into the optic lobe. They provide anterograde signals that induce the development of their targets in the lamina, and select their specific ganglia. During subsequent pupal stages, R-cell axons choose their synaptic target neurons. In the lamina, R1–R6 growth cones derived from each R-cell cluster leave their original bundle and extend laterally in a precise pattern to establish synaptic contacts with adjacent sets of lamina neurons. This leads to the assembly of synaptic units called lamina cartridges (reviewed in Clandinin and Zipursky, 2002; Mast et al., 2006; Ting and Lee, 2007). In the medulla, R7 and R8 axons project initially to temporary layers and then to final recipient layers (M6 and M3), where they connect with target neurons in a layer- and column-specific manner (Ting et al., 2005).
Fig. 1. Glial-cell subtypes and their roles in the larval and adult visual system.
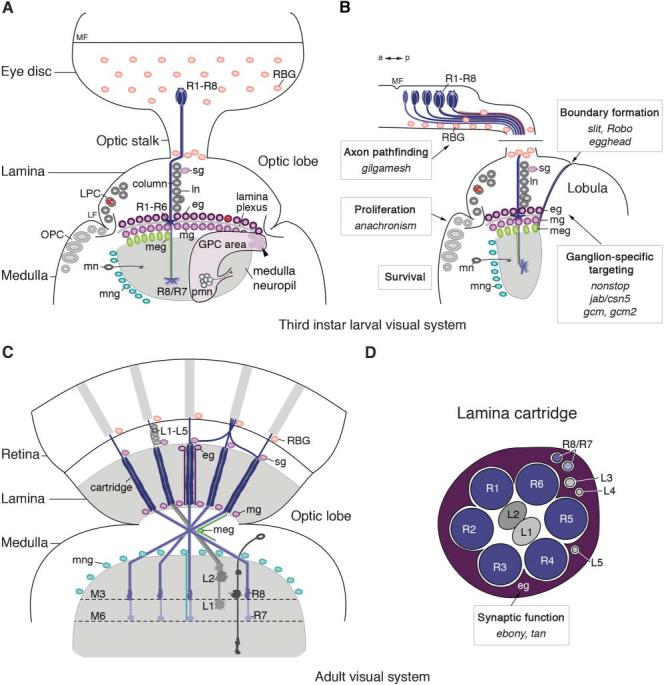
(A,B) Morphology of the 3rd instar larval visual system in frontal (A) and horizontal (B) orientations. R-cell axons (R1–R8) project from the eye imaginal disc through the optic stalk into the optic lobe. Retinal basal glial cells (RBGs) originate in the optic stalk and migrate into the eye disc. Progenitors in the outer proliferation center (OPC) closer to the lamina furrow (LF) give rise to lamina precursor cells (LPCs). These divide to generate lamina neurons (ln). Their cell bodies are organized into columns in close proximity with R-cell axons bundles. Satellite glial cells (sg) are positioned at the level of lamina neuron cell bodies. R1–R6 growth cones terminate between the rows of epithelial (eg) and marginal (mg) glial cells. These are born in glial precursor cell (GPC) areas and share a lineage with proximal medulla neurons (pmn). The arrowhead indicates the position of committed glial precursors adjacent to the R-cell projection field. Epithelial and marginal glial cells migrate to their characteristic positions above and below the lamina plexus. Proximally located progenitors in the OPC give rise to medulla neurons (mn). R7 and R8 axons stop in the medulla neuropil, which is delineated by medulla glial cells (meg) and medulla neuropil glial cells (mng). a, anterior; MF, morphogenetic furrow; p, posterior. (C,D) Morphology of the adult visual system. (C) In the lamina, R1–R6 axons and processes of lamina neurons L1–L5 are organized into cartridges. R7 and R8 axons innervate two neuropil layers (M3 and M6) in the medulla. (D) Schematic drawing of a lamina cartridge cross-section. Boxes in (B) and (D) indicate the developmental processes that are controlled by glial cells, as well as the genes that are associated with these functions.
Most strikingly, from larval stages to adulthood, the fly visual system is endowed with a wealth of different glial cell subtypes. In the adult, these abundantly ensheath neuronal cell bodies, axon bundles and neuropil compartments, and, in the complexity of their morphological features, are a close match to their vertebrate oligodendrocyte and astrocyte glial counterparts (Fig. 2). The design of sophisticated genetic tools has made it possible to determine the requirements of genes, often at the single-cell level, in both neuronal and glial populations in the visual system. This has led to considerable advances in our understanding of the cellular and molecular mechanisms underlying the pathfinding and targeting of R-cell axons (reviewed in Clandinin and Zipursky, 2002; Mast et al., 2006; Ting and Lee, 2007). Here, we focus on the glial cells in the visual system, highlighting recent insights into the mechanisms that regulate their development and migration, and their distinct contributions to the step-by-step assembly and function of neuronal connections.
Fig. 2. Glial cells in the adult visual system of Drosophila.
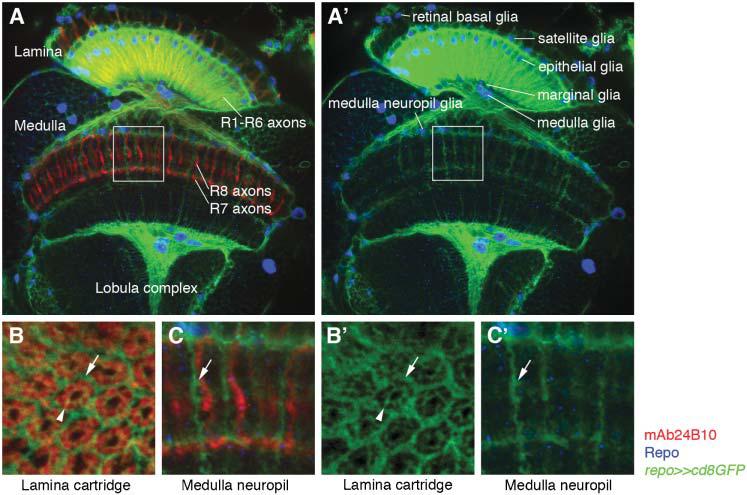
R-cell axons are labeled in red using the R-cell specific marker mAb24B10. Glial nuclei (blue) and their processes (green) are labeled with the glial-specific marker Repo and by using repo-Gal4 to drive expression of membrane-bound green fluorescent protein (GFP), respectively. (A,A') R-cell axons project from the retina to the optic lobe comprising three ganglia, the lamina, medulla and lobula complex. R1–R6 axons stop in the lamina, whereas R7 and R8 axons terminate in the medulla. Different glial cell subtypes are found in each ganglion. Their processes extensively enwrap lamina cartridges and reflect the organization into layers and columns in the medulla. (B,B') High magnification of a cross-section through the lamina. Each lamina cartridge is enwrapped by glial cell processes (arrow). Glial-cell processes also surround individual R1–R6 terminals in each cartridge (arrowheads). (C,C') High magnification of distal medulla neuropil layers (boxes in A and A'). Glial-cell processes extend into medulla neuropil columns in the vicinity of R-cell axons (arrows).
Glial-cell subtypes in the visual system
Previous studies in the embryonic central and peripheral nervous systems (CNS and PNS) of Drosophila have identified four main subtypes of glial cells based on their relative position and morphology: (1) surface glia (perineurial and subperineurial glia), which surround the CNS and peripheral nerves and participate in the formation of the blood–brain barrier; (2) cell body-associated glia, which enwrap neuronal cell bodies in the cortex; (3) neuropil-associated glia, which either form borders between the cortex and the central neuropil or ensheath neuronal compartments and axonal bundles; and (4) nerve-associated glia, which enwrap axons within peripheral nerves (Ito et al., 1995; Hartenstein et al., 1998; Pereanu et al., 2005; reviewed in Freeman and Doherty, 2006).
In the visual system, R-cells differentiate and assemble sequentially into clusters in the eye imaginal disc from the mid-3rd instar larval to early pupal stages (Fig. 1). R-cell axons project basally into the nerve layer, emerging beneath the eye disc epithelium, and turn posteriorly towards the optic stalk. R-cell growth cones then extend through the optic stalk into the optic lobe, where they terminate in either the lamina or the medulla. Within the eye imaginal disc and optic stalk, R-cell axons come into close contact with retinal basal glial cells (RBGs), also called subretinal glia (Winberg et al., 1992; Choi and Benzer, 1994; Rangarajan et al., 1999). Two subtypes of RBGs have so far been identified, surface and wrapping glial cells, which are equivalent to embryonic surface and nerve-associated glia, respectively. Surface glial cell bodies and processes form a single layer that surrounds the basal nerve layer of R-cell axons in the eye disc and the optic stalk. Wrapping glial cells are located in the nerve layer and the optic stalk, and extend fine processes that enwrap R-cell axon bundles from each ommatidial cluster as they mature (Hummel et al., 2002). Thus, newly ingrowing R-cell axons are found at the periphery of the optic stalk and are unwrapped initially. Axon fascicles from older R-cell clusters are located more centrally near Bolwig's nerve, which comprises the axons of larval photoreceptors, and are each surrounded by fine glial processes (Meinertzhagen and Hanson, 1993).
In the 3rd instar larval optic lobe, lamina neurons, the prospective synaptic partners of R1–R6 axons, are organized into columns separated by R-cell axon bundles. Satellite glial cells, equivalent to cell body-associated glia, are in close contact with lamina neurons and ensheath their cell bodies as they mature. With the exception of satellite glia, the other glial-cell subtypes so far identified in the optic lobe are equivalent to neuropil-associated glia. Incoming R1–R6 growth cones stop in the lamina and form a thin stripe of neuropil, called the lamina plexus. They terminate between two rows of cuboidal-shaped glial cells, distal epithelial and proximal marginal glial cells. An underlying third row of oval-shaped medulla glial cells is positioned at the lamina–medulla boundary, each enwrapping fascicles of R7, R8 and target neuron processes. In the adult visual system, epithelial glial cells ensheath each lamina cartridge extensively. Finally, medulla neuropil glial cells separate the medulla cortex from the central neuropil and, in adults, extend fine processes into the medulla neuropil (Figs 1, 2; Table 1) (Winberg et al., 1992; Meinertzhagen and Hanson, 1993; Perez and Steller, 1996a; Tix et al., 1997; Poeck et al., 2001).
Table 1.
Glial cell subtypes and their origins in the visual system
| Subtypes (origin) | Surface glia | Cell body-associated glia | Neuropil-associated glia | Nerve-associated glia |
|---|---|---|---|---|
| Eye (optic stalk) | Retinal basal glia | Retinal basal glia | ||
| -Surface glia | -Wrapping glia | |||
| Optic lobe (GPC area) | Satellite gliaa | Epithelial glia | ||
| Marginal glia | ||||
| Medulla neuropil glia | ||||
| Medulla gliaa |
Origin unknown
Origins of glial-cell subtypes in the eye and optic lobe
In the embryonic CNS, glial cells are derived from either pure glial lineages formed by glioblasts or mixed neuronal/glial lineages formed by neuroglioblasts. The latter divide to give rise to precursors with either neuronal or glial potentials (Type 1) or by asymmetrical division to generate ganglion mother cells (GMCs), which then produce in parallel or sequentially neuronal and glial progeny (Type 2) (reviewed in Egger et al., 2007a).
In the eye imaginal disc, pulse-labeling assays with bromodeoxyuridine have established that RBGs originate from a pure glial lineage in the optic stalk and migrate into the eye imaginal disc (Choi and Benzer, 1994). In the optic lobe, clonal analyses using the ‘FLP-out’ approach (Struhl and Basler, 1993) and mosaic analysis with a repressible cell marker (MARCM) (Lee and Luo, 1999) have identified some of the neuronal and glial lineages in the lamina and medulla (Winberg et al., 1992; Perez and Steller, 1996a; Huang and Kunes, 1996; Huang and Kunes, 1998; Huang et al., 1998; Dearborn and Kunes, 2004; Chotard et al., 2005). Mitotically active progenitors in the outer proliferation center (OPC) are located anteriorly to a groove in the optic lobe surface, called the lamina furrow. These give rise to lamina precursor cells (LPCs) that, in turn, divide once posteriorly of the lamina furrow to produce only lamina neurons and not glia (Selleck and Steller, 1991; Perez and Steller, 1996a). In close association with R-cell axon bundles, lamina neurons are assembled subsequently into columns. OPC progenitors close to the central brain give rise to neuroblasts, which generate GMCs and, finally, medulla neurons (Egger et al., 2007b). Although epithelial and marginal glial cells are adjacent to lamina neurons and are an intricate part of each column, lamina neurons and glial cells have distinct origins. Indeed, epithelial and marginal glial cells are derived from two, superficially located, dorsal and ventral gliogenic domains, called glial precursor cell (GPC) areas (Perez and Steller, 1996a; Huang and Kunes, 1996, 1998; Huang et al., 1998; Dearborn and Kunes, 2004; Chotard et al., 2005). In our current understanding, GPC areas contain multipotent progenitors. Proximally, they produce a subtype of neurons that innervate deeper medulla neuropil layers. Distally, they give rise to committed, mitotically active, glial precursors located at the dorsal and ventral margins of the R-cell projection field. Committed precursors, in turn, generate epithelial and marginal glial cells that migrate to their characteristic positions above and below the lamina plexus, possibly by chain migration (Fig. 1; Table 1) (Perez and Steller, 1996a; Huang and Kunes, 1996, 1998; Dearborn and Kunes, 2004; Chotard et al., 2005). Finally, clonal analysis indicates that medulla neuropil glial cells also originate from parts of the GPC areas (Dearborn and Kunes, 2004). The lineage of medulla glial cells is so far unresolved.
Molecularly, GPC areas are characterized by the combined expression of Wingless (Wg), the Tbx1-related transcription factor Optomotor blind, and the Cadherin family member Dachsous (Dearborn and Kunes, 2004), as well as reporter-gene expression driven by enhancer trap P-element insertions into the lamina-ancestor (lama) locus (Perez and Steller 1996b; Chotard et al., 2005). Committed glial precursors are characterized by the expression of Glial cells missing (Gcm) and Gcm2, two transcriptional regulators with central roles in glial-cell development in the Drosophila embryo (Jones et al., 1995; Hosoya et al., 1995; Vincent et al., 1996; Kammerer and Giangrande, 2001; Alfonso and Jones, 2002; Dearborn and Kunes, 2004; Chotard et al., 2005; Yoshida et al., 2005; Soustelle et al., 2007).
All glial-cell subtypes in the visual system are characterized by the expression of reversed polarity (repo), which encodes a paired-like homeodomain protein (Xiong et al., 1994; Campbell et al., 1994; Halter et al., 1995). Repo expression is used widely as a glial-specific marker in both developing and adult nervous systems of Drosophila, and indeed Repo received its name from findings in the visual system. Loss of repo function interferes with glial differentiation in the eye and optic lobe and, in consequence, leads to a reversal in polarity of electroretinogram (ERG) recordings in response to a light stimulus in adults (Xiong et al., 1994; Campbell et al., 1994; Xiong and Montell, 1995). In addition to their position, some glial cells can be further identified molecularly by subtype-specific markers. For example, P-element insertions and reporter constructs of locomotion defects (loco), encoding a member of the regulator of G-protein signaling (RGS) protein family, label epithelial and marginal glial cells, but not medulla glial cells (Winberg et al., 1992; Granderath et al., 2000; Poeck et al., 2001).
Mechanisms of glial formation and migration in the eye
During the 3rd instar larval stage, the eye imaginal disc displays an apical depression, called the morphogenetic furrow, that sweeps from posterior to anterior across the epithelium. In its wake, R-cells differentiate sequentially and assemble into rows of clusters that extend axons into the underlying basal nerve layer (reviewed in Pappu and Mardon, 2004; Silver and Rebay, 2005). RBGs migrate from the optic stalk to the basal surface of the eye imaginal disc concomitantly with the onset of R-cell differentiation. They remain in close contact with R-cell axons and do not migrate anteriorly past the morphogenetic furrow.
Initiation and propagation of R-cell differentiation posterior to the morphogenetic furrow depends on the combined activity of the early eye-specification factors such as Eyeless (Ey), Sine oculis (So) and Eyes absent (Eya), as well as the morphogens Decapentaplegic (Dpp) and Hedgehog (Hh) (reviewed in Pappu and Mardon, 2004; Silver and Rebay, 2005). The population of RBGs expands throughout the 3rd instar larval stage and undergoes mitotic divisions within the optic stalk and, to some extent, in the eye disc (Rangarajan et al., 2001). Loss- and gain-of-function analyses suggest that Dpp takes part in regulating RBG proliferation (Rangarajan et al., 2001).
In the absence of R-cells in the eye imaginal disc, RBGs fail to migrate and remain in the optic stalk (Choi and Benzer, 1994). However in animals in which R-cells form but fail to extend axonal projections, RBGs are nevertheless able to enter posterior regions of the eye imaginal disc containing differentiated R-cells. They also migrate to ectopic patches of R-cell bodies located anteriorly of the morphogenetic furrow (Rangarajan et al., 1999). This indicates that, although differentiation of R-cells is essential for the migration of RBGs, they do not require contact with extending axons (Choi and Benzer, 1994; Rangarajan et al., 1999). RBGs might, thus, rely on a chemoattractant signal for migration into the eye disc (Rangarajan et al., 1999). Three lines of evidence support the idea that the onset of glial migration is highly regulated. First, in 3rd instar larvae with mutations in either gilgamesh (gish), which encodes casein kinase Iγ, or fear-of-intimacy (foi), which encodes an eight-pass transmembrane domain protein, RBGs migrate beyond posterior parts of the eye disc epithelium, forming streams of cell bodies to ectopic positions anterior of the morphogenetic furrow (Hummel et al., 2002; Pielage et al., 2004). Second, in gish homozygous-mutant animals, RBGs (particularly the surface glial-cell subtype) enter the eye disc before the onset of R-cell differentiation (Hummel et al., 2002). Third, loss of hh, ey and so during early larval development results in RBG-migration defects similar to the phenotype observed in gish mutants (Hummel et al., 2002). Together, these findings indicate that precocious glial migration from the optic stalk into the developing eye disc before R-cell differentiation is prevented by an, as yet, unidentified inhibitory signal from the posterior eye disc. Once R-cells begin to form they might promote RBG migration by either releasing this block or actively increasing glial motility (Hummel et al., 2002).
Mechanisms of glial formation and migration in the optic lobe
As early as 1943, Maxwell Power observed that the development of the optic lobe depends on the compound eye (Power, 1943). We now know that two R-cell-derived, anterograde extrinsic signals control the development of lamina neurons in the optic lobe (Selleck and Steller, 1991; Huang and Kunes, 1996; Huang and Kunes, 1998; Huang et al., 1998). Newly formed R-cell axons project from the eye imaginal disc into the optic lobe, closely adjacent to LPCs. R-cell axons release the first signal Hh to induce cell-cycle progression from G1 to S phase and, consequently, mitotic division of LPCs. Hh also promotes the expression of the retinal determination network member Dachshund (Mardon et al., 1994; Huang and Kunes, 1996) and the basic helix-loop-helix-PAS transcription factor Single-minded (Umetsu et al., 2006). The integration of lamina neurons into columns is mediated by Single-minded (Umetsu et al., 2006), whereas Dachshund is required for the synthesis of the epidermal growth factor (EGF) receptor in LPCs and lamina neurons, which makes them responsive to the second R-cell-derived signal, the EGF-like ligand Spitz (Huang et al., 1998; Chotard et al., 2005). EGF receptor activation controls the subsequent steps of lamina neuron maturation, monitored by the expression of the neuronal differentiation marker Elav and the subtype-specific marker Brain-specific homeoprotein (Huang et al., 1998).
Three observations make a strong case that a third R-cell-derived signal controls the development of lamina glial cells. First, in so or eya homozygous mutant animals, which lack R-cell innervation, the number of Repo-positive glial cells in the lamina is much reduced and expression of late glial-specific markers is abolished. Second, analyses of lineages and GPC-area markers show that glial precursors form before, and independently of, R-cell axon innervation. Third, in the absence of R-cell axons, some glial cells form but they accumulate in ectopic positions within the GPC areas (Winberg et al., 1992; Perez and Steller, 1996a; Huang and Kunes, 1996; Huang and Kunes, 1998). Hence, an R-cell axon-dependent signal does not regulate the initial formation of glial precursors, but is required for continued proliferation, differentiation and migration of glial cells in the lamina (Perez and Steller, 1996a; Huang and Kunes, 1996; Huang and Kunes, 1998). This anterograde signaling mechanism might ensure matching of R-cell projection-field size with the number of glial cells in the lamina.
Epithelial and marginal glial cells express the Hh receptor Patched, suggesting they are responsive to Hh signaling. Nevertheless, in animals, homozygous mutant for the eye-specific allele hh1, in which 11–13 rows of R-cell clusters are formed but do not release Hh, lamina glial cell development is not affected. This shows that their migration does not depend on Hh, but on a third, so far unidentified, R-cell-derived signal (Huang and Kunes, 1998). In animals that are homozygous for jab1/csn5, which encodes the Drosophila homolog of Jun-activation-domain binding protein 1 (Jab1)/subunit 5 of the Arabidopsis COP9 signalosome, many lamina epithelial and marginal glial cells fail to migrate to their characteristic positions above and below the lamina plexus; instead, they accumulate at the dorsal and ventral margins of the R-cell projection field. jab1/csn5 is expressed and genetically required in R-cells, indicating that the R-cell-derived signal that promotes glial migration depends on the function of the COP9 signalosome (Suh et al., 2002). Finally, this or another early R-cell-derived signal is required to promote the outgrowth of an axonal scaffold formed by medulla neuron subtypes within the GPC areas that directs the migration of glial-cell subtypes to their specific locations in the R-cell projection field. Consistent with this model, glial cells in the optic lobe fail to migrate in the absence of these axonal tracts or follow their abnormal trajectories (Dearborn and Kunes, 2004).
In addition to extrinsic regulation, the development of glial cells in the optic lobe is controlled by intrinsic mechanisms. In the Drosophila embryo, Gcm and Gcm2 have a central role in regulating the choice of glial and neuronal fates in offspring of glioblast- and neuroglioblast-derived lineages. In the absence of gcm, precursors do not give rise to glial cells but form neurons, whereas ectopic expression of gcm promotes the formation of glial cells instead of neurons (Jones et al., 1995; Hosoya et al., 1995; Vincent et al., 1996; Kammerer and Giangrande, 2001; Alfonso and Jones, 2002). In the postembryonic optic lobe, gcm and gcm2 mRNA and Gcm protein are expressed in precursors at the border of GPC areas adjacent to the dorsal and ventral margins of the R-cell projection field (Chotard et al., 2005; Soustelle et al., 2007). Furthermore, a P-element enhancer trap insertion into the gcm locus shows perdurance of reporter gene expression in differentiated epithelial and marginal glial cells in addition to these glial precursors, but not in medulla glial cells, confirming the lineage that gives rise to lamina glia (Chotard et al., 2005; Yoshida et al., 2005; Soustelle et al., 2007). Two FLP/FRT based genetic approaches, MARCM and the ey3.5-Gal4, lama-Gal4, UAS-FLP system (ELF) (Chotard et al., 2005) facilitate the analysis of gene functions in target neurons, glial cells and their precursors in the optic lobe. While loss of either gcm or gcm2 alone does not interfere with glial development in the optic lobe, removal of both transcriptional regulators prevents the formation of epithelial and marginal glial cells. This shows that unlike in embryos where gcm by itself has a major role, both genes act redundantly in the postembryonic visual system to promote the formation of lamina glia. This might be due to similar expression levels of both homologs in the optic lobe, while gcm is expressed at higher levels compared to gcm2 in the embryonic CNS (Kammerer and Giangrande, 2001; Alfonso and Jones, 2002; Chotard et al., 2005; Soustelle et al., 2007). Gain-of-function analyses further indicate that Gcm and Gcm2 are sufficient to induce ectopic glial formation in the medulla cortex but not in the lamina (Chotard et al., 2005; Colonques et al., 2007). Consistent with the observation that Dpp is present in dorsal and ventral fields that flank Wg-positive cells and differentiated glial cells within the GPC areas (Kaphingst and Kunes, 1994; Dearborn and Kunes, 2004; Yoshida et al., 2005), loss- and gain-of-function analyses of components of the Dpp pathway in the optic lobe indicate that Dpp signaling acts upstream of gcm in GPC areas and, thus, of lamina glia differentiation (Yoshida et al., 2005).
Analysis in the visual system has further uncovered a cell-autonomous requirement for Gcm and Gcm2 in mediating neuronal development, in addition to their conserved, glial fate-promoting roles. LPCs, the neuronal precursors that give rise to lamina neurons, express gcm and gcm2 mRNA, and Gcm protein (Chotard et al., 2005; Soustelle et al., 2007). Removal of gcm and gcm2 function using the ELF and MARCM approaches (Chotard et al., 2005) and overexpression of a dominant-negative Gcm transgene (Yoshida et al., 2005; Soustelle et al., 2007) in the lamina interferes with LPC proliferation and differentiation, and, consequently, the formation of lamina neurons. Gcm and Gcm2 mediate neurogenesis in the lamina through cooperation with the Hh signaling pathway (Chotard et al., 2005).
Glial cells control R-cell axon pathfinding and targeting
Precise temporal and spatial regulation of glial development and migration in the eye and the optic lobe is crucial for at least two steps of visual circuit assembly (Fig. 1B). During the 3rd instar larval stage, if RBGs are prevented from migrating by over-expressing a dominant-negative form of Ras, R-cells extend axons towards the posterior eye disc, but then stall and fail to enter the optic stalk (Rangarajan et al., 1999). In the absence of gish function, RBGs enter the eye disc prematurely before the onset of R-cell differentiation and migrate to ectopic anterior positions. Strikingly, rather than extending posteriorly, some R-cell axons project anteriorly following the abnormal trajectories of ectopic glial cells in these flies (Hummel et al., 2002). These findings strongly support the idea that RBGs are necessary for R-cell axon pathfinding and have an instructive role by conveying directional information.
In the optic lobe, based on their ideal positions above and below the lamina plexus, epithelial and marginal glia have been suggested to mediate targeting of R1–R6 axons (Winberg et al., 1992; Perez and Steller, 1996a). Because of the lack of specific Gal4 drivers that are active solely in these glial cell subtypes, ablation experiments to demonstrate such a role have so far not been possible. Instead, mutations that affect either migration or formation of glial cells in the optic lobe have been instrumental in providing experimental evidence. In animals mutant for jab1/csn5 and nonstop, encoding a ubiquitin-specific protease, fewer glial cells from dorsal and ventral GPC areas reach their characteristic positions close to the lamina plexus, and instead accumulate at the margins of the R-cell projection field. In consequence, R1–R6 axons frequently do not terminate in the lamina and misproject to the medulla (Poeck et al., 2001; Suh et al., 2002). Removal of gcm and gcm2 function using genetic mosaic analysis interferes with the formation of lamina glial cells and results in similar R1–R6 targeting defects (Chotard et al., 2005). Conversely, in hh1 homozygous mutant animals, in which the formation of lamina neurons is disrupted while glial cells are unaffected, R1–R6 axons terminate correctly in the lamina (Huang and Kunes, 1996, 1998; Poeck et al., 2001; see above). Differentiation of lamina neurons is ongoing during the 3rd instar larval stage and selection of sets of postsynaptic lamina neurons takes place during subsequent pupal stages (reviewed in Mast et al., 2006; Ting and Lee, 2007). Hence, glial cells within the lamina act as intermediate target cells that are required for R1–R6 ganglion-specific targeting, likely by providing one or more stop signals.
During the first half of pupal development, R1–R6 axons leave their bundles of origin in the lamina plexus and project to adjacent sets of lamina neurons in a precise, invariant pattern. Strikingly, glial cell processes form a regular net-like pattern in the lamina and are in close contact with R1–R6 growth cones during the crucial stages of development (Clandinin and Zipursky, 2000). This suggests that glial cells could also provide spatial cues that facilitate afferent–target interactions and, ultimately, support pupal R1–R6 target selection.
Glial cells demarcate ganglion boundaries
Although neurons and glial cells in the developing lamina and lobula cortices are located transiently in close proximity to each other during the 3rd instar larval stage, their cell populations do not mix. Recent studies have begun to shed light on the underlying cellular and molecular mechanisms, demonstrating that glial cells have a crucial role in maintaining these developmental compartment boundaries (Tayler et al., 2004; Fan et al., 2005). Glial cell processes form a sheath-like border that extends from the base of the optic stalk to the level of lamina and medulla glial cells. This boundary separates the lamina from a subtype of lobula neurons called distal cell neurons (Tayler et al., 2004; Fan et al., 2005). In animals that are homozygous mutant for egghead, encoding a glycosyl transferase required for glycosphingolipid biosynthesis (Wandall et al., 2005), the glial interface is disrupted and, in consequence, lobula cells migrate ectopically into the lamina disturbing the positioning of epithelial and marginal glial cells. Eventually, this leads to severe errors in R1–R6 ganglion-specific targeting (Fan et al., 2005). Epithelial, marginal and medulla glial cells at this boundary express the repellent ligand Slit, whereas distal cell neurons in the lobula cortex contain the guidance receptors Robo, Robo2 and Robo3. Loss of either Slit or specific knockdown of Robo function in distal cell neurons causes these neurons to invade the lamina, indicating that glial cells utilize Slit–Robo signaling to maintain the compartment boundary between lamina and lobula (Tayler et al., 2004).
Glial cells modulate neural proliferation and survival
At the end of embryonic development, neuroblasts generally exit the cell cycle and enter a quiescent state. Findings in the visual system have helped to demonstrate that glial cells participate in re-activating neuroblasts during postembryonic development. Indeed, surface glial cells that are close to the OPC express the secreted glycoprotein Anachronism (Ana), which is required to prevent premature neuroblast proliferation in the OPC during early larval stages (Fig. 1B) (Ebens et al., 1993). Moreover, glial cells also mediate neuronal survival in the lamina and medulla. In particular, in flies that are homozygous for the viable hypomorphic allele repo1, lamina glial cells fail to terminally differentiate and undergo apoptosis from late-pupal development onwards. Glial-cell death coincides with apoptosis of lamina neurons (Xiong and Montell, 1995). Similarly, in late-3rd instar larval optic lobes lacking R-cell innervation, the frequency of apoptosis increases for medulla neurons in cortical areas that are devoid of glial cells (Dearborn and Kunes, 2004).
Glial function in the adult visual system
In the adult visual system, R1–R6 axons are organized in a regular array of lamina cartridges in which R-cell axons form synaptic contacts with lamina neurons L1–L3 (Meinertzhagen and O'Neil, 1991). Each synaptic unit is surrounded by processes of epithelial glial cells that extend characteristic deep invaginations, called capitate projections, into R1–R6 terminals within the lamina. Capitate projections also occur in profiles of R7 and R8 axons in the medulla (reviewed in Prokop and Meinertzhagen, 2006), and are likely to be derived from medulla neuropil glial cells. R-cell axons release the neurotransmitter histamine to activate postsynaptic, histamine-gated chloride channels, one of which is encoded by ora transientless (ort)/hclA (reviewed in Stuart, 1999; Gengs et al., 2002; Gisselmann et al., 2002). Because gating of channels at histaminergic synapses leads to hyperpolarization and inhibition of postsynaptic partners, signal transduction depends on the efficient closure of channels and clearance of neurotransmitter from the synaptic cleft (reviewed in Stuart, 1999; Wagner et al., 2007). Strikingly, epithelial and medulla neuropil glial cells contain the β-alanyl biogenic amine synthase Ebony, whereas R-cell axons contain the cysteine peptidase Tan. Both enzymes, in addition to their role in cuticular pigmentation, have central roles in histamine metabolism. In a current model, Ebony functions in glial cells to convert histamine released by R-cells into carcinine (an inactive derivative of histamine), whereas Tan activity is required in R-cell terminals to replenish the histamine pool by hydrolyzing carcinine (Hovemann et al., 1998; Borycz et al., 2002; Richardt et al., 2002; Richardt et al., 2003; True et al., 2005; Wagner et al., 2007). The mechanisms that mediate the transport and uptake of histamine and carcinine by glial-cell processes and R-cell terminals are at present not well understood. Consistent with a role of these two enzymes in promoting histamine recycling and neurotransmission, both ebony and tan homozygous-mutant animals display abnormal ERG responses (e.g. Heisenberg, 1971). Finally, analysis of mutations in endophilin, which encodes an SH3-domain-containing lysophosphatidic acid acyl transferase, has linked glial capitate projections to clathrin-mediated endocytosis and recovery of synaptic vesicles and, possibly, also neurotransmitter uptake at R-cell synapses (Fabian-Fine et al., 2003). Together, these findings suggest that, in addition to their diverse functions during development, glial cells in the mature visual system have a crucial role in synaptic transmission and, thus, in the processing of visual information (Fig. 1D).
CONCLUSIONS
These studies illustrate that the Drosophila visual system is a useful model to investigate the basic mechanisms that regulate glial development in vivo. Clearly, we are just at the beginning and more molecular determinants that control glial formation, specification and migration in the visual system need to be identified. For example, it will be essential to isolate the factors that regulate the separation of neuronal and glial lineages in the optic lobe upstream of Gcm and Gcm2, as well as the factors that control the specification into different subtypes either downstream or independent of these transcription factors. We also will need to identify the R-cell-derived signals that induce glial-cell formation, direct their migration to specific locations at relevant time points in the optic lobe, and, finally, enable them to ensheath the appropriate number of neuronal processes in the optic stalk and lamina cartridges.
Moreover, glial cells in the visual system are clearly not players that wait on the sidelines while big events occur for neurons. From the moment neurons are born, different subtypes of glial cells step in multiple times to assist them in forming a functional visual circuit. Whereas previous studies so far have provided our first insights into the underlying cellular mechanisms, the molecular nature of the glial-derived factor(s), that mediate R-cell axon guidance, is still unclear. The design of systematic approaches, such as forward-genetic screens and micro-array experiments, that will lead to the identification of these glial-specific signals in both the eye and the optic lobe is an important task for the next years to come. In turn, this will help to determine whether glial cells provide diffusible or membrane-bound cues that mediate long-range or local neuron-glial interactions, or both. It is also crucial to understand whether glial cells, in addition to mediating ganglion-specific R-cell axon targeting during larval development, have later functions in regulating target specificity and synapse formation during pupal stages. Given the high degree of conservation of mechanisms that underlie nervous system development and axon guidance uncovered so far, it is safe to say that investigating glial cells from a fly's eye point of view will complement our understanding of glial development and function in vertebrates in the future.
ACKNOWLEDGEMENTS
We thank A. Gould, E. Ober, Z. Ludlow, H. Apitz and W. Joly for helpful discussions and comments on the manuscript. Our research is supported by the Medical Research Council.
REFERENCES
- Alfonso TB, Jones BW. gcm2 promotes glial cell differentiation and is required with glial cells missing for macrophage development in Drosophila. Developmental Biology. 2002;248:369–383. doi: 10.1006/dbio.2002.0740. [DOI] [PubMed] [Google Scholar]
- Borycz J, Borycz JA, Loubani M, Meinertzhagen IA. tan and ebony genes regulate a novel pathway for transmitter metabolism at fly photoreceptor terminals. Journal of Neuroscience. 2002;22:10549–10557. doi: 10.1523/JNEUROSCI.22-24-10549.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell G, Goring H, Lin T, Spana E, Andersson S, Doe CQ, et al. RK2, a glial-specific homeodomain protein required for embryonic nerve cord condensation and viability in Drosophila. Development. 1994;120:2957–2966. doi: 10.1242/dev.120.10.2957. [DOI] [PubMed] [Google Scholar]
- Choe KM, Clandinin TR. Thinking about visual behavior; learning about photoreceptor function. Current Topics in Developmental Biology. 2005;69:187–213. doi: 10.1016/S0070-2153(05)69007-2. [DOI] [PubMed] [Google Scholar]
- Choi KW, Benzer S. Migration of glia along photoreceptor axons in the developing Drosophila eye. Neuron. 1994;12:423–431. doi: 10.1016/0896-6273(94)90282-8. [DOI] [PubMed] [Google Scholar]
- Colonques J, Ceron J, Tejedor FJ. Segregation of postembryonic neuronal and glial lineages inferred from a mosaic analysis of the Drosophila larval brain. Mechanisms of Development. 2007 doi: 10.1016/j.mod.2007.01.004. [Epub ahead of print doi:10.1016/j.mod.2007.01.004] [DOI] [PubMed] [Google Scholar]
- Chotard C, Leung W, Salecker I. glial cells missing and gcm2 cell autonomously regulate both glial and neuronal development in the visual system of Drosophila. Neuron. 2005;48:237–251. doi: 10.1016/j.neuron.2005.09.019. [DOI] [PubMed] [Google Scholar]
- Clandinin TR, Zipursky SL. Afferent growth cone interactions control synaptic specificity in the Drosophila visual system. Neuron. 2000;28:427–436. doi: 10.1016/s0896-6273(00)00122-7. [DOI] [PubMed] [Google Scholar]
- Clandinin TR, Zipursky SL. Making connections in the fly visual system. Neuron. 2002;35:827–841. doi: 10.1016/s0896-6273(02)00876-0. [DOI] [PubMed] [Google Scholar]
- Dearborn R, Jr, Kunes S. An axon scaffold induced by retinal axons directs glia to destinations in the Drosophila optic lobe. Development. 2004;131:2291–2303. doi: 10.1242/dev.01111. [DOI] [PubMed] [Google Scholar]
- Ebens AJ, Garren H, Cheyette BN, Zipursky SL. The Drosophila anachronism locus: a glycoprotein secreted by glia inhibits neuroblast proliferation. Cell. 1993;74:15–27. doi: 10.1016/0092-8674(93)90291-w. [DOI] [PubMed] [Google Scholar]
- Egger B, Chell JM, Brand AH. Insights into neural stem cell biology from flies. Philosophical Transactions of the Royal Society B: Biological Sciences. 2007a doi: 10.1098/rstb.2006.2011. [Epub ahead of print, doi 10.1098/rstb.2006.2011] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger B, Boone JQ, Stevens NR, Brand AH, Doe CQ. Regulation of spindle orientation and neural stem cell fate in the Drosophila optic lobe. Neural Development. 2007b;2:1. doi: 10.1186/1749-8104-2-1. [doi:10.1186/1749-8104-2-1] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian-Fine R, Verstreken P, Hiesinger PR, Horne JA, Kostyleva R, Zhou Y, et al. Endophilin promotes a late step in endocytosis at glial invaginations in Drosophila photoreceptor terminals. Journal of Neuroscience. 2003;23:10732–10744. doi: 10.1523/JNEUROSCI.23-33-10732.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Soller M, Flister S, Hollmann M, Muller M, Bello B, et al. The egghead gene is required for compartmentalization in Drosophila optic lobe development. Developmental Biology. 2005;287:61–73. doi: 10.1016/j.ydbio.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Freeman MR, Doherty J. Glial cell biology in Drosophila and vertebrates. Trends in Neurosciences. 2006;29:82–90. doi: 10.1016/j.tins.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Gengs C, Leung HT, Skingsley DR, Iovchev MI, Yin Z, Semenov EP, et al. The target of Drosophila photoreceptor synaptic transmission is a histamine-gated chloride channel encoded by ort (hclA) Journal of Biological Chemistry. 2002;277:42113–42120. doi: 10.1074/jbc.M207133200. [DOI] [PubMed] [Google Scholar]
- Gisselmann G, Pusch H, Hovemann BT, Hatt H. Two cDNAs coding for histamine-gated ion channels in D. melanogaster. Nature Neuroscience. 2002;5:11–12. doi: 10.1038/nn787. [DOI] [PubMed] [Google Scholar]
- Granderath S, Bunse I, Klambt C. gcm and pointed synergistically control glial transcription of the Drosophila gene loco. Mechanisms of Development. 2000;91:197–208. doi: 10.1016/s0925-4773(99)00304-4. [DOI] [PubMed] [Google Scholar]
- Hartenstein V, Nassif C, Lekven A. Embryonic development of the Drosophila brain. II. Pattern of glial cells. Journal of Comparative Neurology. 1998;402:32–47. [PubMed] [Google Scholar]
- Halter DA, Urban J, Rickert C, Ner SS, Ito K, Travers AA, et al. The homeobox gene repo is required for the differentiation and maintenance of glia function in the embryonic nervous system of Drosophila melanogaster. Development. 1995;121:317–332. doi: 10.1242/dev.121.2.317. [DOI] [PubMed] [Google Scholar]
- Heisenberg M. Separation of receptor and lamina potentials in the electroretinogram of normal and mutant Drosophila. Journal of Experimental Biology. 1971;55:85–100. doi: 10.1242/jeb.55.1.85. [DOI] [PubMed] [Google Scholar]
- Hosoya T, Takizawa K, Nitta K, Hotta Y. glial cells missing a binary switch between neuronal and glial determination in Drosophila. Cell. 1995;82:1025–1036. doi: 10.1016/0092-8674(95)90281-3. [DOI] [PubMed] [Google Scholar]
- Hovemann BT, Ryseck RP, Walldorf U, Stortkuhl KF, Dietzel ID, Dessen E. The Drosophila ebony gene is closely related to microbial peptide synthetases and shows specific cuticle and nervous system expression. Gene. 1998;221:1–9. doi: 10.1016/s0378-1119(98)00440-5. [DOI] [PubMed] [Google Scholar]
- Huang Z, Kunes S. Hedgehog, transmitted along retinal axons, triggers neurogenesis in the developing visual centers of the Drosophila brain. Cell. 1996;86:411–422. doi: 10.1016/s0092-8674(00)80114-2. [DOI] [PubMed] [Google Scholar]
- Huang Z, Kunes S. Signals transmitted along retinal axons in Drosophila: Hedgehog signal reception and the cell circuitry of lamina cartridge assembly. Development. 1998;125:3753–3764. doi: 10.1242/dev.125.19.3753. [DOI] [PubMed] [Google Scholar]
- Huang Z, Shilo BZ, Kunes S. A retinal axon fascicle uses spitz, an EGF receptor ligand, to construct a synaptic cartridge in the brain of Drosophila. Cell. 1998;95:693–703. doi: 10.1016/s0092-8674(00)81639-6. [DOI] [PubMed] [Google Scholar]
- Hummel T, Attix S, Gunning D, Zipursky SL. Temporal control of glial cell migration in the Drosophila eye requires gilgamesh, hedgehog, and eye specification genes. Neuron. 2002;33:193–203. doi: 10.1016/s0896-6273(01)00581-5. [DOI] [PubMed] [Google Scholar]
- Ito K, Urban J, Technau GM. Distribution, classification, and development of Drosophila glial cells in the late embryonic and early larval ventral nerve cord. Roux's Archive in Developmental Biology. 1995;204:284–307. doi: 10.1007/BF02179499. [DOI] [PubMed] [Google Scholar]
- Jones BW, Fetter RD, Tear G, Goodman CS. glial cells missing: a genetic switch that controls glial versus neuronal fate. Cell. 1995;82:1013–1023. doi: 10.1016/0092-8674(95)90280-5. [DOI] [PubMed] [Google Scholar]
- Kaphingst K, Kunes S. Pattern formation in the visual centers of the Drosophila brain: wingless acts via decapentaplegic to specify the dorsoventral axis. Cell. 1994;78:437–448. doi: 10.1016/0092-8674(94)90422-7. [DOI] [PubMed] [Google Scholar]
- Kammerer M, Giangrande A. Glide2, a second glial promoting factor in Drosophila melanogaster. EMBO Journal. 2001;20:4664–4673. doi: 10.1093/emboj/20.17.4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Mardon G, Solomon NM, Rubin GM. dachshund encodes a nuclear protein required for normal eye and leg development in Drosophila. Development. 1994;120:3473–3486. doi: 10.1242/dev.120.12.3473. [DOI] [PubMed] [Google Scholar]
- Mast JD, Prakash S, Chen PL, Clandinin TR. The mechanisms and molecules that connect photoreceptor axons to their targets in Drosophila. Seminars in Cellular and Developmental Biology. 2006;17:42–49. doi: 10.1016/j.semcdb.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Meinertzhagen IA, O'Neil SD. Synaptic organization of columnar elements in the lamina of the wild type in Drosophila melanogaster. Journal of Comparative Neurology. 1991;305:232–263. doi: 10.1002/cne.903050206. [DOI] [PubMed] [Google Scholar]
- Meinertzhagen IA, Hanson TE. The development of the optic lobe. In: Bate M, Martinez-Arias A, editors. The Development of Drosophila melanogaster. Cold Spring Harbor Press; 1993. pp. 1363–1491. [Google Scholar]
- Pappu KS, Mardon G. Genetic control of retinal specification and determination in Drosophila. International Journal of Developmental Biology. 2004;48:913–924. doi: 10.1387/ijdb.041875kp. [DOI] [PubMed] [Google Scholar]
- Pereanu W, Shy D, Hartenstein V. Morphogenesis and proliferation of the larval brain glia in Drosophila. Developmental Biology. 2005;283:191–203. doi: 10.1016/j.ydbio.2005.04.024. [DOI] [PubMed] [Google Scholar]
- Perez SE, Steller H. Migration of glial cells into retinal axon target field in Drosophila melanogaster. Journal of Neurobiolology. 1996a;30:359–373. doi: 10.1002/(SICI)1097-4695(199607)30:3<359::AID-NEU5>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Perez SE, Steller H. Molecular and genetic analyses of lama, an evolutionarily conserved gene expressed in the precursors of the Drosophila first optic ganglion. Mechanisms of Development. 1996b;59:11–27. doi: 10.1016/0925-4773(96)00556-4. [DOI] [PubMed] [Google Scholar]
- Pielage J, Kippert A, Zhu M, Klambt C. The Drosophila transmembrane protein Fear-of-intimacy controls glial cell migration. Developmental Biology. 2004;275:245–257. doi: 10.1016/j.ydbio.2004.07.039. [DOI] [PubMed] [Google Scholar]
- Poeck B, Fischer S, Gunning D, Zipursky SL, Salecker I. Glial cells mediate target layer selection of retinal axons in the developing visual system of Drosophila. Neuron. 2001;29:99–113. doi: 10.1016/s0896-6273(01)00183-0. [DOI] [PubMed] [Google Scholar]
- Power ME. The effect of reduction in numbers of ommatidia upon the brain of Drosophila melanogaster. Journal of Experimental Zoology. 1943;94:33–71. [Google Scholar]
- Prokop A, Meinertzhagen IA. Development and structure of synaptic contacts in Drosophila. Seminars in Cell and Developmental Biology. 2006;17:20–30. doi: 10.1016/j.semcdb.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Ramòn y Cajal S. Recollections of my life. MIT Press; 1937. [Google Scholar]
- Rangarajan R, Courvoisier H, Gaul U. Dpp and Hedgehog mediate neuron-glia interactions in Drosophila eye development by promoting the proliferation and motility of subretinal glia. Mechanisms of Development. 2001;108:93–103. doi: 10.1016/s0925-4773(01)00501-9. [DOI] [PubMed] [Google Scholar]
- Rangarajan R, Gong Q, Gaul U. Migration and function of glia in the developing Drosophila eye. Development. 1999;126:3285–3292. doi: 10.1242/dev.126.15.3285. [DOI] [PubMed] [Google Scholar]
- Richardt A, Rybak J, Stortkuhl KF, Meinertzhagen IA, Hovemann BT. Ebony protein in the Drosophila nervous system: optic neuropile expression in glial cells. Journal of Comparative Neurology. 2002;452:93–102. doi: 10.1002/cne.10360. [DOI] [PubMed] [Google Scholar]
- Richardt A, Kemme T, Wagner S, Schwarzer D, Marahiel MA, Hovemann BT. Ebony, a novel nonribosomal peptide synthetase for beta-alanine conjugation with biogenic amines in Drosophila. Journal of Biological Chemistry. 2003;278:41160–41166. doi: 10.1074/jbc.M304303200. [DOI] [PubMed] [Google Scholar]
- Selleck SB, Steller H. The influence of retinal innervation on neurogenesis in the first optic ganglion of Drosophila. Neuron. 1991;6:83–99. doi: 10.1016/0896-6273(91)90124-i. [DOI] [PubMed] [Google Scholar]
- Silver SJ, Rebay I. Signaling circuitries in development: insights from the retinal determination gene network. Development. 2005;132:3–13. doi: 10.1242/dev.01539. [DOI] [PubMed] [Google Scholar]
- Soustelle L, Trousse F, Jacques C, Ceron J, Cochard P, Soula C, et al. Neurogenic role of Gcm transcription factors is conserved in chicken spinal cord. Development. 2007;134:625–634. doi: 10.1242/dev.02750. [DOI] [PubMed] [Google Scholar]
- Stuart AE. From fruit flies to barnacles, histamine is the neurotransmitter of arthropod photoreceptors. Neuron. 1999;22:431–433. doi: 10.1016/s0896-6273(00)80699-6. [DOI] [PubMed] [Google Scholar]
- Struhl G, Basler K. Organizing activity of wingless protein in Drosophila. Cell. 1993;72:527–540. doi: 10.1016/0092-8674(93)90072-x. [DOI] [PubMed] [Google Scholar]
- Suh GS, Poeck B, Chouard T, Oron E, Segal D, Chamovitz DA, et al. Drosophila JAB1/CSN5 acts in photoreceptor cells to induce glial cells. Neuron. 2002;33:35–46. doi: 10.1016/s0896-6273(01)00576-1. [DOI] [PubMed] [Google Scholar]
- Tayler TD, Robichaux MB, Garrity PA. Compartmentalization of visual centers in the Drosophila brain requires Slit and Robo proteins. Development. 2004;131:5935–5945. doi: 10.1242/dev.01465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting CY, Lee CH. Visual circuit development in Drosophila. Current Opinion in Neurobiology. 2007;17:65–72. doi: 10.1016/j.conb.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Ting CY, Yonekura S, Chung P, Hsu SN, Robertson HM, Chiba A, et al. Drosophila N-cadherin functions in the first stage of the two-stage layer-selection process of R7 photoreceptor afferents. Development. 2005;132:953–963. doi: 10.1242/dev.01661. [DOI] [PubMed] [Google Scholar]
- Tix S, Eule E, Fischbach KF, Benzer S. Glia in the chiasms and medulla of the Drosophila melanogaster optic lobes. Cell and Tissue Research. 1997;289:397–409. doi: 10.1007/s004410050886. [DOI] [PubMed] [Google Scholar]
- True JR, Yeh SD, Hovemann BT, Kemme T, Meinertzhagen IA, Edwards TN, et al. Drosophila tan encodes a novel hydrolase required in pigmentation and vision. PLoS Genetics. 2005;1:e63. doi: 10.1371/journal.pgen.0010063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umetsu D, Murakami S, Sato M, Tabata T. The highly ordered assembly of retinal axons and their synaptic partners is regulated by Hedgehog/Single-minded in the Drosophila visual system. Development. 2006;133:791–800. doi: 10.1242/dev.02253. [DOI] [PubMed] [Google Scholar]
- Vincent S, Vonesch JL, Giangrande A. Glide directs glial fate commitment and cell fate switch between neurones and glia. Development. 1996;122:131–139. doi: 10.1242/dev.122.1.131. [DOI] [PubMed] [Google Scholar]
- Wagner S, Heseding C, Szlachta K, True JR, Prinz H, Hovemann BT. Drosophila photoreceptors express cysteine peptidase tan. Journal of Comparative Neurology. 2007;500:601–611. doi: 10.1002/cne.21138. [DOI] [PubMed] [Google Scholar]
- Wandall HH, Pizette S, Pedersen JW, Eichert H, Levery SB, Mandel U, et al. egghead and brainiac are essential for glycosphingolipid biosynthesis in vivo. Journal of Biological Chemistry. 2005;280:4858–4863. doi: 10.1074/jbc.C400571200. [DOI] [PubMed] [Google Scholar]
- Winberg ML, Perez SE, Steller H. Generation and early differentiation of glial cells in the first optic ganglion of Drosophila melanogaster. Development. 1992;115:903–911. doi: 10.1242/dev.115.4.903. [DOI] [PubMed] [Google Scholar]
- Xiong WC, Montell C. Defective glia induce neuronal apoptosis in the repo visual system of Drosophila. Neuron. 1995;14:581–590. doi: 10.1016/0896-6273(95)90314-3. [DOI] [PubMed] [Google Scholar]
- Xiong WC, Okano H, Patel NH, Blendy JA, Montell C. repo encodes a glial-specific homeo domain protein required in the Drosophila nervous system. Genes and Development. 1994;8:981–994. doi: 10.1101/gad.8.8.981. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Soustelle L, Giangrande A, Umetsu D, Murakami S, Yasugi T, et al. DPP signaling controls development of the lamina glia required for retinal axon targeting in the visual system of Drosophila. Development. 2005;132:4587–4598. doi: 10.1242/dev.02040. [DOI] [PubMed] [Google Scholar]


