Abstract
Toll-like receptors (TLRs) signal through two main pathways: a myeloid differentiation factor (MyD)88-dependent pathway that acts via nuclear factor κB (NF-κB) to induce proinflammatory cytokines such as tumour necrosis factor-α (TNF-α) and a MyD88-independent pathway that acts via type I interferons to increase the expression of interferon-inducible genes. Repeated signalling through TLR4 and a number of other TLRs has been reported to result in a reduction in the subsequent proinflammatory cytokine response, a phenomenon known as TLR tolerance. In this study we have shown that, whilst NF-κB activation and production of TNF-α and interleukin-12 by murine RAW264.7 and J774.2 cells in response to stimulation by TLR4, -5, -7 or -9, was reduced by prior stimulation with TLR4, -5, -7 or -9 ligands, the primary stimulation of TLR3, which does not use the MyD88 pathway, did not reduce the TNF-α or interleukin-12 responses to subsequent TLR stimulation. The response to TLR3 stimulation was not diminished by prior TLR ligand exposure. Furthermore, the production of interferon-β (IFN-β) following stimulation of TLR3 or -4, which is MyD88-independent, was increased by prior activation of TLR4, -5, -7 or -9. In contrast, TLR9 ligand-induced IFN-β production, which is MyD88-dependent, was tolerized by prior TLR stimulation. These results are consistent with differential regulation of MyD88-dependent and MyD88-independent cytokine production following serial activation of TLRs.
Keywords: endotoxin tolerance, intracellular signalling, J774.2, pattern recognition receptors, RAW264.7
Introduction
The toll like receptor (TLR) family of innate pattern recognition receptors recognizes a wide range of conserved microbial components, including cell wall components, unmethylated CpG motif-containing DNA and double- and single-stranded RNA. Stimulation of TLRs results in the activation of innate immune cells such as macrophages and dendritic cells, leading to the production of proinflammatory cytokines and up-regulation of costimulatory molecules. Thus, TLRs provide the ‘danger signals’ required for clearance of infection by innate immune cells and also for the initiation of adaptive immune responses.1 Repeated exposure to a TLR agonist, such as lipopolysaccharide (LPS), results in a diminished response, usually apparent as a reduction in proinflammatory cytokine release.2 This phenomenon, described in vivo and in vitro before the discovery of the TLR family as endotoxin tolerance, is thought to play a part in susceptibility to re-infection in patients treated for sepsis. It is characterized by an initial proinflammatory phase, followed by a hypoimmune state with a diminished ex vivo mononuclear cell response to LPS.3,4 Endotoxin tolerance has also been described in chronic infections such as malaria5 and may be important in maintaining homeostasis in the gut6 and liver,7 which are exposed daily to a range of microbial components from commensal organisms in the gut. TLR tolerance has also been described using ligands such as lipoteichoic acid (TLR2) and unmethylated CpG DNA (CpG, TLR9).8–12
TLRs signal through two main pathways, defined by the adaptor molecules used at the start of each. The best characterized of these is the MyD88-dependent pathway, common to all TLRs except TLR3, which results in rapid activation of the transcription molecules nuclear factor κB (NF-κB), activator protein 1 and Elk-1 and induction of proinflammatory mediators such as tumour necrosis factor-α (TNF-α), interleukin-6 (IL-6) and cyclooxygenase-2.13,14 Activation of the NF-κB p65 and activator protein 1 transcription factors is reduced in endotoxin-tolerant cells.15 TLR7, -8 and -9 can also induce type I interferon (IFN) production in a MyD88-dependent manner, involving the activation of the interferon regulatory factor 5 (IRF-5)16 and IRF-717 transcription factors. The second, less well-studied pathway is thought to be limited to TLR3 and TLR4. TLR signalling via the adaptor molecules Toll/IL-1-receptor-domain-containing adaptor inducing IFN-β (TRIF) and TRIF-related adaptor molecule (TRAM) activates IRF-3 and IRF-7 and, ultimately, IFN-inducible genes such as IFN-γ-inducible protein 10 (IP-10) and RANTES, via type I IFN production and autocrine activation of the JAK/STAT pathway.14,18,19 This pathway also induces late activation of NF-κB.19 A number of negative regulators of TLR signalling have been described, with an emphasis on regulation of the MyD88-NF-κB pathway13 but negative regulators of JAK/STAT activation are also well described.20
Given the shared signalling pathway, it is not surprising that cross-tolerance between a number of MyD88-utilizing TLRs8,9,11 has been described, as well as cross-tolerance between LPS and IL-1, which also uses the MyD88 pathway to activate NF-κB.21 However, to describe TLR tolerance as a hyporesponsive state is an over-simplification. While reductions in the production of proinflammatory cytokines such as IL-1, IL-6 and TNF-α by endotoxin-tolerant cells are well established, other functions may be unaffected or enhanced. Elevated levels of IL-10 have been reported in some in vivo models of endotoxin tolerance, and in hypoimmune septic patients.22,23 Intravenous CpG (TLR9 agonist) protects against LPS-induced airway inflammation24 but is reported to increase IFN-γ and IL-18 levels on re-challenge with LPS.25 Increases in inducible nitric oxide synthase (iNOS)26 and TNF receptor 2 (TNFR-2)27 expression are reported in endotoxin-tolerant macrophages, as is enhanced phagocytosis and clearance of certain bacteria in TLR-tolerized mice,23,28–30 although major histocompatibility complex class II expression31 and antigen presentation32 are reduced on endotoxin-tolerant peripheral blood mononuclear cells. Recently, iNOS expression in macrophages has been linked to IFN-β production via the Src family tyrosine kinases.33
The reported increase in iNOS expression in the endotoxin-tolerized state may be a reflection of differential regulation of the TLR signalling pathways in recurrently exposed cells. In vivo experiments have demonstrated increased apoptosis and lethality on LPS injection following viral infection34,35 or administration of a TLR3 ligand,35 suggesting differences in the response to TLR3 activation from the MyD88-dependent TLRs. The current study was designed to evaluate the effects of sequential exposure to a range of TLR ligands on both MyD88-dependent and MyD88-independent activation of murine macrophages, with particular focus on the interactions of TLR3 ligand stimulation and MyD88-independent IFN-β production. This was achieved by measurement of TNF-α, IL-12 and IFN-β in supernatants and analysis of the transcription factors NF-κB p65 in nuclear lysates in sequentially stimulated cells.
Materials and methods
Cells
Murine RAW264.7 cells (ECACC no. 91062702) and J774.2 cells (ECACC no. 85011428) were cultured in RPMI-1640 (Cambrex, Nottingham, UK) supplemented with 5% fetal calf serum, l-glutamine and no antibiotics. TLR4 and TLR5 expression in both cell lines was confirmed using flow cytometry (data not shown). Cells were passaged by scraping, were seeded at an initial density of 5 × 104 cells/ml and used within 10 passages. Forty-eight-well plates were used for cytokine experiments, and 75-cm2 flasks were used for transcription factor assays (all Corning, Koolhovenlaan, the Netherlands).
Reagents
Poly(I:C), highly purified LPS from Escherichia coli, purified flagellin from Salmonella typhimurium, loxoribine (all Invivogen, San Diego, CA) and unmethylated, phosphorothioate-modified CpG oligodeoxynucleotides (sequence 5′-TCC ATG ACG TTC CTG ACG TT-3′)36 (TAGN Ltd, Tyne and Wear, UK) were reconstituted in accordance with the manufacturers' instructions using Limulus amoebocyte lysate grade pyrogen-free water or sterile physiological salt solution. In some experiments, a control non-stimulatory GpC sequence was used to confirm that the response was sequence-specific, and not the result of contamination (data not shown).
Enzyme-linked immunosorbent assay (ELISA)
Supernatants were collected at appropriate timepoints (6 hr for TNF-α and 24 hr for IL-12 and IFN-β) and stored at − 20°. TNF-α and IL-12 p40 (both Peprotech, London, UK) and IFN-β (PBL Biomedical Laboratories, Piscataway, NJ) were assayed by ELISA according to the manufacturers' instructions.
Western blotting
Nuclear lysates were produced using Pierce NE-PER nuclear and cytoplasmic extraction reagents (Pierce, Rockford, IL) supplemented with protease inhibitors (Roche, Lewes, UK) according to the manufacturer's instructions. Sample protein concentrations were assayed using Pierce Coomassie reagent and were equalized before use. Samples in loading buffer were run on 10% polyacrylamide gels, transferred to nitrocellulose paper and blocked with 5% milk. Membranes were incubated overnight with rabbit anti-NF-κB p65 (Santa Cruz, Santa Cruz, CA) followed by a horseradish peroxidase-conjugated donkey anti-rabbit secondary (Amersham, Bucks, UK). Blots were developed using a Pierce development kit and Kodak paper. Equal loading was confirmed by subsequently copper staining the membranes. Unused samples were stored at − 80°.
NF-κB DNA-binding assays
NF-κB p65 DNA-binding activity was assayed using a Chemicon assay kit in accordance with the manufacturer's instructions (Chemicon, Hampshire, UK). Briefly, 5 μl of each nuclear lysate was mixed with a biotinylated DNA oligonucleotide containing the NF-κB consensus binding site, then transferred to a streptavidin-coated plate. DNA-bound NF-κB p65 was detected using primary and horseradish peroxidase-conjugated secondary antibodies and a colour change was detected using 3,3,5,5-tetramethylbenzidine substrate measured at 450 nm.
Statistical analyses
Individual ELISA experiments were analysed by analysis of variance (anova) and Dunnet's post test, comparing all treatments with stimulated previously naive cells. Standardized data in Table 1 were analysed using one sample Student's t-test.
Table 1.
Summary of outcome of sequential stimulation with TLR ligands
| 1st → 2nd↓ | TLR3 PIC (2·5 μg/ml) | TLR4 LPS (10 ng/ml) | TLR5 Flagellin (100 ng/ml) | TLR7 Loxoribine (100 μm) | TLR9 CpG DNA (0·1 μm) |
|---|---|---|---|---|---|
| TLR4 | **** | *** | * | **** | |
| LPS 10 ng/ml | 1·28 ± 0·18 | 0·18 ± 0·05 | 0·33 ± 0·05 | 0·70 ± 0·09 | 0·46 ± 0·08 |
| TLR5 | * | * | ** | ** | |
| Flagellin 100 ng/ml | 1·14 ± 0·10 | 0·37 ± 0·14 | 0·27 ± 0·16 | 0·49 ± 0·04 | 0·49 ± 0·06 |
| TLR7 | ** | ** | ** | ** | |
| Loxoribine 100 µm | 0·84 ± 0·19 | 0·19 ± 0·11 | 0·12 ± 0·06 | 0·07 ± 0·04 | 0·07 ± 0·06 |
| TLR9 | **** | ** | * | *** | |
| CpG DNA 0·1 µm | 1·32 ± 0·31 | 0·21 ± 0·08 | 0·25 ± 0·07 | 0·18 ± 0·12 | 0·05 ± 0·07 |
J774.2 and RAW264.7 cells were stimulated for 48 hr with the TLR ligands stated above (columns), washed and restimulated with the same or different TLR ligands (rows) and subsequent cytokine production was measured by ELISA. For comparison of data from multiple experiments, TNF-α production in pg/ml was normalized by defining the response of naive cells to each ligand within each experiment as 1 and expressing all other measurements as relative values. Data shown are the mean ± SEM of three to 10 independent experiments, each carried out in triplicate. At least one complete dataset for all TLR ligand combinations is derived from each cell type. No difference was observed between cell types.
P < 0·05
P < 0·01
P < 0·001
P < 0·0001.
Results
Induction of TLR tolerance
Induction of endotoxin tolerance in RAW264.7 and J774.2 murine macrophages was optimized by stimulating with a range of LPS concentrations for 24–48 hr, washing and restimulating with LPS at 100 ng/ml (Fig. 1a,b). A 48-hr primary stimulation was chosen for all subsequent experiments, to minimize the detection of primary stimulation-induced IL-12 following the second stimulation. In both cell types, a dose-dependent reduction in TNF-α and IL-12 was seen on restimulation, although IL-12 production from the primary stimulation continued beyond 48 hr in strongly stimulated cells so 10 ng/ml LPS was used in subsequent experiments (Fig. 1b). Optimal primary and secondary stimulation ligand concentrations were chosen for induction of tolerance with each of the TLRs in subsequent experiments.
Figure 1.
Pre-stimulation with LPS reduces TNF-α (a) and IL-12 (b) production by RAW264.7 cells in response to subsequent LPS stimulation in a dose-dependent manner. Cells were stimulated with LPS at the concentrations stated. After 48 hr, cells were washed then medium was replaced with (grey bars) or without (black bars) the addition of 100 ng/ml LPS. Cytokine production was measured by ELISA of supernatants collected at 6 hr (TNF-α) or 24 hr (IL-12) after restimulation. Results displayed are means ± SEM of triplicate wells, with prestimulated samples compared to the primary response to LPS and have been replicated in J774.2 cells. **P < 0·01.
TLR cross-tolerance
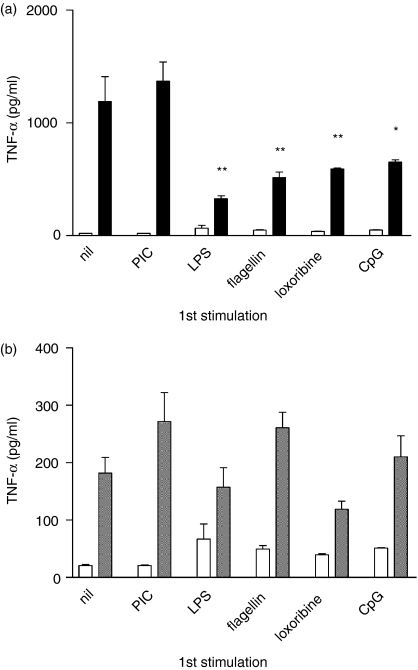
Having established optimal conditions for the induction of TLR tolerance, a series of experiments was performed using both RAW264.7 and J774.2 cells to determine whether a range of TLR ligands could tolerize TLR ligand-induced TNF-α and IL-12 production. Example experiments are shown in Fig. 2(a,b) and Fig. 3 and standardized data from 10 experiments (four using J774.2 cells and six using RAW264.7 cells) are summarized in Table 1. No differences were observed between the J774.2 and RAW264.7 cytokine responses to sequential TLR stimulation. A consistent reduction in LPS-induced TNF-α production by cells pretreated with LPS (P < 0·01), flagellin (P < 0·01), loxoribine (P < 0·01) or CpG (P < 0·05) was seen (Fig. 2a). Cross-tolerance of TNF-α production by prior exposure to LPS, flagellin, loxoribine or CpG was seen in cells subsequently stimulated with either LPS, flagellin, loxoribine or CpG (P ≤ 0·05 in every combination; Table 1). Generally similar results were seen for IL-12 production (P ≤ 0·05 except for LPS and CpG in combination). In a separate experiment (not shown), this finding was confirmed in a primary culture of adherent murine spleen cells.
Figure 2.
Cross-tolerance of LPS-induced TNF-α production (representative example of data summarized in Table 1). (a) RAW264.7 cells were stimulated with TLR ligands as stated. After 48 hr, cells were washed and restimulated with medium alone (open bars) or 10 ng/ml LPS (black bars). (b) PIC-induced TNF-α is not tolerized by prior TLR stimulation. RAW264.7 cells were stimulated with the TLR ligands stated as in (a) then restimulated with medium alone (open bars) or PIC 2·5 μg/ml (grey bars). TNF-α production was assayed by ELISA in supernatants collected at 6 hr following restimulation. Results displayed are mean ± SEM of triplicate wells and prestimulated samples are compared with the primary response to each ligand. Findings were replicated in RAW264.7 and J774.2 cells. *P < 0·05, **P < 0·01.
Figure 3.
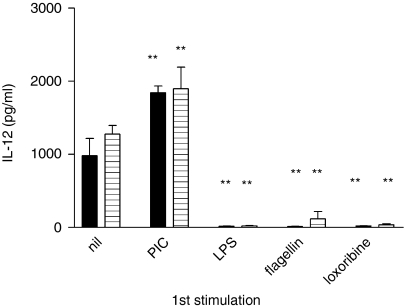
Pretreatment with PIC increases subsequent IL-12 production by RAW264.7 cells in response to LPS (black bars) or flagellin (horizontally striped bars), whilst pretreatment with LPS, flagellin or loxoribine reduces subsequent IL-12 production in response to the same ligands. Results shown are mean ± SEM of triplicate wells and findings have been replicated in RAW264.7 and J774.2 cells. **P < 0·01.
Unlike the other TLR ligands, poly(inosine:cytosine) (PIC, TLR3) activates cells in an entirely MyD88-independent manner.37 Stimulation with PIC resulted in cytokine production (Fig. 2b), which was followed by cell death within 48 hr at PIC concentrations greater than 3 μg/ml. Following stimulation with 2·5 μg/ml PIC, as used in Figs 2 and 3 and Table 1, there were consistently fewer cells in wells exposed to PIC than in unstimulated wells or in those exposed to any of the other ligands (data not shown). CpG also inhibited cell growth, but to a lesser extent. In contrast to the other ligands already described, no tolerance was seen with PIC in combination with any of the ligands tested; overall, prior PIC stimulation resulted in no significant change in TNF-α production on repeat challenge with another TLR ligand, despite the reduced cell numbers (Table 1 and Fig. 2a). IL-12 production in PIC pretreated cells was, in fact, greatly increased on subsequent stimulation (Fig. 3). PIC activation of previously activated cells often resulted in cell death, but where sufficient cells survived, PIC-induced IL-12 and TNF-α levels were not significantly changed (P > 0·05) (Fig. 2b and data not shown).
Nuclear transcription factors
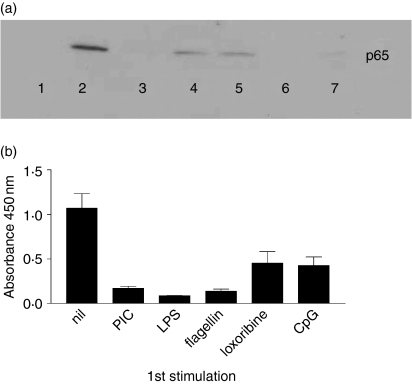
Having demonstrated differences in the cross-tolerance produced by sequential application of a range of TLR ligands, a series of experiments was performed to examine nuclear translocation of NF-κB p65 by Western blotting of nuclear lysates. Early (30 min) nuclear accumulation of NF-κB p65 in response to LPS stimulation was reduced in cells previously stimulated with LPS, flagellin, loxoribine, CpG and PIC (Fig. 4a). Cross-tolerance of NF-κB p65 was also seen in cells pre-exposed to LPS that were then restimulated with PIC, LPS, flagellin, loxoribine or CpG, and similar results were seen when cells prestimulated with the panel of ligands were restimulated with CpG (data not shown). DNA binding of NF-κB was confirmed using a Chemicon ELISA-based assay which detects activated p65 bound to a biotinylated DNA oligonucleotide containing the NF-κB consensus binding sequence (Fig. 4b). This confirmed the potential of PIC, LPS, flagellin, loxoribine and CpG to reduce NF-κB accumulation following subsequent stimulation with LPS.
Figure 4.
(a) Tolerization of LPS-induced NF-κB by prior TLR stimulation. RAW264.7 cells were prestimulated with TLR ligands as described below. After 48 hr, cells were restimulated with LPS for 30 min. NF-κB p65 was detected by Western blotting of nuclear lysates. Lane 1, unstimulated; lane 2, primary stimulation with LPS (no prestimulation); lane 3, prestimulation with PIC, restimulation with LPS; lane 4, prestimulation with LPS, restimulation with LPS; lane 5, prestimulation with flagellin, restimulation with LPS; lane 6, prestimulation with loxoribine, restimulation with LPS; lane 7, prestimulation with CpG, restimulation with LPS. (b) Reduced DNA binding of NF-κB p65 in tolerized cells. Cells were prestimulated with the TLR ligands listed and 48 hr later were restimulated with LPS for 30 min. Nuclear lysates were incubated with a biotinylated oligonucleotide containing the NF-κB consensus binding sequence in streptavidin-coated wells. DNA-bound NF-κB p65 was detected using a horseradish peroxidase-conjugated antibody and 3,3,5,5-tetramethylbenzidine substrate as in a conventional ELISA.
Interferon-β production is increased on repeat TLR3 or TLR4 stimulation
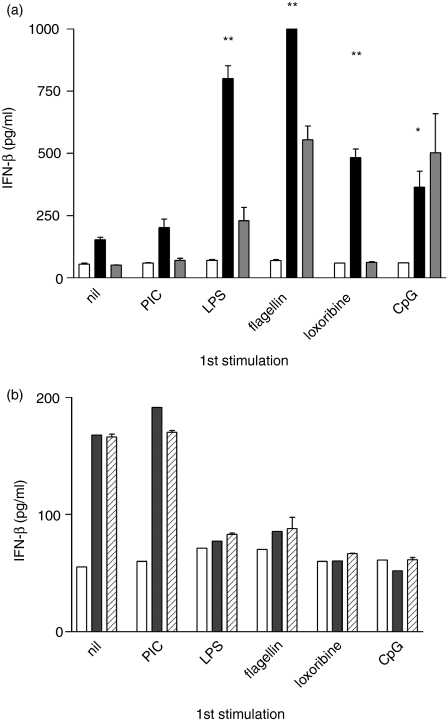
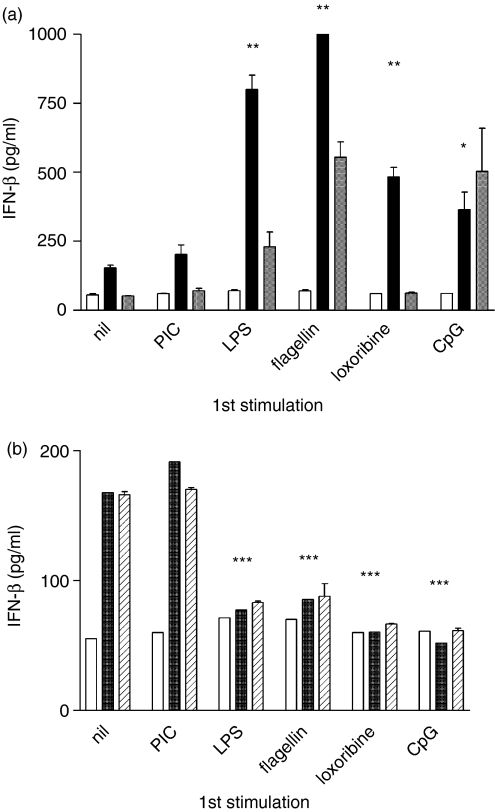
Having shown that NF-κB activation, and production of TNF-α and IL-12 are differentially regulated by prior TLR stimulation, a further series of experiments was performed to examine the effect of IFN-β production. Stimulation of naive cells with PIC resulted in the production of large amounts of IFN-β (not shown). PIC- or LPS-induced IFN-β production was significantly greater in cells previously stimulated with LPS (P < 0·01), flagellin (P < 0·01), loxoribine (P < 0·01) or CpG (P < 0·05) (Fig. 5a). There was no significant change in PIC- or LPS-induced IFN-β production in cells pretreated with PIC. Loxoribine or CpG-induced IFN-β appeared reduced by prior stimulation with LPS, flagellin, loxoribine or CpG (P < 0·05) and was unaffected by prior PIC stimulation (Fig. 5b). Thus, prior stimulation with a TLR3 ligand (MyD88-independent) does not tolerize cells to subsequent TLR stimulation, and prior signalling via the MyD88-dependent TLR4, -5, -7 or -9 results in a tolerized MyD88-dependent TNF-α or IFN-β response but an enhanced MyD88-independent IFN-β response to subsequent stimulation.
Figure 5.
(a) Enhancement of PIC-induced and LPS-induced IFN-β production by prior TLR ligand exposure. RAW264.7 cells were stimulated with each of the TLR ligands shown. After 48 hr, cells were washed and restimulated with 0·1 μg/ml PIC (grey bars), LPS (black bars) or medium alone (open bars). IFN-β was measured by ELISA in supernatants collected at 6 hr. Although higher concentrations of PIC induce a potent IFN-β response in naive cells, a low concentration (0·1 μg/ml) that did not induce detectable IFN-β in naive cells was used in the experiment shown to demonstrate the enhancement of the response to PIC within the limit of detection of the assay. *P < 0·05, **P < 0·01. (b) Tolerization of loxoribine- or CpG-induced IFN-β production by prior TLR ligand exposure. Cells were stimulated as in (a) and restimulated with loxoribine (diagonal lines), CpG (grey bars) or medium alone (open bars). Results are expressed as mean ± SEM of duplicate wells. ***P < 0·001. Data shown are representative of at least two independent experiments.
Discussion
In this study we have confirmed the attenuation of NF-κB activation and MyD88-dependent cytokine responses to repeated TLR stimulation by demonstrating a reduction in TLR4-, TLR5-, TLR7- and TLR9-dependent TNF-α production in both RAW264.7 and J774.2 cells. Cross-tolerance of IL-12 production between TLR4, -5, -7 and -9 was consistently observed in all combinations except TLR4 and TLR9 and NF-κB activation was reduced in tolerized cells. In vitro cross-tolerance of TNF-α and IL-6 production between TLR4 and TLR9 has been described previously.9 In conjunction with previous reports of cross-tolerance between TLR2 and TLR4,8,11 these findings suggest a consistent auto-regulation of the MyD88-dependent signalling pathway. The large number of regulatory proteins identified from the cell surface to the nucleus along the MyD88–NF-κB pathway is added evidence of the importance of controlling this powerful response to inflammatory stimuli.38
We have demonstrated, however, interesting differences between the modification of ‘classical’ MyD88-dependent TLR responses and those responses that have been reported to be MyD88-independent, namely TLR4-induced type 1 IFN production and responses to TLR3 ligand stimulation. In contrast to the tolerization seen with other TLR combinations, there was an enhanced TNF-α and IL-12 response in PIC-prestimulated cells, despite an often reduced number of cells. PIC stimulation of previously stimulated cells frequently resulted in cell death. This is consistent with in vivo data showing that prior viral infection/TLR3 activation results in increased apoptosis and mortality on subsequent LPS challenge.34,35 TLR3 does not signal via MyD88; instead it signals via TRIF, resulting in late activation of NF-κB, and production of type I interferons via IRF-3. This is a pathway that is shared with TLR4 but not with the other TLRs.
PIC-induced or LPS-induced IFN-β release was enhanced by prior stimulation with MyD88-activating TLR ligands including LPS (TLR4) but appeared to be unaffected by prior PIC exposure. It is possible that an unchanged total represents an increased production at the level of the individual cell, given the relative reduction in cell numbers. The LPS-induced expression of iNOS by RAW264.7 cells is dependent on IFN-β production via the activation of Src-family tyrosine kinases.33 The enhanced LPS-induced IFN-β production in these current experiments may therefore explain the enhanced iNOS activity previously reported in endotoxin-tolerant macrophages.26 IFN-β production in response to TLR9 stimulation was tolerized by prior TLR4, -5, -7 or -9 activation, as shown with TNF-α. This is perhaps not surprising because TLR9 induces IFN-β production via IRF-7 in a MyD88-dependent manner.17 While activation of the MyD88 pathway resulted in a reduction in subsequent MyD88-dependent effects, therefore, MyD88-independent IFN-β production was enhanced in the same cells. MyD88-independent IFN-β production was not tolerized by prior MyD88-independent signalling and MyD88-dependent effects were enhanced by prior TLR3 activation.
It is interesting to speculate about the possible implications of this observation. MyD88-dependent activation of NF-κB is common to the majority of TLRs and the IL-1 and IL-18 receptors.39 The MyD88-dependent pathway activates an acute inflammatory response that is necessary for the clearance of many pathogens40–43 but that can also be harmful in excess. Like TLR3, TLR7, -8 and -9 are located in endosomal compartments, recognize RNA and/or DNA and all can induce type I interferons, although via MyD88 and IRF-5 and IRF-7 rather than TRIF and IRF-3. The MyD88-dependence explains why the attenuation of cytokine responses to and by loxoribine and CpG resembled those seen with flagellin rather than PIC.
The IRF3 pathway is used by TLR3 and TLR4 and recently, further cytosolic antiviral signalling proteins capable of activating IRF3 have been identified.44 TLR3 detects PIC/double-stranded RNA of extracellular origin, whereas other mechanisms may detect the same ligands within the cytosol.45 These proteins alert a cell that it has been infected and IRF3 induces production of type I interferons, a crucial part of antiviral defence. It is possible, then, that the observed tolerization of MyD88-dependent responses and the enhancement of IRF3-dependent IFN-β reflect the different requirements of an acute, predominantly innate response, with resolution following clearance, versus a potentially more prolonged antiviral response often requiring activation of the adaptive immune system. A number of studies have shown that viruses that cause persistent infection, such as hepatitis C virus, disrupt IRF3-mediated type I interferon production.46–48 In using the TRIF–IRF3 pathway, TLR3 and TLR4 may have ‘hijacked’ a primarily intracellular antiviral pathway. That TLR4 activates IRF3 may seem contradictory to this model when it is considered as the receptor for LPS. However, there is a body of evidence suggesting that TLR4 recognizes a number of endogenous ligands;49,50 it may be that it is in this role of detecting cellular damage, rather than particular extracellular pathogens, that enhanced IFN production is most important.
In conclusion, rather than the simple down-regulation suggested by TLR tolerance, we have demonstrated a more complex ‘TLR adaptation’ characterized by differential regulation of the downstream pathways. Macrophage phenotype and response to stimuli can adapt sequentially to different environments, changing between immunostimulatory and tolerogenic phenotypes depending on the cytokine milieu.51 Understanding the complex relationships between the different signalling pathways should therefore ultimately enable development of therapies aimed at modulating the immune response so that adequate function can be restored in post-septic hypoimmune patients.
Acknowledgments
This work was funded by the Medical Research Council and by the Newcastle University Hospitals Special Trustees
Glossary
Abbreviations:
- CpG
unmethylated CpG-motif containing DNA
- ELISA
enzyme-linked immunosorbent assay
- IFN
interferon
- IL
interleukin
- iNOS
inducible nitric oxide synthase
- IRF
interferon regulatory factor
- LPS
lipopolysaccharide
- MyD88
myeloid differentiation factor-88
- NF-κB
nuclear factor κB
- PIC
poly(inosine) cytosine
- TLR
Toll-like receptor
- TNF
tumour necrosis factor
- TRAM
TRIF-related adaptor molecule
- TRIF
Toll/IL-1 receptor domain containing adaptor inducing IFN-β
References
- 1.Matzinger P. An innate sense of danger. Ann NY Acad Sci. 2002;961:341–2. doi: 10.1111/j.1749-6632.2002.tb03118.x. [DOI] [PubMed] [Google Scholar]
- 2.Wysocka M, Robertson S, Riemann H, et al. IL-12 suppression during experimental endotoxin tolerance: dendritic cell loss and macrophage hyporesponsiveness. J Immunol. 2001;166:7504–13. doi: 10.4049/jimmunol.166.12.7504. [DOI] [PubMed] [Google Scholar]
- 3.Wilson CS, Seatter SC, Rodriguez JL, Bellingham J, Laurel C, West MA. In vivo endotoxin tolerance: impaired LPS-stimulated TNF-release of monocytes from patients with sepsis but not SIRS. J Surg Res. 1997;69:101–6. doi: 10.1006/jsre.1997.5040. [DOI] [PubMed] [Google Scholar]
- 4.Hotchkiss RS, Karl IE. The pathophysiology and treatment of sepsis. N Engl J Med. 2003;348(2):138–50. doi: 10.1056/NEJMra021333. [DOI] [PubMed] [Google Scholar]
- 5.Perry JA, Olver CS, Burnett RC, Avery AC. Cutting edge. The acquisition TLR tolerance during malaria infection impacts on T cell activation. J Immunol. 2005;174:5921–5. doi: 10.4049/jimmunol.174.10.5921. [DOI] [PubMed] [Google Scholar]
- 6.Abreu MT, Fukata M, Arditi M. TLR signaling in the gut in health and disease. J Immunol. 2005;174:4453–60. doi: 10.4049/jimmunol.174.8.4453. [DOI] [PubMed] [Google Scholar]
- 7.Uhrig A, Banafsche R, Kremer M, et al. Development and functional consequences of LPS tolerance in sinusoidal endothelial cells of the liver. J Leukoc Biol. 2005;77:626–33. doi: 10.1189/jlb.0604332. [DOI] [PubMed] [Google Scholar]
- 8.Dobrovolskaia MA, Medvedev AE, Thomas KE, et al. Induction of in vitro reprogramming by Toll-like receptor (TLR) 2 and TLR4 agonists in murine macrophages: effects of TLR ‘homotolerance’ versus ‘heterotolerance’ on NF-κB signaling pathway components. J Immunol. 2003;170:508–19. doi: 10.4049/jimmunol.170.1.508. [DOI] [PubMed] [Google Scholar]
- 9.Yeo S-J, Yoon J-G, Hong S-C, Yi A-K. CpG DNA induces self and cross-hyporesponsiveness of RAW264.7 cells in response to CpG DNA and lipopolysaccharide: alterations in IL-1 receptor-associated kinase expression. J Immunol. 2003;170:1052–61. doi: 10.4049/jimmunol.170.2.1052. [DOI] [PubMed] [Google Scholar]
- 10.Bafica A, Scanga CA, Equils O, Sher A. The induction of Toll-like receptor tolerance enhances rather than suppresses HIV-1 gene expression in transgenic mice. J Leukoc Biol. 2004;75:460–6. doi: 10.1189/jlb.0803388. [DOI] [PubMed] [Google Scholar]
- 11.Sato S, Nomura F, Kawai T, Takeuchi O, Muhlradt PF, Takeda K, Akira S. Synergy and cross-tolerance between Toll-like receptor (TLR) 2- and TLR4-mediated signaling pathways. J Immunol. 2000;165:7096–101. doi: 10.4049/jimmunol.165.12.7096. [DOI] [PubMed] [Google Scholar]
- 12.Wang JH, Doyle M, Manning BJ, Di Wu Q, Blankson S, Redmond HP. Induction of bacterial lipoprotein tolerance is associated with suppression of Toll-like receptor 2 expression. J Biol Chem. 2002;277(39):36068–75. doi: 10.1074/jbc.M205584200. [DOI] [PubMed] [Google Scholar]
- 13.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 14.Moynagh PN. TLR signalling and activation of IRFs: revisiting old friends from the NF-κB pathway. Trends Immunol. 2005;26(9):469–76. doi: 10.1016/j.it.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 15.Medvedev AE, Kopydlowski KM, Vogel SN. Inhibition of lipopolysaccharide-induced signal transduction in endotoxin-tolerized mouse macrophages: dysregulation of cytokine, chemokine, and Toll-like receptor 2 and 4 gene expression. J Immunol. 2000;164:5564–74. doi: 10.4049/jimmunol.164.11.5564. [DOI] [PubMed] [Google Scholar]
- 16.Schoenemeyer A, Barnes BJ, Mancl ME, Latz E, Goutagny N, Pitha PM, Fitzgerald KA, Golenbock DT. The interferon regulatory factor, IRF5, is a central mediator of Toll-like receptor 7 signaling. J Biol Chem. 2005;280(17):17005–12. doi: 10.1074/jbc.M412584200. [DOI] [PubMed] [Google Scholar]
- 17.Gautier G, Humbert M, Deauvieau F, et al. A type I interferon autocrine-paracrine loop is involved in Toll-like receptor-induced interleukin-12p70 secretion by dendritic cells. J Exp Med. 2005;201:1435–46. doi: 10.1084/jem.20041964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirotani T, Yamamoto M, Kumagai Y, Uematsu S, Kawase I, Takeuchi O, Akira S. Regulation of lipopolysaccharide-inducible genes by MyD88 and Toll/IL-1 domain containing adaptor inducing IFN-β. Biochem Biophys Res Commun. 2005;328:383–92. doi: 10.1016/j.bbrc.2004.12.184. [DOI] [PubMed] [Google Scholar]
- 19.Fitzgerald KA, Rowe DC, Barnes BJ, et al. LPS-TLR4 signaling to IRF-3/7 and NF-κB involves the Toll adapters TRAM and TRIF. J Exp Med. 2003;198:1043–55. doi: 10.1084/jem.20031023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim KI, Malakhova OA, Hoebe K, Yan M, Beutler B, Zhang D-E. Enhanced antibacterial potential in UBP43-deficient mice against Salmonella typhimurium infection by up-regulating type I IFN signaling. J Immunol. 2005;175:847–54. doi: 10.4049/jimmunol.175.2.847. [DOI] [PubMed] [Google Scholar]
- 21.Alves-Rosa F, Vulcano M, Beigier-Bompadre M, Fernandez G, Palermo M, Isturiz MA. Interleukin-1beta induces in vivo tolerance to lipopolysaccharide in mice. Clin Exp Immunol. 2002;128:221–8. doi: 10.1046/j.1365-2249.2002.01828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grutz G. New insights into the molecular mechanism of interleukin-10-mediated immunosuppression. J Leukoc Biol. 2005;77:3–15. doi: 10.1189/jlb.0904484. [DOI] [PubMed] [Google Scholar]
- 23.Varma TK, Durham M, Murphey ED, Cui W, Huang Z, Lin CY, Toliver-Kinsky T, Sherwood ER. Endotoxin priming improves clearance of Pseudomonas aeruginosa in wild-type and interleukin-10 knockout mice. Infect Immun. 2005;73:7340–7. doi: 10.1128/IAI.73.11.7340-7347.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwartz DA, Wohlford-Lenane CL, Quinn TJ, Krieg AM. Bacterial DNA or oligonucleotides containing unmethylated CpG motifs can minimize lipopolysaccharide-induced inflammation in the lower respiratory tract through an IL-12-dependent pathway. J Immunol. 1999;163:224–31. [PubMed] [Google Scholar]
- 25.Gould MP, Greene JA, Bhoj V, DeVecchio JL, Heinzel FP. Distinct modulatory effects of LPS and CpG on IL-18-dependent IFN-γ synthesis. J Immunol. 2004;172:1754–62. doi: 10.4049/jimmunol.172.3.1754. [DOI] [PubMed] [Google Scholar]
- 26.Boyte W, Meals E, English B. Acquired hyporesponsiveness to bacterial lipopolysaccharide and interferon-gamma in RAW 264.7 macrophages. Shock. 1996;6:218–22. [PubMed] [Google Scholar]
- 27.Henricson B, Manthey C, Perera P, Hamilton T, Vogel S. Dissociation of lipopolysaccharide (LPS) -inducible gene expression in murine macrophages pretreated with smooth LPS versus monophosphoryl lipid A. Infect Immun. 1993;61:2325. doi: 10.1128/iai.61.6.2325-2333.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Brien GC, Wang JH, Redmond HP. Bacterial lipoprotein induces resistance to Gram-negative sepsis in TLR4-deficient mice via enhanced bacterial clearance. J Immunol. 2005;174:1020–6. doi: 10.4049/jimmunol.174.2.1020. [DOI] [PubMed] [Google Scholar]
- 29.Lehner MD, Ittner J, Bundschuh DS, van Rooijen N, Wendel A, Hartung T. Improved innate immunity of endotoxin-tolerant mice increases resistance to Salmonella enterica serovar typhimurium infection despite attenuated cytokine response. Infect Immun. 2001;69:463–71. doi: 10.1128/IAI.69.1.463-471.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang JH, Doyle M, Manning BJ, Blankson S, Wu QD, Power C, Cahill R, Redmond HP. Cutting edge: bacterial lipoprotein induces endotoxin-independent tolerance to septic shock. J Immunol. 2003;170:14–18. doi: 10.4049/jimmunol.170.1.14. [DOI] [PubMed] [Google Scholar]
- 31.Wolk K, Kunz S, Crompton NEA, Volk H-D, Sabat R. Multiple mechanisms of reduced major histocompatibility complex class II expression in endotoxin tolerance. J Biol Chem. 2003;278(20):18030–6. doi: 10.1074/jbc.M207714200. [DOI] [PubMed] [Google Scholar]
- 32.Wolk K, Docke W-D, von Baehr V, Volk H-D, Sabat R. Impaired antigen presentation by human monocytes during endotoxin tolerance. Blood. 2000;96:218–23. [PubMed] [Google Scholar]
- 33.Lee JY, Lowell CA, Lemay DG, et al. The regulation of the expression of inducible nitric oxide synthase by Src-family tyrosine kinases mediated through MyD88-independent signaling pathways of Toll-like receptor 4. Biochem Pharmacol. 2005;70:1231–40. doi: 10.1016/j.bcp.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 34.Fejer G, Szalay K, Gyory I, et al. Adenovirus infection dramatically augments lipopolysaccharide-induced TNF production and sensitizes to lethal shock. J Immunol. 2005;175:1498–506. doi: 10.4049/jimmunol.175.3.1498. [DOI] [PubMed] [Google Scholar]
- 35.Nansen A, Randrup Thomsen A. Viral infection causes rapid sensitization to lipopolysaccharide. Central role of IFN-αβ. J Immunol. 2001;166:982–8. doi: 10.4049/jimmunol.166.2.982. [DOI] [PubMed] [Google Scholar]
- 36.Wongratanacheewin S, Kespichayawattana W, Intachote P, Pichyangkul S, Sermswan RW, Krieg AM, Sirisinha S. Immunostimulatory CpG oligodeoxynucleotide confers protection in a murine model of infection with Burkholderia pseudomallei. Infect Immun. 2004;72:4494–502. doi: 10.1128/IAI.72.8.4494-4502.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cusson-Hermance N, Khurana S, Lee TH, Fitzgerald KA, Kelliher MA. Rip1 mediates the Trif-dependent Toll-like receptor 3- and 4-induced NF-κB activation but does not contribute to interferon regulatory factor 3 activation. J Biol Chem. 2005;280(44):36560–6. doi: 10.1074/jbc.M506831200. [DOI] [PubMed] [Google Scholar]
- 38.Takeda K, Akira S. Toll-like receptors in innate immunity. Int Immunol. 2005;17:1–14. doi: 10.1093/intimm/dxh186. [DOI] [PubMed] [Google Scholar]
- 39.Dunne A, O'Neill LAJ. The interleukin-1 receptor/Toll-like receptor superfamily: signal transduction during inflammation and host defense. Science STKE. 2003;171:re3. doi: 10.1126/stke.2003.171.re3. [DOI] [PubMed] [Google Scholar]
- 40.Naiki Y, Michelsen KS, Schroder NWJ, et al. MyD88 is pivotal for the early inflammatory response and subsequent bacterial clearance and survival in a mouse model of Chlamydia pneumoniae pneumonia. J Biol Chem. 2005;280(32):29242–9. doi: 10.1074/jbc.M503225200. [DOI] [PubMed] [Google Scholar]
- 41.Power MR, Peng Y, Maydanski E, Marshall JS, Lin T-J. The development of early host response to Pseudomonas aeruginosa lung infection is critically dependent on myeloid differentiation factor 88 in mice. J Biol Chem. 2004;279(47):49315–22. doi: 10.1074/jbc.M402111200. [DOI] [PubMed] [Google Scholar]
- 42.Yauch LE, Mansour MK, Shoham S, Rottman JB, Levitz SM. Involvement of CD14, Toll-like receptors 2 and 4, and MyD88 in the host response to the fungal pathogen Cryptococcus neoformans in vivo. Infect Immun. 2004;72:5373–82. doi: 10.1128/IAI.72.9.5373-5382.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wieland CW, Florquin S, Maris NA, Hoebe K, Beutler B, Takeda K, Akira S, van der Poll T. The MyD88-dependent, but not the MyD88-independent, pathway of TLR4 signaling is important in clearing nontypeable Haemophilus influenzae from the mouse lung. J Immunol. 2005;175:6042–9. doi: 10.4049/jimmunol.175.9.6042. [DOI] [PubMed] [Google Scholar]
- 44.Seth RB, Sun L, Ea C-K, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NFκB and IRF3. Cell. 2005;122:669–82. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 45.Li K, Chen Z, Kato N, Gale M, Jr, Lemon SM. Distinct poly (I-C) and virus-activated signaling pathways leading to interferon-β production in hepatocytes. J Biol Chem. 2005;280(17):16739–47. doi: 10.1074/jbc.M414139200. [DOI] [PubMed] [Google Scholar]
- 46.Foy E, Li K, Sumpter R, Jr, et al. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. PNAS. 2005;102:2986–91. doi: 10.1073/pnas.0408707102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li K, Foy E, Ferreon JC, et al. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. PNAS. 2005;102:2992–7. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li X-D, Sun L, Seth RB, Pineda G, Chen ZJ. From the cover: hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. PNAS. 2005;102(49):17717–22. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quintana FJ, Cohen IR. Heat shock proteins as endogenous adjuvants in sterile and septic inflammation. J Immunol. 2005;175:2777–82. doi: 10.4049/jimmunol.175.5.2777. [DOI] [PubMed] [Google Scholar]
- 50.Johnson GB, Brunn GJ, Platt JL. Cutting edge: an endogenous pathway to systemic inflammatory response syndrome (SIRS)-like reactions through Toll-like receptor 4. J Immunol. 2004;172:20–4. doi: 10.4049/jimmunol.172.1.20. [DOI] [PubMed] [Google Scholar]
- 51.Stout RD, Jiang C, Matta B, Tietzel I, Watkins SK, Suttles J. Macrophages sequentially change their functional phenotype in response to changes in microenvironmental influences. J Immunol. 2005;175:342–9. doi: 10.4049/jimmunol.175.1.342. [DOI] [PubMed] [Google Scholar]