Abstract
The genetic pathways that control development of the early mammalian embryo have remained poorly understood, in part because the systematic mutant screens that have been so successful in the identification of genes and pathways that direct embryonic development in Drosophila, Caenorhabditis elegans, and zebrafish have not been applied to mammalian embryogenesis. Here we demonstrate that chemical mutagenesis with ethylnitrosourea can be combined with the resources of mouse genomics to identify new genes that are essential for mammalian embryogenesis. A pilot screen for abnormal morphological phenotypes of midgestation embryos identified five mutant lines; the phenotypes of four of the lines are caused by recessive traits that map to single regions of the genome. Three mutant lines display defects in neural tube closure: one is caused by an allele of the open brain (opb) locus, one defines a previously unknown locus, and one has a complex genetic basis. Two mutations produce novel early phenotypes and map to regions of the genome not previously implicated in embryonic patterning.
Many of the mutations that affect development of the mouse embryo have been generated by targeted disruption of previously cloned genes (1). These mutations have defined a number of important processes in mammalian embryogenesis; however, it has been estimated that targeted mutations currently sample fewer than 1% of the genes in the mouse genome (2). To gain a deeper understanding of the events that program mammalian embryogenesis, it will be necessary to identify and characterize the function of additional genes required for mouse development.
Recent advances in positional cloning have made the molecular identification of mammalian genes defined by point mutations practical, as demonstrated by the positional cloning of genes defined by ethylnitrosourea (ENU)-induced mutations including the circadian rhythm gene Clock (3, 4) and the developmental regulatory gene eed (5). The unique strength of ENU mutagenesis is its great efficiency: the most efficient ENU dosage regime gave an average of one new mutation per gene in 700 F1 progeny in the specific locus test (6). This high rate of mutagenesis should make it possible to identify that small fraction of genes in the genome that affects a particular process of interest. For example, in 100 ENU-treated lines, 5,000–10,000 different new mutations can be screened (6, 7). If the desired phenotypes can be identified efficiently, ENU mutagenesis will provide the means to identify genes that play essential roles in the process to be studied.
ENU mutagenesis has been used successfully to identify recessive lethal mutations in defined regions of the mouse genome (8–11). Recent progress in the mouse genome project (12) should make it practical to screen the entire genome for recessive mutations that affect any process of interest because the segregation of any new mutation can be followed on the basis of linkage to polymorphic DNA markers. We therefore carried out a pilot screen to test the feasibility of identifying recessive mutations that affect the morphology of early mouse embryos and mapped the mutations with respect to polymorphic DNA markers. The pilot screen showed that phenotype-based screens are a practical way to identify genes important in early mouse development.
MATERIALS AND METHODS
Animals and Mutagenesis.
C57BL/6J, C3H/HeJ, CBA/CaJ, and C57BL/6J-Twisttm1Bhr mice were obtained from The Jackson Laboratory; opb heterozygous animals were a gift from Marilyn Fisher (University of Virginia). ENU mutagenesis was performed essentially as described (6). ENU, obtained from Sigma (N-3385) in Isopac bottles containing approximately 1 g of ENU, was dissolved in 10 ml of 95% ethanol. After 1 hr, 90 ml of phosphate–citrate buffer (0.1 M Na2HPO4/0.05 M citrate, pH 5.0) was added and the resulting 10 mg/ml ENU solution promptly was injected i.p. into male mice 9–10 weeks old at the time of the first injection. C57BL/6J males received ENU in either a single dose of 150 mg/kg body weight, or in three doses of 100 mg/kg body weight administered once a week for 3 weeks. Only C57BL/6J males were used to establish lines. Ten percent saturated ceric ammonium nitrate (Sigma) was used to inactivate ENU. For most lines, C3H/HeJ females were mated with mutagenized males; in a small number of lines the females were CBA/CaJ. Fertility was recovered starting approximately 7 weeks after treatment for both dose regimens. For the mice receiving the 3 × 100 mg/kg dose, 8 of 20 C57BL6/J males, 3 of 5 B6CBAF1 males, and 0 of 10 CBA/CaJ males treated recovered fertility. Eight of 10 C57BL6/J males receiving the 150 mg/kg dose recovered fertility. C57BL/6J and C3H/HeJ strains were chosen because both had been typed for the MIT microsatellite markers (12), and crosses between these strains gave reasonably large litters. For all lines except line 5, the mother of the founder male was C3H/HeJ; for line 5, the mother of the founder was CBA/CaJ. No more than 10 F1 males from each mutagenized male were used to establish lines to minimize the probability recovering multiple mutations that were identical by descent (6).
Screening and Recovery of Mutations.
F1 sons of mutagenized mice were crossed as shown in Fig. 1 to produce lines of mice for screening. Fifty-three lines were derived from the single, 150-mg/kg injection protocol and 77 were derived from the triple, 100-mg/kg injection protocol. Each F1 male was mated to 4 C3H/HeJ females, and only the 86 lines in which 5 or more second-generation (G2) females were obtained were screened. Five to 10 G2 females per line were mated; there were an average of 8.15 embryos per pregnant G2 female. Lines with two or more litters containing embryos with similar abnormal morphology were considered potentially mutant. G2 males and females from these lines were produced and intercrossed, and litters were screened at 9.5 days post coitum (dpc) to confirm the heritability of the phenotype and to identify G2 males that carried the mutation. Lines were maintained initially by outcrossing the carrier males to C3H/HeJ, intercrossing the resulting progeny, and examining embryos to identify new male carriers. After the mutation responsible for the phenotype was mapped, carriers of both sexes were identified by PCR as those carrying the C57BL/6J alleles of markers flanking the induced mutation. Line 25 and wsnp were derived from the triple, 100-mg/kg dose and lines 105 (opm), 118 (opb2), and bnb were from the single, 150-mg/kg dose. Thus, although the sample size here was small, the data suggest that the single, 150-mg/kg dose was approximately as mutagenic as the 3 × 100-mg/kg regime.
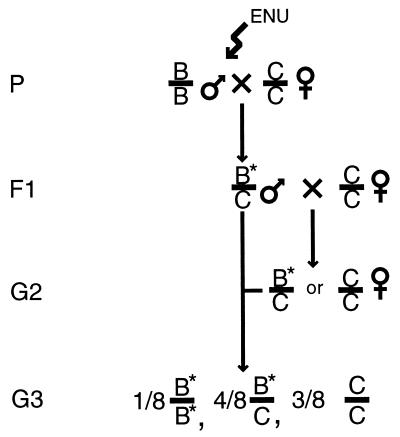
Figure 1.
Screen design. C57BL/6J male mice were mutagenized and mated to C3H/HeJ females to produce F1 male mice carrying induced mutations associated with C57BL/6J DNA. C57BL/6J chromosomes, C3H/HeJ chromosomes, and induced mutations are indicated as B, C, and ∗, respectively. F1 males were crossed to C3H/HeJ females to produce G2 mice, half of which were heterozygous for any particular induced mutation. Female G2 mice were mated to their father and sacrificed at 9.5 dpc to examine the phenotype of G3 embryos. If a mutation of interest was induced in a line, half of the litters should have contained one-quarter homozygous mutant embryos. Linkage of the induced mutation to C57BL/6J alleles of microsatellite markers was used for mapping.
Mapping.
Mutations were mapped by using liver or tail DNA from at least 20 adult mice presumed to be heterozygous for the mutation because they were the parents of mutant embryos. The markers listed below were used in an initial genome scan at 40-centimorgan (cM) intervals; if no clear linkage was observed, additional markers spaced at 20 cM intervals were tested. Linkage was confirmed with additional carrier animals and mutant embryos, and the region including the gene was defined by using more closely spaced markers. In all cases, linkage was determined by computing pairwise LOD (logarithm of odds) scores, with a LOD of 5 indicating confirmed linkage. The primer pairs chosen for mapping give PCR products 100–250 bp long, and the C57BL/6J and C3H/HeJ alleles can be distinguished reliably on ethidium-stained agarose gels because they differ in size by at least 10 bp. The markers used in the initial 40-cM scan were: D1Mit213, D1Mit191, D1Mit166, D2Mit249, D2Mit226, D3Mit151, D3Mit106, D4Mit108, D4Mit187, D5Mit148, D5Mit188, D6Mit74, D6Mit366, D7Mit82, D7Mit332, D8Mit294, D8Mit156, D9Mit130, D9Mit12, D10Mit3, D10Mit180, D11Mit349, D11Mit61, D12Mit147, D12Mit263, D13Mit179, D13Mit151, D14Mit149, D14Mit166, D15Mit209, D15Mit161, D16Mit13, D16Mit152, D17Mit34, D17Mit187, D18Mit70, D18Mit153, D19Mit61, and D19Mit137. Markers for testing suggested linkage were selected from the following: D1Mit3, D1Mit181, D1Mit227, D1Mit362, D2Mit1, D2Mit237, D2Mit307, D2Mit229, D3Mit164, D3Mit137, D3Mit19, D4Mit235, D4Mit297, D4Mit226, D5Mit346, D5Mit205, D5Mit292, D6Mit83, D6Mit213, D6Mit201, D7Mit191, D7Mit238, D8Mit124, D8Mit113, D9Mit89, D9Mit262, D10Mit189, D10Mit42, D11Mit2, D11Mit41, D12Mit37, D12Mit214, D13Mit57, D13Mit159, D14Mit99, D14Mit30, D15Mit175, D15Mit42, D16Mit154, D17Mit238, D18Mit124, and D19Mit1.
Mapping of the exencephalic phenotype seen in line 105 was complex, apparently because more than one mutation that promoted exencephaly was present in the founder male. After six generations of outcrossing and selecting for the phenotype, it became clear that a segment of the proximal region of the C57BL/6J chromosome 12 had been selected along with the phenotype, even though only 18/37 exencephalic embryos genotyped were homozygous for that segment of chromosome 12. Test crosses confirmed that homozygosity for proximal chromosome 12 was sufficient to produce exencephaly with an approximately 88% penetrance (see Table 1); the chromosome 12 locus was named open mind (opm). The additional loci promoting exencephaly in this line have not yet been mapped.
Table 1.
Mutants isolated
| Mutant | Mutant/total embryos, % | Map position
|
% Penetrance (homozygotes, n) | ||
|---|---|---|---|---|---|
| Chromosome | Closet marker (cM), no. nonrecombinants | Interval, cM | |||
| 25 | 50/292, 17.1 | — | — | — | — |
| 105, opm | 84/364, 23.1 | 12 | D12Mit69 (28.0), 68* | 6.0–28.0 | 88 (8) |
| 118, opb2 | 39/140, 27.9 | 1 | D1Mit318 (18.5), 82 | 17.0–20.2 | 94 (18) |
| bnb | 89/361, 24.7 | 6 | D6Mit159 (7.0), 61 | 3.0–15.5 | 100 (8) |
| wsnp | 126/509, 24.8 | 16 | D16Mit59 (27.8), 39 | 21.5–32.0 | 100 (5) |
The second column shows the total number of phenotypically mutant embryos per total embryos in mutant-containing litters observed. Closest marker is the marker that showed no recombination with the mutation (except for 105, see*) [Mouse Genome Database (MGD) map position]. No. nonrecombinants, number of independent meiotic products in which the chromosomes carried both the mutation and the closest marker, including data from both heterozygous adult carriers and homozygous mutant embryos. Interval includes the gene responsible for the phenotype, defined by flanking markers that recombine with the mutation (MGD map position). The last column shows direct measurement of penetrance. For each line, all the embryos from several litters were typed and embryos that were not recombinant between flanking markers were scored. Penetrance was defined as the number of homozygous embryos that showed the mutant phenotype out of the total number of homozygous embryos in these litters. The high penetrance of opb2, bnb, and wsnp measured directly is consistent with the 25% mutant embryos seen in mutant litters in these lines (column 2). For opb2, bnb, and wsnp, only embryos that were homozygous for the region showed the mutant phenotype. The opm mutation is not completely penetrant: 7/8 embryos homozygous for the region of chromosomes 12 were exencephalic. In addition, some exencephalic embryos in line 105 litters were not homozygous for opm (see *).
There are exencephalic embryos in line 105 that are not homozygous for the chromosome 12 opm mutation, apparently because of other mutations induced in the line. We observed that this marker was linked to the opm mutation in 68/88 independent meiotic products genotyped; some of the cases where linkage was not observed are presumably recombinants, whereas others represent embryos that were presumably exencephalic because of mutations at other genomic loci.
DNA Preparation and PCR.
DNA was isolated as described from liver, tail, and fixed embryonic tissues (13). PCR was carried out in 10-μl volumes with 50 ng genomic DNA template in 1× PCR Buffer II (Perkin–Elmer) and 1.5 mM MgCl2. Taq Gold polymerase (Perkin–Elmer) was used in the following PCR: 94°C for 12 min, then either 55 or 75 cycles of 94°C for 20 s, 55°C for 30 s, and 72°C for 45 s, followed by 7 min at 72°C. Reactions were run on an MJ Research PTC-200 thermal cycler (Cambridge, MA), analyzed on 3.5% agarose gels [2.5% Ultrapure (GIBCO) + 1% Nusieve GTG (FMC)], and stained with ethidium bromide. After initial linkage was established, DNA from 10.5-dpc embryonic yolk sacs and adult ear punches also was used; these DNAs were digested overnight at 55°C with 290 μg/ml Proteinase K (Boehringer Mannheim) in 100 μl of 1× PCR Buffer II, heated to 99°C for 10 min, vortexed, cooled, and added to PCRs at a 1:20 dilution.
Histology and Staining.
Embryos were fixed in 4% paraformaldehyde in PBS for in situ hybridization and in 4% paraformaldehyde in PBS, 10% formalin, or Bouin’s fixative for paraffin sectioning at 8 μm and hematoxylin/eosin staining. For staining of endogenous alkaline phosphatase activity (14), embryos were fixed in 4% paraformaldehyde and stained with NBT (4-nitroblue tetrazolium chloride) and BCIP (5-bromo-4-chloro-3-indolyl phosphate) (Boehringer Mannheim) according to the manufacturer’s instructions. Fixation, histology, and in situ hybridization were performed according to standard protocols (13). PECAM-1 (15) staining followed standard protocols (12), but with fixation overnight in 4% paraformaldehyde and detection with diaminobenzidine and peroxidase-conjugated goat anti-rat IgG (H+L) (Jackson ImmunoResearch, 112–035-003).
RESULTS
Identification of Recessive Mutations that Affect the Morphology of the Midgestation Mouse Embryo.
We modified mutagenesis protocols and breeding schemes that have been used to identify lethal mutations in localized regions of the mouse genome (9–11) to screen the entire autosomal genome for recessive mutations that specifically disrupt the organization of the midgestation embryo. C57BL/6J male mice were treated with the point mutagen ENU (6) and crossed to C3H/HeJ females. Two additional generations of crosses were carried out such that approximately 1/8 of the third generation (G3) embryos produced by each line would be homozygous for any newly induced mutation on any autosome (Materials and Methods; Fig. 1). G3 embryos were examined at 9.5 dpc, because at this stage embryos that have defects in early postimplantation development can be identified in the dissecting microscope by abnormal morphology, but would not have been resorbed completely. Mutants were identified at high frequency: from 86 lines screened, 5 lines were found in which approximately 25% of the embryos in mutant-containing litters showed a characteristic set of defects, and the trait was inherited for at least four more generations.
The mutations responsible for the embryonic phenotypes were mapped with respect to simple sequence-length polymorphisms between the mutagenized C57BL/6J strain and the C3H/HeJ strain used in all outcrosses (12) to identify those regions of the C57BL/6J genome linked to the mutation (Materials and Methods). Four mutations segregated as simple recessive traits and mapped to single loci (Table 1), whereas the phenotype of one line (25) could not be mapped to a single locus and therefore appeared to have a complex genetic basis. The genotypes of individual embryos showed that those embryos homozygous for the region of interest showed the mutant phenotype with high penetrance (Table 1).
Mutants in three lines (25, 105, and 118) displayed exencephaly, the failure to close the cranial neural tube (Figs. 2 and 3). The high frequency of mutations causing neural tube defects in this sample suggests that a large number of genes are required for normal closure of the neural tube, which is consistent with previous fortuitous identification of mutations in almost two dozen mouse genes that cause neural tube defects (ref. 16; http://www.jax.org/resources/documents/NeuroNews.html). Mutants in two other lines, which we named bent body (bnb) and wing-shaped neural plate (wsnp), had early global defects in many tissues (see below; Figs. 4 and 5).
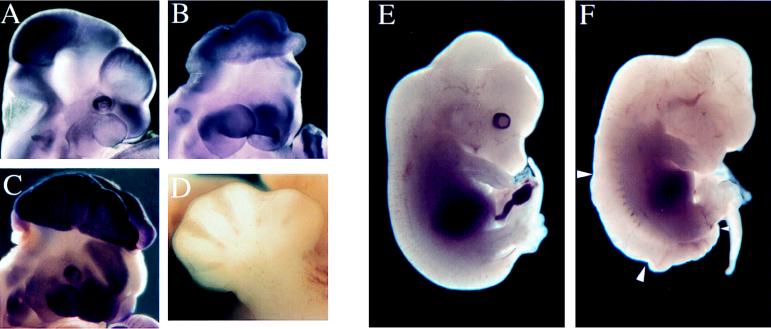
Figure 2.
Exencephalic mutants. (A–C) Embryos stained for endogenous alkaline phosphatase activity. (A) Head of 10.5-dpc wild-type embryo. (B) Exencephalic 10.5-dpc 118 (opb2) embryo lacking visible eyes. (C) Exencephalic 11.5-dpc line 25 embryo. (D) Most line 118 embryos died between 12.5 and 14.5 dpc, but those that could be scored had an extra anterior digit, shown here on the left forelimb of a 13.5-dpc 118 embryo. (E and F) Littermates (13.5 day) from a 118/+ X opb/+ cross. (E) Embryo with wild-type phenotype and genotype. (F) Genotypically 118/opb embryo: normal eyes were not present, the dorsally exposed neural tube between the large arrowheads was like that of opb homozygous embryos (18), and the more posterior neural tube had not closed (between the large and small arrowheads). The genotype of these embryos was determined based on the alleles of flanking DNA polymorphisms present in yolk sac DNA. A similar recurved tail was seen in some 118 homozygous embryos.
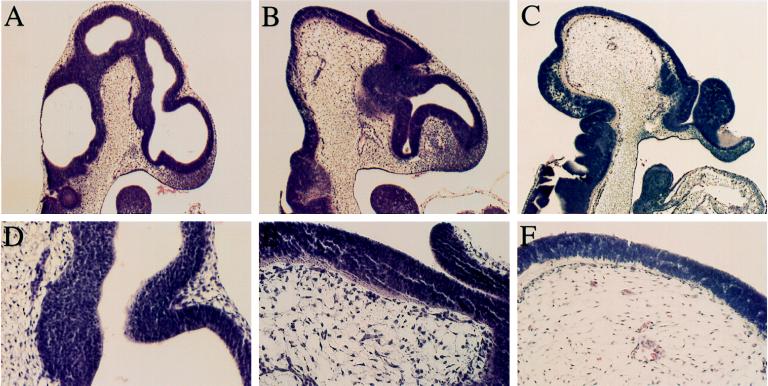
Figure 3.
Histology of exencephalic mutants. Sagittal sections of wild-type (A and D), 118 (opb2) (B and E), and opm (homozygous for the C57BL/6J region of proximal chromosome 12 of line 105) (C and F) 10.5-dpc embryos. The cranial mesenchyme cells of opm appeared small, spindle-shaped, and widely spaced.
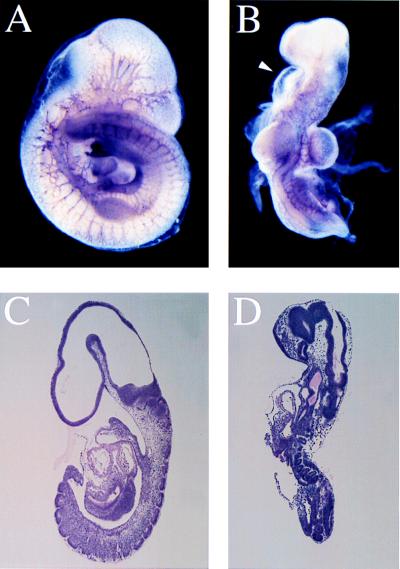
Figure 4.
The bent body (bnb) phenotype. Staining for the PECAM-1/CD31 antigen in wild-type (A) and bnb (B) littermates to visualize the developing vascular system. Note the relatively large forelimb buds of the mutant. Arrowhead points out the pericardial edema. Sagittal sections of 9.5-dpc wild-type (C) and bnb (D) littermates. The small, immature somites, pericardial edema, and twisted body axis of the mutant can be seen.
Figure 5.
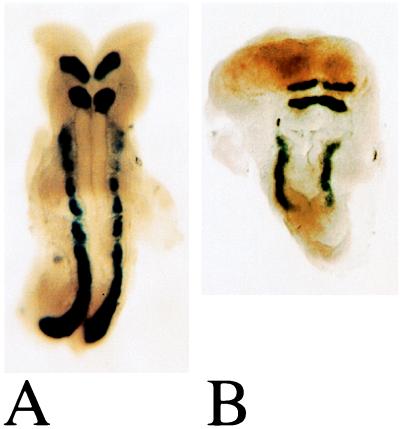
The wing-shaped neural plate (wsnp) phenotype. Double in situ hybridization showing the localization of Krox20 and Mox1 RNAs in wild-type (A) and line wsnp (B) 8.5-dpc littermates. The position of the two hindbrain Krox20 stripes suggests that anterior patterning was relatively normal in the mutant and that the body axis of the mutant was truncated posteriorly. In wsnp, Mox1 was expressed in the paraxial mesoderm in shortened, unsegmented stripes.
Mutations Causing Neural Tube Defects.
In addition to exencephaly (Figs. 2B and 3 B and E), mutant embryos from line 118 showed a syndrome of defects in other tissues, including failure to close the posterior neural tube, absence or abnormalities of one or both eyes, and a supernumerary anterior digit on all four limbs (Fig. 2 B and D). This syndrome is similar to that produced by the spontaneous mutation open brain (opb) (17). The mutation responsible for the 118 phenotype maps to the same 3.2-cM interval as does opb (ref. 18; Table 1). We crossed 118 and opb heterozygous animals and found that the two mutations failed to complement (Fig. 2F); therefore, we designate the line 118 mutation opb2. The exencephaly in opb embryos is associated with ventralization of the pattern of cell types in the neural tube (17); the other opb phenotypes also appear to reflect patterning defects. The identification of this additional mutant allele of opb will aid in the molecular identification of this gene and characterization of its role in embryonic patterning.
Previous studies have shown that one cause of exencephaly is abnormal cranial mesenchyme (19, 20). In line 105 mutant embryos, cells in the mesenchyme underlying the exencephalic neuroepithelium appeared small and widely spaced (Fig. 3 C and F). Defects in cranial mesenchyme also are seen in embryos homozygous for a targeted mutation in Twist, which maps to the same 20-cM region of chromosome 12 that includes the line 105 mutation (21). However, in complementation tests, 10.5- to 11.5-dpc embryos carrying the 105 mutation in trans to the targeted Twist allele did not show exencephaly or any other morphological defect (n = 7). The mutation in line 105 therefore defines a new locus on chromosome 12 required for normal cranial mesenchyme and neural tube closure, which we named open mind (opm).
Mutations Causing Early Defects.
At 9.5 dpc, bent body embryos were small and failed to complete embryonic turning, but had closed neural tubes, beating hearts, and chorioallantoic connections (Fig. 4). The bnb mutants had pericardial edema, hemorrhages in mesenchymal regions of tail, limb buds and head, and disproportionately large forelimb buds. Although the pericardial edema and hemorrhages seen in bnb embryos could be the result of vascular defects, the vasculature of the mutants appeared to initiate normally, as visualized by staining for the PECAM-1/CD31 antigen (15) (Fig. 4 A and B). The small size, failure to turn, pericardial edema, and peripheral hemorrhages of bnb also are observed in no turning (nt) mutants (22). Like nt embryos, bnb embryos have defects in laterality: although all bnb embryos failed to complete turning, in a sample of 25 mutant embryos, 13 had begun to turn to the right (the normal direction) and 12 had begun to turn to the left. Cardiac looping in bnb was abnormal, but in the majority of embryos no clear looping to the left or right was seen, in contrast to nt, where heart looping was reversed in 50% of the mutant embryos (22). The nt gene has not been mapped, so it is possible that bnb and nt are allelic. However, the neural tubes are closed in bnb and open in some nt embryos, and the heart-looping abnormalities appear to be different in the two mutants, suggesting that nt and bnb are mutations in different genes that affect common processes. No genes mapped to the region of chromosome 6 that includes bnb have been implicated previously in embryonic development.
The wing-shaped neural plate (wsnp) mutant embryos had striking abnormalities in both neural and mesodermal tissues by 8.5 dpc. At that stage, wsnp embryos were small, had a completely open neural plate, and lacked visible somites (Fig. 5). Within the neural plate, some normal anterior–posterior patterning had taken place: Krox20 was expressed in wsnp embryos in two hindbrain stripes similar to those of wild-type siblings (23). In the paraxial mesoderm of wsnp embryos, Mox1 (24) was expressed in shortened stripes and was not segmented into somites. This reduction in paraxial mesoderm and failure of somite formation is similar to that seen in embryos homozygous for a targeted mutation in fibronectin (25, 26). However, embryos that lack fibronectin form a neural tube, suggesting that wsnp has a more global role in morphogenesis. No genes in the region of chromosome 16 where wsnp maps have been implicated previously in embryonic morphogenesis.
DISCUSSION
The results presented here demonstrate that chemical mutagenesis and direct phenotypic screening are efficient means of isolating new alleles of known genes and defining new genes and their functions in postimplantation mammalian development. This approach could have failed to identify new mutations of interest if too few embryos with a reliable phenotype were obtained in individual lines, if phenotypes were caused by mutations in multiple different loci, or if the class of phenotypes sought was too rare to be seen without screening large numbers of lines. None of these potential pitfalls was encountered. The ENU treatment did not decrease litter size significantly, so it was possible to obtain enough embryos to identify phenotypes from a modest number of matings. The phenotypes obtained were constant enough between embryos in a line to make it possible to detect mutations in the relatively small number of embryos examined. Despite the large number of mutations per genome induced by ENU, most of the phenotypes identified proved to be caused by mutations that mapped to single loci. Clear morphological abnormalities at midgestation were obtained in more than 1 of 20 lines in this experiment, and mutant phenotypes were seen in more than 1 of 10 lines in more recent screenings (data not shown), so it is possible to identify mutations affecting early development in a modest-scale screen.
Although many common molecular processes undoubtedly underlie the development of all vertebrate embryos, the morphological phenotypes of the mutants identified here do not correspond in any simple way to those of zebrafish embryonic lethal mutations (27, 28). Some differences in phenotypes reflect underlying differences in the biology of mouse and zebrafish embryogenesis; for example, the zebrafish neural tube forms by cavitation of a rod of cells (29) rather than by rolling of the neural plate into a tube. Other differences in phenotypes may reflect the different maternal and zygotic contributions to development: early development in the zebrafish relies on stored maternal components, whereas the mouse embryo depends on the zygotic genome for early growth, patterning, and establishment of connections to maternal sources of nutrition.
Based on the efficiency of ENU mutagenesis in the mouse (6) and the results of our experiments, mutations in the majority of the genes that can mutate to give clear phenotypes in the midgestation embryo should be detected in a screen of only 1,000 lines. Such a screen should identify a few hundred mutants and define the constellation of pathways that control early postimplantation development of the mouse, as well as identify sets of genes that control specific developmental processes. The map positions of the mutations will identify candidate genes to test for allelism or define new genes that play novel roles in early embryogenesis. Establishment of congenic lines using methods that make it possible to produce more than eight generations per year (30, 31) will facilitate rapid mapping of mutations to single loci. Information from the mouse and human genome projects, which has greatly simplified positional cloning, together with phenotype-based mutant screens, should provide an important dimension in the identification and characterization of regulatory pathways that control mammalian embryogenesis.
Acknowledgments
We extend special thanks to Rosa Beddington and the members of her lab for hosting K.V.A. while on sabbatical and for continued discussions and encouragement. We thank Klaus Schughart, Ralf Spoerle, and Marilyn Fisher for making the opb mice available to us and for providing helpful information, and Tim Bestor, Jonathan Eggenschwiler, and Lee Niswander for helpful comments on the manuscript. We thank David Kingsley, Monica Justice, and Brigid Hogan for helpful discussions, Ed Espinoza, Pearl Chang, and Jonathan Eggenschwiler for help with PCR, Ed Espinoza for help with the figures, and Brian Lau and Jennifer Bryan for help with the mouse maintenance. The Krox20 clone was a gift from Richard Behringer, and the Mox1 clone was a gift from Chris Wright. This work was supported by grants from March of Dimes (97–0575) and the National Institutes of Health (R55 HD/OD35455) to K.V.A. and by the Memorial Sloan–Kettering Cancer Center Support Grant; preliminary studies were supported by the Hawn fund of the University of California. A.K. was a National Science Foundation Predoctoral Fellow.
ABBREVIATIONS
- G2
second generation
- dpc
days post coitum
- ENU
ethylnitrosourea
- cM
centimorgan
References
- 1.Tam P P L, Behringer R R. Mech Dev. 1997;68:3–25. doi: 10.1016/s0925-4773(97)00123-8. [DOI] [PubMed] [Google Scholar]
- 2.Evans M J, Carlton M B L, Russ A P. Trends Genet. 1997;13:370–374. doi: 10.1016/s0168-9525(97)01240-7. [DOI] [PubMed] [Google Scholar]
- 3.King D P, Zhao Y, Sangoram A M, Wilsbacher L D, Tanaka M, Antoch M P, Steeves T D, Vitaterna M H, Kornhauser J M, Lowrey P L, et al. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antoch M P, Song E J, Chang A M, Vitaterna M H, Zhao Y, Wilsbacher L D, Sangoram A M, King D P, Pinto L H, Takahashi J S. Cell. 1997;89:655–667. doi: 10.1016/s0092-8674(00)80246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schumacher A, Faust C, Magnuson T. Nature (London) 1996;383:250–253. doi: 10.1038/383250a0. [DOI] [PubMed] [Google Scholar]
- 6.Hitotsumachi S, Carpenter D A, Russell W L. Proc Natl Acad Sci USA. 1985;82:6619–6621. doi: 10.1073/pnas.82.19.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silver L M. Mouse Genetics: Concepts and Applications. New York: Oxford Univ. Press; 1995. [Google Scholar]
- 8.Davis A P, Justice M J. Genetics. 1998;148:7–12. doi: 10.1093/genetics/148.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bode V C. Genetics. 1984;108:457–470. doi: 10.1093/genetics/108.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shedlovsky A, Guénet J-L, Johnson L L, Dove W F. Genet Res. 1986;47:135–142. doi: 10.1017/s0016672300022977. [DOI] [PubMed] [Google Scholar]
- 11.Rinchik E M, Carpenter D A. Mamm Genome. 1993;4:349–353. doi: 10.1007/BF00360583. [DOI] [PubMed] [Google Scholar]
- 12.Dietrich W F, Miller J, Steen R, Merchant M A, Damron-Boles D, Husain Z, Dredge R, Daly M J, Ingalls K A, O’Connor T J, et al. Nature (London) 1996;380:149–152. doi: 10.1038/380149a0. [DOI] [PubMed] [Google Scholar]
- 13.Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 14.Kwong W H, Tam P P L. J Embryol Exp Morphol. 1984;82:241–251. [PubMed] [Google Scholar]
- 15.Baldwin H S, Shen H M, Yan H C, DeLisser H M, Chung A, Mickanin C, Trask T, Kirschbaum N E, Newman P J, Albelda S M, et al. Development. 1994;120:2539–2953. doi: 10.1242/dev.120.9.2539. [DOI] [PubMed] [Google Scholar]
- 16.Copp A J. CIBA Found Symp. 1994;181:118–143. doi: 10.1002/9780470514559.ch8. [DOI] [PubMed] [Google Scholar]
- 17.Günther T, Struwe M, Aguzzi A, Schughart K. Development (Cambridge, UK) 1994;120:3119–3130. doi: 10.1242/dev.120.11.3119. [DOI] [PubMed] [Google Scholar]
- 18.Günther T, Spörle R, Schughart K. Mamm Genome. 1997;8:583–585. doi: 10.1007/s003359900509. [DOI] [PubMed] [Google Scholar]
- 19.Chen Z F, Behringer R R. Genes Dev. 1995;9:686–699. doi: 10.1101/gad.9.6.686. [DOI] [PubMed] [Google Scholar]
- 20.Zhao Q, Behringer R R, de Crombrugghe B. Nat Genet. 1996;13:275–283. doi: 10.1038/ng0796-275. [DOI] [PubMed] [Google Scholar]
- 21.Lyu M S, Park D J, Rhee S G, Kozak C A. Mamm Genome. 1996;7:501–504. doi: 10.1007/s003359900151. [DOI] [PubMed] [Google Scholar]
- 22.Melloy P G, Ewart J L, Cohen M F, Desmond M E, Kuehn M R, Lo C W. Dev Biol. 1998;193:77–89. doi: 10.1006/dbio.1997.8787. [DOI] [PubMed] [Google Scholar]
- 23.Wilkinson D G, Bhatt S, Chavier P, Bravo R, Charnay P. Nature (London) 1989;337:461–464. doi: 10.1038/337461a0. [DOI] [PubMed] [Google Scholar]
- 24.Candia A F, Hu J, Crosby J, Lalley P A, Noden D, Nadeau J H, Wright C V. Development (Cambridge, UK) 1992;116:1123–1136. doi: 10.1242/dev.116.4.1123. [DOI] [PubMed] [Google Scholar]
- 25.George E L, Georges-Labouesse E N, Patel-King R S, Rayburn H, Hynes R O. Development (Cambridge, UK) 1993;119:1079–1091. doi: 10.1242/dev.119.4.1079. [DOI] [PubMed] [Google Scholar]
- 26.Georges-Labouesse E N, George E L, Rayburn H, Hynes R O. Dev Dyn. 1996;207:145–156. doi: 10.1002/(SICI)1097-0177(199610)207:2<145::AID-AJA3>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 27.Haffter P, Granato M, Brand M, Mullins M C, Hammerschmidt M, Kane D A, Odenthal J, van Eeden F J, Jiang Y J, Heisenberg C P, et al. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- 28.Driever W, Solnica-Krezel L, Schier A F, Neuhauss S C, Malicki J, Stemple D L, Stainier D Y, Zwartkruis F, Abdelilah S, Rangini Z, et al. Development. 1996;123:37–46. doi: 10.1242/dev.123.1.37. [DOI] [PubMed] [Google Scholar]
- 29.Pappan C, Campos-Ortega J A. Roux’s Arch Dev Biol. 1994;203:178–186. doi: 10.1007/BF00636333. [DOI] [PubMed] [Google Scholar]
- 30.Markel P, Shu P, Ebeling C, Carlson G A, Nagle D L, Smutko J S, Moore K J. Nat Genet. 1997;17:280–284. doi: 10.1038/ng1197-280. [DOI] [PubMed] [Google Scholar]
- 31.Behringer R. Nat Genet. 1998;18:108. doi: 10.1038/ng0298-108. [DOI] [PubMed] [Google Scholar]