Abstract
Porcine circovirus type 2 (PCV2) infection of natural interferon producing cells (NIPCs) impairs the induction of interferon (IFN)-α and tumour necrosis factor (TNF)-α by cytosine-phosphorothioate-guanine (CpG) oligodeoxynucleotides (ODNs), thereby preventing both their autocrine maturation and the paracrine maturation of myeloid dendritic cells (DCs). The present study shows that the PCV2-mediated inhibition of NIPCs was mediated by viral DNA, although it was independent of virus replication. The inhibitory effect of PCV2 DNA was more diversified than if it had simply targeted CpG-ODN-induced cytokines (IFN-α, TNF-α, interleukin-6, IL-12). A broad spectrum inhibition was noted, affecting responses induced by toll-like receptor (TLR)-7 and TLR9 agonists, as well as viruses including pseudorabies virus, transmissible gastroenteritis virus and classical swine fever virus. From these results, it would appear that PCV2 DNA can induce a dominant negative signal influencing independent pattern recognition receptor-induced activation cascades. Despite a concomitant internalization of PCV2 DNA and CpG-ODNs, no colocalization was observed, indicating that PCV2 DNA and CPG-ODNs may not target the same receptor. This study describes a novel modulation of the innate immune response, which would render the host more susceptible to secondary or concomitant microbial infections.
Keywords: natural interferon producing cells, DNA, porcine circovirus type 2, inhibition, toll-like receptors
Introduction
Recent investigations into natural interferon producing cells (NIPCs), also called plasmacytoid dendritic cells (pDCs), have highlighted their critical role in both innate and adaptive immunity. These important sentinels of immune defences sense ‘intruders’, and shape the strength, duration and quality of NK, T and B cell responses (reviews1,2). Indeed, NIPCs respond efficiently and rapidly to invading pathogens by producing particularly high amounts of type 1 interferon (IFN) through recognition of bacterial and viral components via pattern recognition receptors (PRRs), including toll-like receptors (TLRs).3,4 It is to this property that NIPCs owe their nomenclature, and their additional title of ‘professional interferon-producing cells’. NIPCs also produce important quantities of other cytokines, including tumour necrosis factor (TNF)-α and interleukin (IL)-6.2 These characteristics have also been identified in porcine NIPCs.5,6 Indeed, the now well characterized phenotypic and functional properties of porcine NIPCs provide a valuable model with which to study the biology of these cells, including the influence of virus infections.
NIPCs respond to synthetic oligodeoxynucleotides (ODNs) containing unmethylated cytosine-phosphorothioate-guanine (CpG) motifs in particular base contexts called CpG-ODNs, which are recognized by TLR9.7 Viral DNA can contain such motifs, as witnessed by the involvement of TLR9 in herpesviruses-induced NIPC activation, indicating that this pathway may be important for NIPC interaction with and in response to DNA viruses.8,9 Conversely, viral elements counteracting IFN type 1 pathways are important for the successful evolution of viruses.10 Yet despite the importance of NIPCs in virus-induced IFN responses, viruses able to counteract the activation of NIPCs have only recently been identified for respiratory syncitial virus, measles virus and porcine circovirus type 2 (PCV2).11,12 Neither the viral nor the cellular elements involved in this inhibition have been described.
PCV2, a small non-enveloped virus possessing a circular single-stranded DNA genome of 1759 bp,13 is the causative agent of post-weaning multisystemic wasting syndrome (PMWS)14,15 and other pathological syndromes collectively referred to as PCV2-associated diseases (PCVD).16–18 The virus can persist in pigs, mainly asymptomatically19,20 and coinfections – bacterial or viral – or other cofactors are required for disease manifestation.21–24 This has led to the hypothesis that immunosuppressive events elicited following PCV2 infection may play a role in the development of PCVD, a concept supported by the demonstration of PCV2-induced lymphopenia in symptomatic animals.25
Recently, we demonstrated that PCV2 can be endocytosed by myeloid DCs without any evidence of virus replication or functional impairment of cell activity.12,26 The virus immunomodulatory capacity appeared to focus on a selective impairment of NIPCs, in terms of their IFN-α and TNF-α response to CpG-ODNs.12 Based on this work, the present study sought to elaborate on the underlying mechanisms of PCV2-dependent inhibition of IFN-α production by NIPCs, particularly with respect to the viral elements responsible for the functional disruption of the cells.
Materials and methods
Animals
Swiss White Landrace specific pathogen-free (SPF) pigs, previously shown to be seronegative for PCV2, were used throughout this study.
Viruses
A previously characterized PCV2 isolate (strain 1010 Stoon) from Canada was used to generate the virus stock pools, as described elsewhere.27 Briefly, the porcine kidney cell line PK15A, free of PCV1 and PCV2, was cultured to prepare virus pools and mock lysates. PCV2 viral titres were determined by end-point titration in PK15A cells; the presence of PCV2 antigen in the cells was detected using a monoclonal antibody (mAb) against the ORF2-encoded capsid protein (7G5-G4-A1; see below under ‘Antibodies, flow cytometry and confocal microscopy’). The titres were expressed in 50% tissue culture infectivity doses (TCID50/ml).28
In some experiments, the PCV2 inoculum was pretreated with DNAse I (Ambion, Cambridgeshire, UK) at 400 U/ml for 30 min at 37°. Control inocula comprising UV-inactivated PCV2 or mock lysate (104J/m2 for 20 min) were also used. Classical swine fever virus (CSFV; strain Brescia) was prepared in SK-6 strain as described.29 Pseudorabies virus (PRV; strain E12215) was propagated in Madin–Darby bovine kidney (MDBK) cells.30 Transmissible gastroenteritis virus (TGEV; strain Perdue 115) was propagated in swine testicular (ST) cells, and virus preparations were inactivated prior to use, as described previously.31
Viral DNA preparation
Plasmids containing double stranded (ds) DNA forms of the complete PCV2 genome (pGEM7-PCV2, kindly provided by Dr Brian Meehan, Queen's University of Belfast, Belfast, UK; Meehan etal. 199813) or PCV1 genome (kindly provided by Dr Annette Mankertz, Robert Koch Institute, Berlin, Germany) were amplified and purified using a Qiagen Maxi Plasmid Kit (Qiagen AG, Hombrechtikon, Switzerland). The inserts of PCV2 or PCV1 were then isolated by an EcoRI or PstI digestion, respectively, followed by two rounds of agarose gel purification of the insert (QIAquick Gel Extraction Kit; Qiagen AG).
NIPC isolation and stimulation for cytokine responses
Peripheral blood mononuclear cells (PBMCs) were prepared by Ficoll−Paque differential centrifugation.32 These PBMCs were then enriched by CD172a or CD4 sorting using magnetic antibody cell sorting (MACS; Miltenyi Biotec GMbH, Bergish Gladbach, Germany) as previously described.5,6
For certain experiments, particularly those in which monocyte exclusion was critical, the PBMCs were fractionated on a 55% (v/v) Percoll density gradient prior to a CD4 sorting as above.33 This method is referred to in the text as the ‘Percoll/CD4 protocol’.
The sorted CD172+ or CD4+ cells were stimulated as specified in the text with CSFV or PRV or TLR7/9 agonists (R837 (1 µg/ml), CpG D32 10 µg/ml (Biosource Int., Camarillo, CA) Supernatants from activated NIPC cultures were tested for IFN-α and IL-12 as described5,6 and TNF-α or IL-6 by enzyme-linked immunosorbent assay (ELISA kits; Biosource Int.).
Antibodies, flow cytometry and confocal microscopy
Anti-SWC3 (CD172a, 74-22-15) and anti-CD4 (74-12-4) were prepared from hybridomas provided by Dr J. Lunney (Beltsville, MD) and anti-CD4 (PT90A) was purchased from Veterinary Medical Research & Development (VMRD) (Pullman, WA). Anti-IFN-α mAbs F17 and K9 were kindly provided by Dr Bernard Charley (INRA, Jouy-en-Josas, France. Anti-PCV2 capsid protein (7G5-G4-A1; kindly provided by Francis McNeilly, Department of Agriculture and Rural Development for Northern Ireland, Veterinary Sciences Division, Belfast, UK) was used to detect intracellular PCV2 virus.
For flow cytometry analysis, reacted mAbs were revealed using fluorescein isothiocyanate (FITC), phycoerythrin (PE) or biotin-conjugated goat F(ab′)2 antimouse isotype-specific immunoglobulins (Southern Technology, Birmingham, UK). PE-Cy5 conjugated streptavidin (Dako, Zug, Switzerland) was finally added to detect the biotinylated conjugate. For intracellular staining of PCV2 antigen or IFN-α, the cells were first incubated with antibodies against cell surface molecules and then fixed and permeabilized using a cell permeabilization kit (Harlan Sera-Laboratory, Crawley Down, UK), followed by detection using the above conjugates. The data were acquired using a FACScalibur and analysed using CellQuest Pro Software (BD Biosciences, Mountain View, CA).
For confocal microscopy, Alexa fluorochrome-labelled antimouse secondary antibodies (Molecular Probes, Eugene, OR) were employed. Data were acquired using a Leica TCS-SL spectral confocal microscope and Leica LCS software (Leica Microsystems AG, Glattbrugg, Switzerland).
Nucleic acid labelling and cell uptake studies
CpG-ODNs and purified viral DNA inserts were labelled using the Label IT® TrackerTM CX-Rhodamine and CyTM3 Kits (Mirus, VWR International, Switzerland) according to the manufacturer's instructions. Briefly, CpG-ODNs and purified viral DNA were incubated for 1 hr at 37° with Label IT® TrackerTM reagent in the appropriate buffer, and precipitated with 100% ethanol for 1 hr at − 70°. Labelled DNA was washed twice with 70% ethanol and resuspended in H2O.
CD4+ cells were enriched from PBMCs by magnetic sorting, and incubated with 5 µg labelled-CpG-ODN or viral DNA for 1–3 hr at 39°. The cells were then washed twice with cold phosphate-buffered saline (PBS) (4°), before staining for surface CD4 for 20 min on ice. Labelled CD4 and internalized ODNs or DNA were visualized by confocal microscopy as described above.
Results
PCV2 inhibits IFN-α induction in NIPCs independently of viral replication
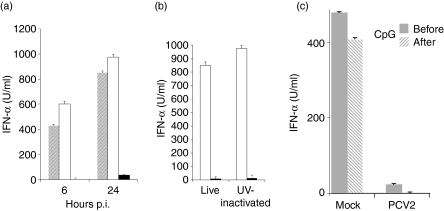
We have previously shown that the IFN-α response of CD172a+ enriched NIPCs to CpG-ODNs was inhibited when cells were infected concomitantly with PCV2 at a multiplicity of infection (MOI) of 3 TCID50/cell.12 In order to investigate if this effect was longlasting, this inhibition experiment was repeated and supernatants were compared for the presence of IFN-α at 6 hr and 24 hr (Fig. 1a). The ability of the virus to block IFN-α induction was both rapid and pronounced, observed at both 6 hr and 24 hr (Fig. 1a). To ascertain whether viral replication was prerequisite for the response elicited, UV-inactivated PCV2 – corresponding to an MOI of 3 TCID50/cell – was applied in similar experiments in place of the live virus. The results showed that the UV-treated virus was as effective at inhibiting CpG-induced IFN-α production (Fig. 1b), indicating that viral replication was not essential.
Figure 1.
Porcine circovirus type 2 (PCV2)-mediated inhibition of interferon (IFN)-α production by natural interferon producing cells (NIPCs) in response to cytosine-phosphorothioate-guanine oligodeoxynucleotides (CpG-ODNs) is replication-independent.(a)Enriched NIPCs (CD172a+) were incubated with CpG-ODN D32 (10 µg/ml) alone (grey bars), or in the presence of mock lysate (white bars) or PCV2 at 3 50% tissue culture infectivity doses (TCID50)/cell (black bars). Cell-free supernatants were tested for IFN-α after 6 hr and 24 hr by enzyme-linked immunosorbent assay.(b)Enriched NIPCs (CD172a+) were stimulated with CpG-ODN D32 (10 µg/ml) in the presence of mock lysate (white bars) or PCV2 (3 TCID50/cell or equivalent, black bars) present as either live virus or UV-inactivated PCV2. Culture supernatants were tested for IFN-α after 24 hr.(c)Time-dependence of PCV2-induced IFN-α inhibition. Enriched NIPCs (CD172+) were stimulated with either CpG-ODN D32 (10 µg/ml) for 2 hr before or after addition of mock lysate or PCV2 at 3 TCID50/cell. Culture supernatants were tested for IFN-α after 24 hr.
Efficiency of PCV2-mediated NIPC inhibition relative to time post-simulation
The above results demonstrated PCV2 inhibition of NIPC cytokine responsiveness when the virus was added simultaneously with the stimulant. It was considered important to analyse this PCV2 inhibitory effect when the virus was added after the stimulation event. Adding the virus either 2 hr before or 2 hr after the CpG-ODNs did not alter this strong inhibition (Fig. 1c). It was not until the CpG stimulation of the NIPCs was applied 4 hr prior to the addition of PCV2 that the IFN-α response resumed the level seen with the mock controls (data not shown).
PCV2 inhibits IFN-α production induced via TLR-dependent and independent signalling
The inhibitory potential PCV2 was further characterized with respect to its effect on NIPC activation by various agents including the TLR7 ligand R837 and three different viruses. The latter were:
TGEV, a porcine coronavirus stimulating NIPCs through viral glycoproteins independent of viral nucleic acid;34
CSFV, a pestivirus representing a virus replication-dependent induction of IFN-α in NIPCs,33 and
PRV, related to human herpesvirus-1 and activating NIPCs via TLR9-dependent and independent pathways.35
For this work NIPCs were enriched using the Percoll/CD4 protocol, which excludes the presence of contaminating monocytes to avoid their potential contribution to the cytokine responses.
The mock-treated NIPCs displayed the levels of IFN-α production expected in response to all the above stimuli (Fig. 2a). In contrast, PCV2 clearly impaired these IFN-α responses to all stimuli (Fig. 2a). This was not due to a toxic effect of the virus preparation on NIPCs, because cell survival was identical in the mock- and PCV2-treated cells (data not shown).
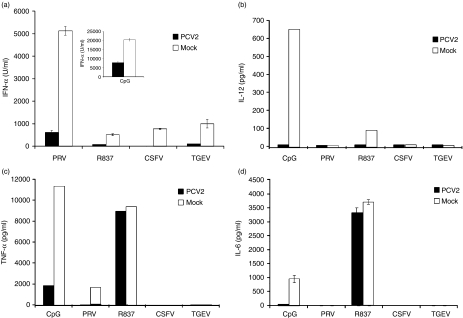
Figure 2.
Porcine circovirus type 2 (PCV2)-mediated inhibition of natural interferon producing cell (NIPC) cytokine induction. Enriched NIPCs (Percoll/CD4 protocol) received PCV2 at 3 50% tissue culture infectivity doses (TCID50)/cell 1 hr before addition of cytosine-phosphorothioate-guanine (CpG) (10 µg/ml), R837 (10 µg/ml), pseudorabies virus (PRV) (TCID50/cell), classical swine fever virus (CSFV) (0·1 TCID50/cell) or transmissible gastroenteritis virus (TGEV) (0·1 TCID50/cell). Supernatants were harvested from the cultures after 24 hr and tested for(a)interferon (IFN)-α,(b)interleukin (IL)-12,(c)tumour necrosis factor (TNF)-α and(d)IL-6. The results represent three independent experiments.
The extent of this PCV2-dependent inhibition was analysed with reference to cytokines other than IFN-α. Supernatants from the stimulated mock- and PCV2-treated cells were further tested for the presence of IL-12 (Fig. 2b), TNF-α (Fig. 2c) and IL-6 (Fig. 2d). None of the stimulants induced all these cytokines. Nevertheless, the PCV2 did suppress all cytokine responses to each stimulus, with the exception of the TNF-α and IL-6 response to the TLR7 ligand R837 (Fig. 2c, d).
Relationship of PCV2 inhibition with MOI and capsid uptake
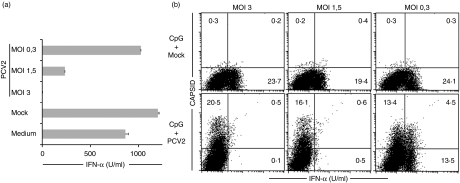
In order to determine which viral component was responsible, we first examined the relationships of PCV2 inhibition with the MOI and capsid uptake. When enriched NIPCs were treated for 24 hr with the CpG-ODNs and PCV2 at various MOIs (3, 1·5 or 0·3 TCID50/cell), the lower virus MOIs were clearly less efficient at inhibiting IFN-α-induction (Fig. 3a). This decrease in efficiency was dependent on the MOI used. In order to determine the relationship to capsid uptake and IFN-α inhibition, intracellular staining for IFN-α was employed with enriched NIPCs. The PCV2-dependent block of IFN-α-induction was also observed in the intracellular compartment (Fig. 3b). Mock-treated NIPCs responded to CpG-ODNs with clear IFN-α production (Fig. 3b, upper panels), whereas no IFN-α expressing cells were detectable after infection with PCV2 at an MOI of 3 or 1·5 TCID50/cell (Fig. 3b, lower panels). Interestingly, a minority of NIPCs were double-positive for PCV2 capsid and IFN-α at the MOI of 0·3 TCID50/cell (Fig. 3b, lower right panel). These double-positive cells, as well as the single IFN-α positive cells, disappeared with increasing MOI, demonstrating that the inhibitory effect of PCV2 on IFN-α induction in NIPCs was dependent on the amount of virus present. The fact that IFN-secreting cells could be observed carrying viral capsid – at the lower MOI – suggested that this inhibition of IFN-α induction was not directly related to the presence of viral capsid protein.
Figure 3.
Relationship of porcine circovirus type 2 (PCV2) inhibition with multiplicity of infection (MOI) and capsid uptake.(a)Enriched natural interferon producing cells (NIPCs) (CD172a+) were treated with cytosine-phosphorothioate-guanine oligodeoxynucleotide (CpG-ODN) D32 (10 µg/ml) alone (‘Medium’), or in the presence of mock lysate (‘Mock’) or PCV2 at 3, 1·5 and 0·3 50% tissue culture infectivity doses (TCID50)/cell for 24 hr, and supernatants tested for interferon (IFN)-α.(b)Enriched NIPCs (CD4+ sorting) were incubated with CpG-ODN D32 (10 µg/ml) in the presence of mock lysate or PCV2 at 3, 1·5 and 0·3 TCID50/cell for 6 hr. Cells were labelled with monoclonal antibody against surface CD4, then fixed and permeabilized for intracellular detection of IFN-α and PCV2 capsid antigen. Only CD4+ FSChigh gated cells representing NIPCs are shown. Supernatants were tested for the presence of IFN-α by enzyme-linked immunosorbent assay 6 hr post-stimulation. The results represent three independent experiments.
Role of viral DNA in PCV2-mediated inhibition of IFN-α induction in NIPCs
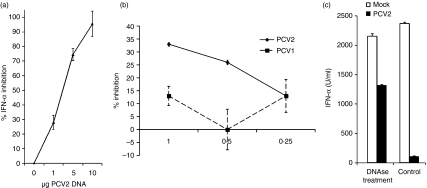
To investigate the role of specific viral components required for PCV2-mediated inhibition of IFN-α induction in NIPCs, purified linearized PCV2 DNA – representing a replicative intermediate form13– was added simultaneously with CpG-ODNs to enriched NIPCs. When the culture supernatants were analysed after 24 hr for the presence of IFN-α, the PCV2 DNA was seen to mediate inhibition of the IFN-α-induced by CpG-ODNs (Fig. 4a). Moreover, this occurred in a dose-dependent manner.
Figure 4.
Viral DNA inhibits interferon (IFN)-α production by natural interferon producing cells (NIPCs).(a)Enriched NIPCs (Percoll/CD4protocol) were stimulated with cytosine-phosphorothioate-guanine oligodeoxynucleotide (CpG-ODN) D32 (10 µg/ml) for 24 hr in the presence of increasing amounts of the porcine circovirus type 2 (PCV2) genome in a linear double-stranded form (PCV2 DNA). The y-axis shows the percentage of IFN-α inhibition in the presence of the PCV2 DNA, relative to the levels obtained with the CpG-ODN (‘0% inhibition’). Means of three independent experiments are shown, with error bars indicating the standard deviation from the three experiments.(b)Enriched NIPCs (Percoll/CD4 protocol) were stimulated with CpG-ODN D32 (10 µg/ml) for 24 hr in the presence of increasing amounts of the PCV2 or PCV1 genomic DNA, both in a linear double-stranded form. The y-axis shows the percentage of IFN-α inhibition in the presence of the PCV DNA, relative to the levels obtained with the CpG-ODN alone.(c)Enriched NIPCs (CD172a+) were stimulated with transmissible gastroenteritis virus (TGEV) for 24 h in the presence of mock lysate or PCV2. The mock and PCV2 were either untreated (‘control’) or DNAse-treated (PCV2 at 3 50% tissue culture infectivity doses/cell, pretreated with 400 U/ml DNAse I for 30 min at 37 °. Culture supernatants were tested for the presence of IFN-α by enzyme-linked immunosorbent assay.
The activity of PCV2 DNA was compared with DNA from the closely related PCV1, a non-pathogenic virus with significant sequence homology to PCV2.36,37Figure 4(b) demonstrates that PCV1 DNA was less inhibitory to IFN-α production by NIPCs in response to CpG-ODNs. Moreover, the low-level effect of the PCV1 DNA was not dose-dependent, unlike the inhibition observed with PCV2 DNA.
Inhibition of NIPCs is partly mediated by free PCV2 DNA
The PCV2 preparations used in this work were derived by lysis of infected PK15A cells associated with the release of free viral DNA, then present in the lysates.13 Considering the role of PCV2 DNA in NIPC inhibition, we analyse the role played by this free PCV2 DNA by pretreating the PCV2 inoculum with deoxyribonuclease (DNAse) I. The efficiency of DNAse digestion was controlled on an agarose gel (data not shown). In comparison with untreated virus lysates, DNAse-treated PCV2 preparations were less efficient at inhibiting IFN-α production (Fig. 4c), although the PCV2 infectious titres of the digested preparations were not reduced (data not shown). Nevertheless, the levels of IFN-α did not return to those seen in the absence of PCV2, suggesting that encapsulated viral DNA may also play a role within the NIPCs.
PCV2 DNA does not impair CpG-ODN uptake in NIPCs
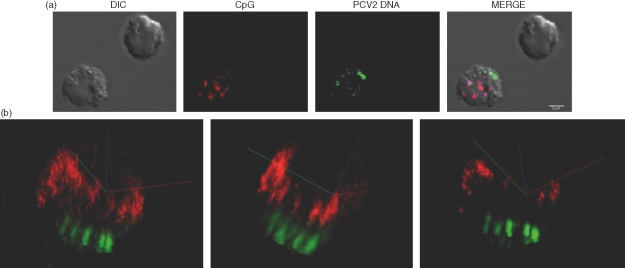
In order to determine if PCV2 DNA could inhibit CpG-ODN entry, the cellular uptake of both PCV2 DNA and CpG-ODNs in NIPCs was analysed. Figure 5(a) shows clear internalization of both nucleic acids – viral genome (PCV2 DNA, Cy5 green) and CpG-ODN (CpG, CX-rhodamine red) at 3 hr after addition of the nucleic acids. A similar distribution was already detectable after 1 h (data not shown). Detailed analysis of the double positive cells using imaris colocalization software revealed that the PCV2 genomic DNA and CpG-ODN did not colocalize following addition of the nucleic acids. This is represented in Fig. 5(b) by a 3-dimensional image taken at 3 hr post-interaction. These results demonstrate that free viral DNA is certainly efficiently internalized by NIPCs and that PCV2 DNA does not influence CpG-ODN internalization. With the two nucleic acids appearing in distinct cytoplasmic localizations, it would appear that the PCV2 DNA and CpG-ODNs do not interact with the same receptor.
Figure 5.
Localization of cytosine-phosphorothioate-guanine oligodeoxynucleotide (CpG-ODN) and purified porcine circovirus type 2 (PCV2) DNA in natural interferon producing cells (NIPCs). Enriched NIPCs (Percoll/CD4 protocol) were incubated with 5 µg CpG-ODN CX-rhodamine-labelled (red) and 5 µg PCV2 DNA Cy5-labelled (green) for 3 hr.(a)Confocal and differential interference contrast (DIC) images of representative cells.(b)Three-dimensional views created using the colocalization software imaris.
Discussion
Professional IFN type 1 producing cells – NIPCs/pDCs – play an important role in antiviral immune defence. They respond to ‘danger’ signals by producing a number of cytokines, but it is their capacity to secrete elevated quantities of type 1 IFN that endows these cells with their particular characteristics.2,38 Moreover, the action and interaction of NIPCs on both neighbouring cells and systemically is an important factor in the development of innate and adaptive immune defences.2 In addition to antiviral effects, the cytokines secreted by NIPCs – particularly IFN-α– will alert immune defences to danger and assist in initiating adaptive effector and memory immune responses. If a virus evolves mechanisms that counteract the capacity of NIPCs to function, this presents a major barrier to immune defence development. PCV2 is one of only a few viruses described as directly influencing NIPC activity12 and thus may potentially enable us to understand more about this virus-mediated subversion of immune defences.
PCV2 was found to impair NIPC responsiveness to the danger signal provided by stimulatory CpG-ODNs.12 This occurs with a virus that does not display a need to replicate in DCs, but which will persist in these cells without apparent destruction of its infectivity26– a similar lack of replication has been noted for other monocytic cells.26 Akin to this situation with myeloid DCs, the present study shows that PCV2 inhibition of NIPC responsiveness to CpG-ODNs does not require virus replication. By contrast, the presence of viral DNA, even in an inactivated form, is critical.
The PCV2 DNA can still inhibit CpG-ODN activity when added 2 hr after the ODN, which itself requires 4–6 hr to activate NIPCs into detectable IFN-α production.5 Thus, Yamada etal. (2002) suggested that CpG-induced immune activation is an ongoing process that can be inhibited even after the stimulatory signal has been delivered.39
It is known that NIPCs are capable of responding against inactivated viruses through recognition of viral nucleic acid via receptors such as TLR9.2 The PCV2 genome also possesses sequences that may be immunomodulatory for NIPCs. Based on criteria suggested by Krieg etal.,40 the PCV2 genome was predicted to have 60% inhibitory and 40% stimulatory CpG motifs.41 Five synthetic ODNs based on the PCV2 sequence permitted the identification of an inhibitory CpG motif when tested in vitro.41 Such observations would suggest that the results from our present study were due to dominating inhibitory CpG motifs in the PCV2 genome. Although observations made with synthetic sequences may not be directly translatable to the whole genome, it is clear that the PCV2 genome possesses the capacity to interfere with CpG-ODN induced IFN-α production by NIPCs.
The fact that the PCV2 DNA inhibition of stimulatory CpG-ODN signalling was found with preparations from virus-infected cells indicates that the activity does have a biological significance. Moreover, the PCV2 impairment of CpG-ODN-induced IFN-α responses does not require artificial modes of entry into the cell, such as lipofection. This contrasts with the effect obtained using the synthetic inhibitory ODNs, which were only active when the stimulatory CpG-ODNs were lipofected.41 Certainly, there are CpG-ODNs which will be internalized by NIPCs without assistance (such as D32) It seems likely that NIPCs may efficiently internalize PCV2 DNA, as they do for a number of ODNs which they readily recognize as signifying danger. In this sense, the NIPCs would be responding to PCV2 DNA as presenting a danger, a role for which they have evolved. However, rather than being stimulated by PCV2 DNA, the cells are rendered unresponsive to TLR9-mediated signalling.
Interestingly, the inhibition of NIPCs by PCV2 DNA was clearly not restricted to TLR9 agonists. Indeed, PCV2 interfered with the IFN-α response of NIPCs induced by a TLR7 ligand and several viruses – PRV, CSFV and TGEV – that differ fundamentally in their signalling. The result with PRV may relate to direct interference with the TLR9 pathway, because this virus is a member of the α-herpesviruses closely related to herpes simplex virus (HSV). Conversely, this does not explain the interference with CSFV and TGEV activation of NIPCs. Both are RNA viruses. Moreover, TGEV stimulates NIPCs via a viral glycoprotein-dependent, chloroquine-insensitive pathway independent of an involvement for nucleic acid.42 It is likely that TGEV stimulation of NIPCs occurs via a cell surface receptor. With CSFV, this is likely to be due to the dsRNA replicative intermediates generated during virus replication.33 This is in agreement with the demonstration that inhibitory CpG-ODN can prevent IFN-α responses induced by HIV-1, a virus proposed to trigger NIPCs through an RNA-dependent pathway.43
Our results suggest that both free viral DNA as well as virus-particle mediated delivery of DNA can inhibit NIPC function. In an in vivo situation, association of PCV2 DNA with the viral particle would protect the DNA until delivery into the cell. However, it has been reported that high levels of PCV2 DNA are found in the serum of infected animals.20
Proposals for the mechanism behind the immunomodulatory activity of PCV2 DNA would tend to favour interaction with TLR9, which is the only known endocytic DNA receptor. In addition, inhibitory CpG-ODN motifs binding to TLR9 have been described.44 However, the present study demonstrates that PCV2 DNA and CpG-ODNs did not show detectable colocalization in NIPCs. It was noted that the PCV2 DNA and CpG-ODNs were in clearly distinct intracytoplasmic compartments of NIPCs. The CpG-ODNs appeared to be more perinuclear, whereas the PCV2 DNA remained more apical or basolateral. This may reflect the use of different receptors by the two DNAs.
These results support the suggestion that PCV2 inhibition of NIPC capacity to produce IFN-α does not reflect a simple receptor competition with the stimulatory CpG-ODN binding to TLR9. The PCV2 DNA must either interact with a dominant inhibitory receptor or influence a downstream element of the signalling pathways initiated by various NIPC pattern recognition receptors. This proposal is further supported by the observation that induction of IFN-α by the TLR7 ligand R837 is inhibited by PCV2 DNA, whereas TNF-α and IL-6 induction by this same ligand is unaffected. It is known that several pathways of cytokine activation through TLR receptors use different downstream elements.4,45 Our results suggest that the pathway associated with TLR7 ligation-dependent IFN-α induction is inhibited by PCV2 DNA, whereas an alternative pathway for TLR7-associated TNF-α and IL-6 induction must be impervious to PCV2 DNA activity. In its entirety, the present work underlines the presence of a potent and dominant inhibitory pathway operative in NIPCs, and supports the suggestion that this pathway can be targeted by viruses to escape innate immune responses mediated by NIPCs. Considering the broad effect of PCV2 on various danger signals – ODNs and viruses from different families triggering NIPCs through DNA−, RNA− and glycoprotein−receptor interactions – this presents the immunomodulatory capacity of PCV2 as a major problem for innate defence recognition. Indeed, the important role played by NIPCs in antiviral innate immunity may indicate that viral inhibitory activity is a key event in the pathogenesis of PCV2 diseases. In this respect, it is important to note that PCV2 alone in pigs does not usually result in pronounced clinical disease, but when concomitant bacterial or other viral infections are present, disease can develop.16,22,23 Such in vivo relevance gains credence from our observation that in DNA form, non-pathogenic PCV1 does not mediate inhibition of NIPC responsiveness. It is also likely that a number of pathogenic viruses will display this capacity to interfere with NIPCs. Indeed, it is now known that measles and respiratory syncytial viruses can interfere with IFN-α production in NIPCs.11
Acknowledgments
This work was supported by the Swiss Federal Office for Education and Science (#99·0588) through an EU Framework 5 project (#QLK2-CT-1999-00445) and by the EU Framework 6 project PCVD (#513928). The authors thank Annette Mankertz (Robert Koch Institute, Berlin, Germany) for the PCV1 plasmid and Marco Alves (Institute of Virology and Immunoprophylaxis, IVI) for critical discussion. The authors also thank Brigitte Herrmann (IVI) for excellent technical assistance, Francis McNeilly (Department of Agriculture and Rural Development for Northern Ireland, Veterinary Sciences Division) for PCV2 stock and monoclonal antibody, Heidi Gerber (IVI) for confocal microscopy help and the animal handlers for taking care of the blood donor pigs and for routine bleeding.
Glossary
Abbreviations:
- CpG
cytosine-phosphorothioate-guanine
- CSFV
classical swine fever virus
- IFN
interferon
- IL
interleukin
- NIPC
natural interferon producing cell
- ODN
oligodeoxynucleotide
- PBMC
peripheral blood mononuclear cell
- PCVD
PCV2-associated diseases
- PCV2
porcine circovirus type 2
- pDC
plasmacytoid dendritic cell
- PMWS
post-weaning multisystemic wasting syndrome
- PRV
pseudorabies virus
- TGEV
transmissible gastroenteritis virus
- TLR
toll-like receptor
- TNF
tumour necrosis factor
References
- 1.Colonna MG, Trinchieri Liu Y-J. Plasmacytoid dendritic cells in immunity. Nat Immunol. 2004;5:1219–26. doi: 10.1038/ni1141. [DOI] [PubMed] [Google Scholar]
- 2.Liu Y-J. IPC: professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Ann Rev Immunol. 2005;23:275–306. doi: 10.1146/annurev.immunol.23.021704.115633. [DOI] [PubMed] [Google Scholar]
- 3.Janeway CA, Medzhitov R. Innate immune recognition. Ann Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 4.Takeda K, Kaisho T, Akira S. Toll-like receptors. Ann Rev Immunol. 2003;21:335–76. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 5.Guzylack-Piriou L, Balmelli C, McCullough KC, Summerfield AS. Type A CpG oligonucleotides activate exclusively porcine natural interferon-producing cells to secrete interferon-α, tumour necrosis factor-a and interleukin-12. Immunology. 2004;112:28–37. doi: 10.1111/j.1365-2567.2004.01856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Summerfield A, Guzylack-Piriou L, Schaub A, Carrasco CP, Tâche V, Charley B, McCullough KC. Porcine peripheral blood dendritic cells and natural interferon-producing cells. Immunology. 2003;110:440–9. doi: 10.1111/j.1365-2567.2003.01755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hemmi H, Takeuchi O, Kawai T, et al. A toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–5. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 8.Krug A, Luker GD, Barchet W, Leib DA, Akira S, Colonna M. Herpes simplex virus type 1 activates murine natural interferon-producing cells through toll-like receptor 9. Blood. 2004;103:1433–7. doi: 10.1182/blood-2003-08-2674. [DOI] [PubMed] [Google Scholar]
- 9.Lund J, Sato A, Akira S, Medzhitov R, Iwasaki A. Toll-like receptor 9-mediated recognition of herpes simplex virus-2 by plasmacytoid dendritic cells. J Exp Med. 2003;198:513–20. doi: 10.1084/jem.20030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haller O, Kochs G, Weber F. The interferon response circuit: induction and suppression by pathogenic viruses. Virology. 2006;344:119–30. doi: 10.1016/j.virol.2005.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schlender J, Hornung V, Finke S, et al. Inhibition of toll-like receptor 7- and 9-mediated α/β interferon production in human plasmacytoid dendritic cells by respiratory syncytial virus and measles virus. J Virol. 2005;79:5507–15. doi: 10.1128/JVI.79.9.5507-5515.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vincent IE, Carrasco CP, Guzylack-Piriou L, McCullough KC, Summerfield A. Subset-dependent modulation of dendritic cell activity by circovirus type 2. Immunology. 2005;115:388–98. doi: 10.1111/j.1365-2567.2005.02165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meehan BM, McNeilly F, Todd D, et al. Characterization of novel circovirus DNAs associated with wasting syndromes in pigs. J Gen Virol. 1998;79:2171–9. doi: 10.1099/0022-1317-79-9-2171. [DOI] [PubMed] [Google Scholar]
- 14.Allan GM, McNeilly F, Kennedy S, et al. Isolation of porcine circovirus-like viruses from pigs with a wasting disease in the USA and Europe. J Vet Diagn Invest. 1998;10:3–10. doi: 10.1177/104063879801000102. [DOI] [PubMed] [Google Scholar]
- 15.Clark EG. Pathology of the post-weaning multisystemic syndrome of pigs. Proc West Can Assoc Swine Pract. 1996;Abstracts:22–5. [Google Scholar]
- 16.Allan GM, McNeilly F, Kennedy S, Meehan BM, Ellis JA, Krakowa S. Immunostimulation, PCV2 and PMWS. Vet Rec. 2000;147:170–1. [PubMed] [Google Scholar]
- 17.Chae C. A review of porcine circovirus 2-associated syndrome and diseases. Vet J. 2005;169:326–36. doi: 10.1016/j.tvjl.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 18.Harding JC. The clinical expression and emergence of porcine circovirus type 2. Vet Microbiol. 2004;98:131–5. doi: 10.1016/j.vetmic.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 19.Resendes A, Segales J, Balasch M, et al. Lack of effect of a commercial vaccine adjuvant on the development of post-weaning multisystemic wasting syndrome (PMWS) in porcine circovirus type 2 (PCV2) experimentally infected conventional pigs. Vet Res. 2004;35:83–90. doi: 10.1051/vetres:2003039. [DOI] [PubMed] [Google Scholar]
- 20.Segales J, Calsamiglia M, Olvera A, Sibila M, Badiella L, Domingo M. Quantification of porcine circovirus (PCV2) DNA in serum and tonsillar, nasal, tracheo-bronchial, urinary and faecal swabs of pigs with and without post-weaning multisystemic wasting syndrome (PMWS) Vet Microbiol. 2005;111:223–9. doi: 10.1016/j.vetmic.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 21.Allan GM, Ellis JA. Porcine circoviruses: a review. J Vet Diagn Invest. 2000;12:3–14. doi: 10.1177/104063870001200102. [DOI] [PubMed] [Google Scholar]
- 22.Kim J, Chung HK, Chae C. Association of porcine circovirus 2 with porcine respiratory disease complex. Vet J. 2003;166:251–56. doi: 10.1016/s1090-0233(02)00257-5. [DOI] [PubMed] [Google Scholar]
- 23.Rovira A, Balasch M, Segales J, et al. Experimental inoculation of conventional pigs with porcine reproductive syndrome virus and porcine circovirus type 2. J Virol. 2002;76:3232–9. doi: 10.1128/JVI.76.7.3232-3239.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wellenberg GJ, Stockhofe-Zurwieden N, Boersma WJ, De Jong MF, Elbers AR. The presence of co-infection in pigs with cervical signs of PMWS in the Netherlands: a case-control study. Res Vet Sci. 2004;77:177–84. doi: 10.1016/j.rvsc.2004.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nielsen J, Vincent IE, Botner A, Ladekjaer-Mikkelsen A-S, Allan G, Summerfield A, McCullough KC. Association of lymphopenia with the porcine circovirus type 2 induced post-weaning multisystemic wasting syndrome. Vet Immunol Immunopathol. 2003;92:97–111. doi: 10.1016/s0165-2427(03)00031-x. [DOI] [PubMed] [Google Scholar]
- 26.Vincent IE, Carrasco CP, Hermann B, Meehan BM, Allan GM, Summerfield A, McCullough KC. Dendritic cells harbour infectious PCV2 in the absence of apparent cell modulation or replication of the virus. J Virol. 2003;77:13288–300. doi: 10.1128/JVI.77.24.13288-13300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allan GM, McNeilly F, Foster JC, Adair B. Infection of leucocyte cell cultures derived from different species with pig circovirus. Vet Microbiol. 1994;41:267–79. doi: 10.1016/0378-1135(94)90107-4. [DOI] [PubMed] [Google Scholar]
- 28.McNeilly F, McNair L, Mackie DP, et al. Production, characterization and applications of monoclonal antibodies to porcine circovirus type 2. Arch Virol. 2001;146:909–22. doi: 10.1007/s007050170124. [DOI] [PubMed] [Google Scholar]
- 29.Knoetig SM, Summerfield A, Spagnuolo-Weaver M, McCullough KC. Immunopathogenesis of classical swine fever: role of monocytic cells. Immunology. 1999;97:359–66. doi: 10.1046/j.1365-2567.1999.00775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Summerfield AS, Rziha HJ. Functional characterization of porcine CD4+ and CD8+ extrathymic T lymphocytes. Cell Immunol. 1996;168:291–6. doi: 10.1006/cimm.1996.0078. [DOI] [PubMed] [Google Scholar]
- 31.Charley B, Lavenant L, Lefevre F. Coronavirus transmissible gastroenteritis virus mediated induction of IFN-a-mRNA in porcine leucocytes requires prior synthesis of soluble proteins. Vet Res. 1994;25:29–36. [PubMed] [Google Scholar]
- 32.McCullough KC, Schaffner R, Fraefel W, Kihm U. The relative density of CD44-positive porcine monocytic cell populations varies between isolations and upon culture and influences susceptibility to infection by African swine fever virus. Immunol Lett. 1993;37:83–9. doi: 10.1016/0165-2478(93)90136-p. [DOI] [PubMed] [Google Scholar]
- 33.Balmelli C, Vincent IE, Rau H, Guzylack-Piriou L, McCullough KC, Summerfield A. FcgRII-dependent sensitization of natural interferon-producing cells for viral infection and interferon-α responses. Eur J Immunol. 2005;35:2406–15. doi: 10.1002/eji.200525998. [DOI] [PubMed] [Google Scholar]
- 34.Baudoux P, Carrat C, Besnardeau L, Charley B, Laude H. Coronavirus pseudoparticles formed with recombinant M and E proteins induce α interferon synthesis by leucocytes. J Virol. 1998;72:8636–43. doi: 10.1128/jvi.72.11.8636-8643.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malmgaard L, Melchjorsen J, Bowie AG, Mogensen SC, Paludan SR. Viral activation of macrophages through TLR-dependent and independent pathways. J Immunol. 2004;173:6890–8. doi: 10.4049/jimmunol.173.11.6890. [DOI] [PubMed] [Google Scholar]
- 36.Allan GM, McNeilly F, Cassidy JP, Reilly GA, Adair B, Ellis WA, McNulty MS. Pathogenesis of porcine circovirus: experimental infections of colostrum deprived piglets and examination of pig foetal material. Vet Microbiol. 1995;44:49–64. doi: 10.1016/0378-1135(94)00136-k. [DOI] [PubMed] [Google Scholar]
- 37.Tischer I, Mields W, Wolff D, Vagt M, Griem W. Studies on epidemiology and pathogenecity of porcine circovirus. Arch Virol. 1986;91:271–6. doi: 10.1007/BF01314286. [DOI] [PubMed] [Google Scholar]
- 38.Fitzgerald-Bocarsky P. Natural interferon-α producing cells: the plasmacytoid dendritic cells. Bio Techniques. 2002;33(Suppl):16–29. [PubMed] [Google Scholar]
- 39.Yamada H, Gursel I, Takeshita F, Conover J, Ishii KJ, Gursel M, Takeshita S, Klinman DM. Effect of suppressive DNA on CpG-induced immune activation. J Immunol. 2002;169:5590–4. doi: 10.4049/jimmunol.169.10.5590. [DOI] [PubMed] [Google Scholar]
- 40.Krieg AM, Wu T, Weeratnar R, et al. Sequence motifs in adenoviral DNA block immune activation by stimulatory CpG motifs. PNAS. 1998;95:12631–6. doi: 10.1073/pnas.95.21.12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hasslung FC, Berg M, Allan GM, Meehan BM, McNeilly F, Fossum C. Identification of a sequence from the genome of porcine circovirus type 2 with an inhibitory effect on IFN-α production by porcine PBMCs. J Gen Virol. 2003;84:2937–45. doi: 10.1099/vir.0.19362-0. [DOI] [PubMed] [Google Scholar]
- 42.Charley B, Levenant L, Delmas B. Glycosylation is required for coronavirus TGEV to induce an efficient production of IFN-α by blood mononuclear cells. Scand J Immunol. 1991;33:435–40. doi: 10.1111/j.1365-3083.1991.tb01792.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beignon AS, McKenna K, Skoberne M, et al. Endocytosis of HIV-1 activates plasmacytoid dendritic cells via toll-like receptor–viral RNA interactions. J Clin Invest. 2005;115:3265–75. doi: 10.1172/JCI26032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Latz E, Schoenemeyer A, Visintin A, et al. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat Immunol. 2004;5:190–8. doi: 10.1038/ni1028. [DOI] [PubMed] [Google Scholar]
- 45.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]