Abstract
Influenza A virus causes worldwide epidemics and pandemics and the investigation of memory T helper (Th) cells that help maintain serological memory following infection is important for vaccine design. In this study we investigated CD94 and NKG2 gene expression in memory CD4 T-cell clones established from the spleens of C57BL/10 (H-2b) and BALB/c (H-2d) mice infected with influenza A virus (H3N2). CD94 and NKG2A/C/E proteins form heterodimeric membrane receptors that are involved in virus recognition. CD94 and NKG2 expression have been well characterized in natural killer (NK) and cytotoxic T cells. Despite CD94 being potentially an important marker for Th1 cells involved in virus infection, however, there has been little investigation of its expression or function in the CD4 T-cell lineage and no studies have looked at in-vivo-generated Th cells or memory cells. We show in this study that in-vivo-generated CD4 Th1 cells, but not Th2 cells, exhibited full-length CD94 and NKG2A gene expression following activation with viral peptide. For NKG2A, a novel ‘short’ (possibly redundant) truncated isoform was detectable in a Th2 cell clone. Another member of the NK receptor family, NKG2D, but not NKG2C or E, was also differentially expressed in Th1 cells. We show here that CD94 and NKG2A may exist as multiple isoforms with the potential to distinguish helper T-cell subsets.
Keywords: helper T cells, influenza virus type A, memory cells, natural killer receptors
Introduction
Influenza A virus causes worldwide epidemics and pandemics.1 Symptoms in man can vary from a mild respiratory disease to viral pneumonia. Vaccines are recommended for vulnerable groups and are designed each year based on monitoring of emerging virus subtypes worldwide.
Adaptive immune responses are directed against the two major surface glycoproteins, haemagglutinin (HA) and neuraminidase, and to internal proteins.2–9 The murine response is diverse, with T-cell clones from individual mice recognizing different HA epitopes10–13 and with clear major histocompatibility complex (MHC) patterns emerging,14,15 all of which are important considerations in future vaccine design.
Previous studies by our group found an influence of MHC background on CD4 T helper (Th) cell cytokine profiles following influenza infection14(Table 1). To look at differential gene expression in T cells, we undertook a study16 using subtractive hybridization for two C57BL/10 CD4+ T-cell clones isolated from a single donor. These two clones had identical T-cell receptor sequences and recognized the same region of HA (site B, p186–205), a site recently shown to be important for vaccine design,14 but differed in their cytokine secretion profile, either interferon-γ (IFN-γ), interleukin-4 (IL-4), IL-5 (clone Bpp19) or IL-4, IL-5, IL-10 (clone Bpp9).
Table 1.
Summary of T-cell clone cytokine profiles and influenza virus HA specificity
| Clone | Haplotype | HA1 specificity | Cytokine profile |
|---|---|---|---|
| Bpp19 | b | p186–205 | IFN-γ, IL-4, IL-5 |
| Bpp9 | b | p186–205 | IL-4, IL-5, IL-10 |
| Bpp17 | b | p186–205 | IFN-γ, IL-4, IL-5 |
| Bpp23 | b | p186–205 | IFN-γ, IL-4, IL-5 |
| BA5T-6 | d | p177–199 | IFN-γ |
| ML7P-66 | d | p177–199 | IFN-γ |
| BA5-6 | d | p56–76 | IFN-γ |
| BA6-1 | d | p56–76 | IFN-γ |
Evident from this study was that the membrane heterodimeric receptor CD94/NKG2A [documented predominantly in natural killer (NK) and CD8+ T cells] was expressed in the activated Bpp19 (Th1-type) cell, but not in the Bpp9 (Th2-type) cell. We decided, therefore to look in more detail at expression of this receptor family in CD4 T cells, the results of that study are reported here.
CD94 (Klrd1) and NKG2A (Klrc1) are both members of the C-type lectin Group V NK receptor family17–21 that was originally found in NK cells.22,23 In NK cells and CD8 T cells these proteins together form a novel inhibitory receptor with a role in innate and adaptive immunity.24–26 Apart from binding to NKG2A, CD94 protein can also associate with alternative NKG2 isoforms (NKG2C or NKG2E) to produce stimulatory molecules that complex with the MHC class I ligands HLA-E and Qa-1b in humans and mice, respectively,27–30 to recognize infected cells. Interestingly, another member of the family, NKG2D, forms a homodimeric receptor and its ligand, the stress-inducible MHC class I chain-related protein A (MICA), is expressed by tumour cells and leads to activation of NK cells and macrophages.20,31–33
Although the CD94/NKG2A heterodimer is termed an NK-cell receptor, it has been described in a subset of both murine and human CD8+ T cells,34–38 in which it identifies a subpopulation of effector and memory cells.39,40 In addition to our in-vivo-generated CD4 T cells,16 recent studies have shown that CD94 and NKG2 were also found in CD4 T cells in humans41 or mice42in vitro. In CD4 T cells, this heterodimer was shown to costimulate Th1 effector function,42 in contrast with its well-characterized inhibitory function on NK cells.
In mice, the genes for CD94 and NKG2A are present on chromosome 6 and are within a 200-kilobase region known as the NK complex that includes other members of the C-type lectin family20,43,44(Fig. 1). Recent papers have investigated what regulates CD94 gene expression in NK and CD8+ T cells.45–48 In cultured murine splenocytes, CD94 was found to be under the control of dual promoters located in either exon 1a (distal) or exon 1b (proximal).47 This control led to the synthesis of two types of transcript that differed at the 5′ end. In primary NK and CD8+ T cells, only the proximal promoter was found to be active. In addition for human NK and CD8 T cells, both promoters show differential sensitivity to IL-2 and IL-15, which are cytokines that increase CD94 expression in NK cells.46 CD94 promoter usage may also vary with cell type, that is NK or T cell.47
Figure 1.
Relative position of genes in NK cluster on mouse chromosome 6. **CD94, NKG2D, NKG2C/E and NKG2A/B/2A2, are located in the region at 130·25–130·35 Mb. Data from http://www.ensembl.org.
For CD94 and NKG2A, recent papers have concentrated on receptor expression in NK cells and CD8+ T cells, and only two papers41,42 have looked at CD94 and NKG2A expression in CD4 T cells, both were in vitro studies. Here we look at the expression of CD94 and NKG2A in CD4 T cells, including promoter usage, based on in vivo studies and extend our investigations to look at T cells of two haplotypes that recognize different sites on influenza virus HA and to other members of the NK family.
Materials and methods
Generation and isolation of Th clones
Th clones were generated from mice infected intranasally with influenza virus and isolated as described previously.14,16
cDNA preparation
In-vivo-generated T cells were cultured as described previously.16 For activation, cells were stimulated for 2 hr with influenza HA peptide p186–205, as described elsewhere.16 Cells were disrupted using a QIA shredder (Qiagen, Hilden, Germany). RNA was extracted using the RNeasy kit (Qiagen, Hilden, Germany), DNase treated, and reverse transcribed as described previously.16
In-vitro-generated T cells (Th1, Th2) were freshly isolated T cells and were grouped as either resting (unstimulated) cells or as stimulated cells, stimulated with either phorbol 12-myristate 13-acetate (40 ng/ml) or ionomycin (100 ng/ml), RNA was extracted and cDNA was prepared as described previously.49,50
Primers
For CD94 and NKG2A expression, primers were designed to produce full-length gene transcripts (primer pairs CD94-F2/CD94-R2 and NKG2A-F2/NKG2A-R2) using Primer3 software (http://www.frodo.wi.mit.edu). In addition, primers were designed to distinguish NKG2A/2A2/B and NKG2C/E isoforms according to published sequences20,48 (see Table 2). For Bpp19 CD94 transcripts off the distal promoter, primers were synthesized according to published sequences46,47 (primer pair CD94-F3/CD94-R2).
Table 2.
Primers used for T-cell amplification
| Primer | Sequence (5′ to 3′) |
|---|---|
| CD94-F11 | ATGCTGTGTTTGCCTGGAC |
| CD94-R11 | AACGCTTTTGCTTGGACTGT |
| CD94-F2 | ATGGCAGTTTCTAGGATCACTCG |
| CD94-R2 | CATTTAAATAGGCAGTTTCTTACAGATGTA |
| CD94-F3 | AACATCAACATCCCACACTTGTATGAC |
| CD94-F42 | CACCTTCTCCAACCACCACT |
| CD94-R42 | TCTGCGAAGCACAGAAATC |
| NKG2A-F11 | CGAAGCAAAGGCACAGA |
| NKG2A-R11 | ATGGCACAGTTACATTCATCAT |
| NKG2A-F2 | GACATGAGTAATGAACGCGTCACC |
| NKG2A-R2 | TCAGATGGGGAATTTACACTTAC |
| NKG2A-F32 | AGGTTTTCCATCTCCTCCAGAGA |
| NKG2A-R32 | CACAGCGACAATTAAGACAAAACA |
| NKG2C-F11 | GCTGAACTGAAGAAGCAGATCC |
| NKG2C/E-R11 | TGGGGAATTTACACTTACAAAG |
| NKG2D-F11 | ACAAGGAAGTCCCAGTTTCC |
| NKG2D-R11 | TTACACCGCCCTTTTCATGCAGATGTA |
| NKG2E-F11 | TATTACATTGGCATGGAAAGA |
Real-time quantitative polymerase chain reaction (PCR) primers were designed according to ABI specifications using Primer Express software and were provided by GenoSys (Sigma–GenoSys, Pampisford, Cambs, UK) (see Table 2).
RT-PCR and real-time quantitative PCR
The resulting cDNAs were subjected to reverse transcription PCR (RT-PCR): 33 cycles of 30 s at 95°, 30 s at 60° and 2 min at 68° using Advantage 2 Polymerase Mix and Advantage 2 PCR buffer (BD Clontech, Mountview, CA), dNTPs (Amersham, Little Chalfont, UK) and the relevant primers. The resulting products were run on a 1·2% agarose gel against known molecular weight markers to check specificity.
In addition, cDNA was analysed for the expression of CD94 and NKG2A by real-time quantitative PCR assay using an ABI Prism 7000 Sequence Detection System (Applied Biosystems, Foster City, CA). Target gene mRNA expression was quantified using SYBR Green (Applied Biosystems) and was normalized to ubiquitin cDNA levels. The cDNA from Bpp19 and Bpp9 stimulated with HA peptide were included as controls.
Sequencing
PCR products were sequenced directly or following cloning into plasmids. PCR products were cloned using the Clontech Advantage PCR Cloning kit (BD Biosciences, Erembodegem, Belgium). Escherichia coli competent cells were transformed and spread onto luteinizing broth/agar plates. Following incubation at 37°, colonies were picked arbitrarily for analysis.
Plasmids were isolated using Wizard Plus Minipreps (Promega, Madison, WI). The presence of plasmid inserts was confirmed following digestion with the EcoRI restriction enzyme and by 1·2% agarose gel. Inserts were then sequenced with a DNA Sequencing Kit (Applied Biosystems) using M13 primers and run on an ABI Prism 377 DNA Sequencer (Applied Biosystems) or at the Advanced Biotechnology Centre (http://www.bm-abc01.cx.med.ic.ac.uk).
Data analysis
Sequencing data were mapped to the Ensembl database (http://www.ensembl.org) and aligned using ClustalW software (http://www.ebi.ac.uk).
Rapid amplification of cDNA ends (RACE)
RACE was carried out using the SMART RACE cDNA amplification kit (BD Clontech).
Results
Sequence of differentially expressed Bpp19 (Th1) CD94 is consistent with the known sequence
The mouse CD94 gene contains six exons and codes for a transcript 848 base pairs (bp) in length including untranslated regions, of which 543 bp translates a 181-amino acid (aa) type II protein.36 Four differentially expressed CD94 cDNA library fragments from Bpp19 were sequenced and had a readable length of around 650 bp. These fragments were replicates that covered the whole of the CD94 gene extending 39 bp into the 5′ untranslated region and 87 bp into the 3′ untranslated region. Sequences were aligned against an EBI published gene sequence (ENSMUSG00000030165). The difference products were almost identical to the known transcript (ENSMUST00000088068), differing only by an in-frame substitution at the exon 4/exon 5 junction. CD94 from the Bpp19 difference products was found to be identical to another CD94 transcript (ENSMUSESTT00000016770) at this junction.20
Sequences of differentially expressed Bpp19 NKG2A align to the 3′ end of the gene
The NKG2A gene is composed of seven exons that are approximately 2000 bp in length in total including the untranslated regions, of which 732 bp translates a 244-aa protein. Five differentially expressed NKG2A fragments varied in length from around 200 to 400 bp. Transcripts aligned with three separate regions of the published gene sequence (ENS0000030167) at 672–925, 1027–1417 and 1523–1917 bp. These transcripts spanned the final 60 bp of the translated region and part of the 3′ untranslated region.
Full-length CD94 and NKG2A is expressed in Bpp19 (Th1) but not Bpp9 (Th2)
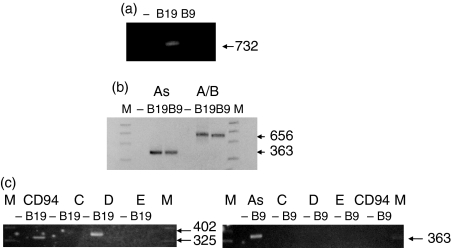
Following investigation of the differentially expressed products, we decided to look at the expression of CD94 and NKG2A in total RNA. As anticipated, full-length NKG2A (bp 1–732, ATG start to stop sequence) was expressed in Bpp19 but not Bpp9 (Fig. 2a). However, PCR amplification of Bpp9 DNA covering the first six exons, the region not spanned by the difference products, identified a ‘short’ form of NKG2A (bp 1–656) in Bpp9 cells (Fig. 2b). Specific primers and PCR sequencing confirmed that the product was the NKG2A isoform (ENSMUS00000030167) (As, A/B, Fig. 2b) and not the NKG2A2 isoform (minus exon 2, ENSMUST00000032271) or NKG2B isoform (minus exon 3, ENSMUST00000036061) (data not shown). To determine if the Bpp9 NKG2A ‘short’ form extended 3′ of bp 656, repeated attempts were made to synthesize a product using 5′ RACE, but these failed to give any positive results, which may suggest the introduction of a stop codon in the region from 657 to 671 bp, rather than alternative splicing to 3′ of exon 8.
Figure 2.
CD94 and NKG2A expression in Bpp19 and Bpp9 from total RNA. (a) Primers were designed to cover the full-length 1–732 bp translated product. Full-length NKG2A is expressed in Bpp19 but not Bpp9. (b) Primers were used that covered bp 1–656 and bp 293–656, respectively, to distinguished the isoform NKG2A from isoforms NKG2A2 and NKG2B, which would not be amplified. Positive product showed that NKG2A (bp 1–656), not NKG2A2 or NKG2B, is expressed in both Bpp19 and Bpp9. In Bpp9 Th2 cells, the NKG2A product is a non-full-length product (but covers at least the first 656 bp), designated as the ‘short’ form, whereas the Bpp19 Th1 product is part of the full-length product. (c) Primers were used to determine if Bpp19 and Bpp9 expressed NKG2D or NKG2C and NKG2E isoforms. Bpp19 expresses CD94 (as shown in a) and NKG2D, but not NKG2C or NKG2E; Bpp9 expresses NKG2A ‘short’ form (As) only (as shown in b), i.e. not NKG2C, NKG2D or NKG2E.
In contrast, CD94 was expressed only in Bpp19 (Fig. 2c). The CD94 full-length sequence was identical to that found for the differentially expressed Bpp19 cDNA.
Bpp19 expresses NKG2D, but not NKG2C or NKG2E
The NKG2C gene consists of six exons, with a transcript length of 714 bp and translates a 238-aa protein. NKG2E, an isoform of NKG2C, is almost identical to NKG2C, but is missing exon 3 and translates a 221-aa protein. NKG2D has eight exons with a length of 1049 bp and translates a 232-aa protein. NKG2D was expressed in Bpp19 but not Bpp9 T cells (Fig. 2c). Interestingly neither NKG2C nor NKG2E were expressed in Bpp19 or in Bpp9 cells (Fig. 2c).
CD94 in Bpp19 can be expressed off the distal promoter
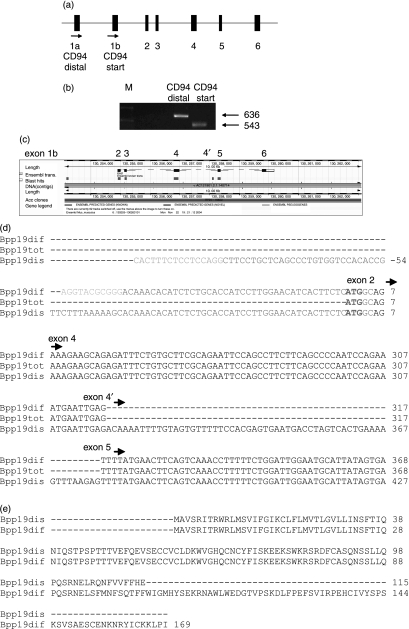
A CD94 product from Bpp19 could be expressed off the distal promoter (exon 1a) in addition to the product from the start codon at the end of exon 1b (Figs 3a,b). Alignment of the CD94 product off the distal promoter onto the Ensembl database revealed an insert between exons 4 and 5 (designated exon 4′) (Fig. 3c). This product was identical to the Bpp19 full-length sequence, except for the addition of this 59-bp insertion in the middle of the transcript (Fig. 3d). The insertion, however, had introduced a stop codon in this region and may translate a product of only 115 amino acids (Fig. 3e). This protein, if synthesized, would encompass the whole of the signalling and transmembrane region, but only part of the carbohydrate-binding site.
Figure 3.
Bpp19 CD94 expression off the distal promoter. (a) The mouse CD94 gene contains six exons and codes for a transcript 848 bp in length including untranslated regions, of which 543 bp translates a 181-aa type II protein. Exons 1a and most of exon 1b encode the 5′ untranslated region, and exon 2 encodes the transmembrane domain. Exon 3 encodes the stalk region and exons 4–6 encode the carbohydrate-recognition domain (CRD).36 (b) Primers were used to see if a CD94 Bpp19 Th1 transcript could be expressed off the distal promoter exon 1a. The gel shows the distal primer product (636 bp) compared to the full-length product (start) (543 bp). (c) The product off the distal promoter was aligned (http://www.ensembl.org) and this showed that this isoform included a new exon, designated here as 4′. (d) Sequencing of the distal promoter isoform showed that this included a 59-bp insertion that introduces a stop codon. Arrows indicate the start of each exon. Exon 1b is coloured blue. For Bpp19 sequences, splice sites are indicated in orange (Bpp19 distal promoter) and in green (Bpp19 difference product). Start codon ATG at the end of exon 1 is in bold. Bpp19dif, Bpp19 difference product; Bpp19tot, Bpp19 start sequence; Bpp19dis, Bpp19 product distal promoter. (e) The Bpp19 difference and distal promoter products were converted to their proposed amino acid sequences and aligned (http://www.ebi.ac.uk). Translation of the distal promoter product would result in an amino acid with only part of the carbohydrate recognition domain.
CD94 and NKG2A are expressed in other H-2b and H-2d Th1 clones to influenza HA
We wished to extend the scope of our studies to establish if CD94 and NKG2A were expressed in influenza HA-specific clones of more than one haplotype and cytokine secretion profile (Table 1) and in other types of CD4 T cells.
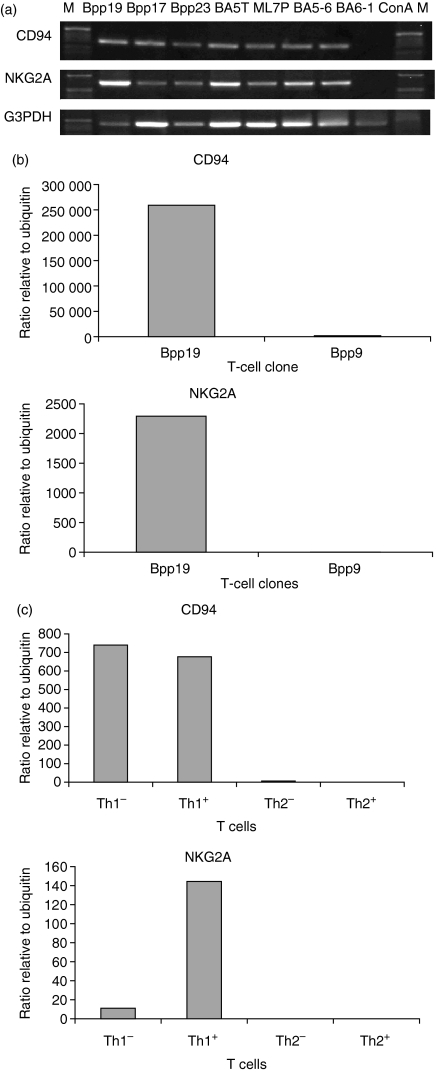
Two C57BL/10 (H-2b) clones (Bpp17, Bpp23), and four BALB/c (H-2d) clones (BA5T-6, ML7P-66, BA5-6, BA6-1), all of which secreted IFN-γ and were specific for influenza HA (Table 1), were included in the study. All clones produced transcripts for full-length CD94 and NKG2A (Fig. 4a). C57BL/10 spleen cells stimulated with concanavalin A were also included in the experiment, but in this case no CD94 or NKG2A products were found (Fig. 4a). This finding is consistent with the lack of expression of CD94/NKG2A in unstimulated CD4 T cells. In addition, Bpp17 and Bpp23 products were sequenced and found to be identical to those from Bpp19 (data not shown).
Figure 4.
CD94 and NKG2A expression in T cells. (a) PCR with full-length primers was carried out to investigate the expression of CD94 and NKG2A in other C57BL/10 (H-2b) and BALB/c (H-2d) Th1 clones to influenza HA. Positive product was found for all clones, but not for concanavalin A (ConA)-stimulated spleen cells; (b) Real-time quantitative PCR showed that CD94 and NKG2A expression were up-regulated in the activated Th1 memory clone Bpp19, but not in Bpp9. (c) Real-time quantitative PCR showed that CD94 and NKG2A are up-regulated in in-vitro-isolated Th1 cells. +, stimulated; –, unstimulated.
CD94 and NKG2A are expressed in Th1 cells – real-time quantitative PCR
For the memory T-cell clones Bpp19 and Bpp9, CD94 are up-regulated in the stimulated Th1 clone but not the Th2 clone (Fig. 4b), which is consistent with the previous findings. For the in-vitro-derived T cells, CD94 was up-regulated in both the stimulated (+) and the unstimulated (–) Th1 cells, CD94 is known to be expressed initially as a homodimer. NKG2A was up-regulated in stimulated Th1 cells only (Fig. 4c). Expression levels were much higher in the in-vivo-isolated memory Th cells relative to the in-vitro-generated Th cells.
Discussion
In this study we have shown that CD94 and NKG2A expression are markers for a panel of IFN-γ-secreting, activated CD4 memory Th cells generated by influenza virus infection. In addition, expression of a novel CD94 isoform highlights sequence variability and the effect of promoter usage. The findings for CD4 Th cells are consistent with our earlier in vivo studies16 and with the in vitro studies of Hartt Meyers and colleagues,42 who described CD94/NKG2A expression on CD4 T cells stimulated in vitro with ovalbumin 323–339 peptide and polarized to Th1 and Th2 cells. There is also some agreement with a study by Romero and colleagues41 who found that human CD4 T cells expressed CD94 and NKG2A in vitro after 15 days of CD3-mediated stimulation.
In our previous study, looking at differential expression between Bpp19 (Th1) and Bpp9 (Th2) difference products for NKG2A, confirmed by sequencing, were found for the Bpp19 clone only, leading us to believe that NKG2A was expressed only in Bpp19. However, on alignment against the published sequence the difference products covered only the region 672–1917 bp, i.e. the last 60 bp of the translated region and part of the 3′ untranslated region. This suggested that there might be a truncated product expressed in Bpp9 that covered bp 1–671, which we found here and have designated as the ‘short’ form of NKG2A. Primers that covered bp 1–656 showed that this truncated product was indeed present in Bpp9 and these primers also amplified this portion of the 732 bp full-length product for Bpp19. The ‘short’ form in Bpp9 was confirmed by sequencing and by specific primers to be NKG2A. RACE of the NKG2A Bpp9 product was repeatedly unsuccessful, which may suggest the termination of the sequence in the 657–671-bp region, rather than alternative splicing to 3′ of exon 8 and bp1917.
In this study, NKG2A was not expressed on resting Th1 cells. CD94/NKG2A is known to be expressed constitutively in NK cells, but in T cells the receptor shows variable expression: being induced in naive T cells, following viral and bacterial infection, and persistent after pathogen clearance35 initiated by T-cell-receptor-mediated signals.51–53 Classically, on NK cells CD94/NKG2A is an inhibitory receptor. In T cells, CD94/NKG2A receptor function is not fully understood and this heterodimer may have several different roles. On CD4 T cells CD94 may act as a costimulatory molecule.42 Persistent stimulation by virus antigens may prevent CD94/NKG2A receptor down-regulation18,25,51,53–60 and result in the expression on cytotoxic cells that is seen in chronic virus or bacterial infections or it may modulate the response.61,62
It has been shown that cytokines increase CD94 expression in NK cells (IL-15) and CD8+ T cells (IL-2, IL-15),63,64 or expression of both CD94 and NKG2A in CD8+ T cells (transforming growth factor-β,65,66 IL-1267). IFN-γ may also regulate NK T-cell function using CD94/NKG2.68 In this study, CD94/NKG2A-positive transcripts were found in IFN-γ-secreting clones specific for several sites on HA and from both C57BL/10 and BALB/c haplotypes. Hartt Meyers and colleagues42 showed that cross-linking the CD3 and CD94/NKG2 receptors resulted in IFN-γ and tumour necrosis factor-α (TNF-α) production and cell proliferation. In contrast, in humans, however, co-ligation of NKG2A and CD94 resulted in the inhibition of IFN-γ and TNF-α production.42
CD94 is known be expressed off two promoters – distal (from exon 1a) and proximal (from exon 1b) – and the choice of promoter is known to vary with either cell type (NK or CD8+ T cell) or cytokine (IL-2 or IL-15) stimulation. Human CD94 promoters both contain the STAT family protein-binding site GAS (IFN-γ-activated site) with differential sensitivity to IL-2 and IL-15 as well as a potential EBS (Ets-binding site);46 however, the exon 1a promoter region is not conserved between humans and mice.47 Interestingly, STAT proteins are known to induce numerous lymphocyte genes and the Ets family regulates cell processes including growth control and activation.46 It has been suggested that the novel 5′ UTR sequence created from the CD94 exon 1a distal promoter may change the secondary structure of mRNA and influence the translation of various transcript forms.47 This theory is supported by our finding here of a novel isoform produced off the exon 1a CD94 distal promoter. Although this isoform would translate a truncated protein, this protein might still be functionally expressed because the expressed RNA contains the signalling and transmembrane regions. It is interesting that, for CD4 T cells, the product off the distal promoter is not the form found differentially expressed in Bpp19 (IFN-γ-positive) compared with Bpp9 (IFN-γ-negative) cells.
For CD4+ T cells there is recent evidence for differing types of memory cells, small resting CD62low CCR7low effector memory T cells in the lung and CD62, CCR7, MEL14– CD4+ T cells (central memory T cells) that home to the lymph nodes or spleen, respectively.8,69–71 Also, CD94 can form heterodimeric receptors not only with NKG2A, but also with two other members of the NKG family, NKG2C and NKG2E and we included these in this study. Neither the Bpp19 nor the Bpp9 clone produced transcripts for NKG2C or NKG2E. Our results with memory T cells isolated from the spleen concur with a study by Arlettaz et al.72 for murine CD8+ T cells, in which expression of receptors CD94/NKG2A and CD94/NKG2C was mutually exclusive. Furthermore, Arlettaz and colleagues found that CD94/NKG2C and CD94/NKG2A receptors were expressed by distinct subsets (CCR7+ memory cells and CCR7– effector memory T cells) of committed CD8+ T-cell receptor αβ lymphocytes.
The NKG2D gene is adjacent to CD94 on chromosome 6 and classically on NK cells the NKG2D receptor is a stimulatory molecule. In addition, we show here that Bpp19, a CD94+ Th1 cell, can also express NKG2D; no NKG2D transcript was found in the Th2 clone. These findings may be typical of activated CD4 memory T cells in vivo, but although interesting were not the main focus of this paper and were not investigated further.
Interestingly, NKG2D has been found to be up-regulated on hepatitis B virus-specific IL-7/IL-15 expanded CD4 T cells and its engagement promoted expansion and IFN-γ production.73 In contrast to our finding, a previous study in the human41 showed that NKG2A/C/D and E could all be expressed at one time. It may be therefore that Romero's group highlights differences between human and mouse expression or that the human study may reflect a mixed population.
In summary, we find that full-length CD94/NKG2A expression is restricted to the Th1, but not Th2, memory cells generated in vivo, substantiating our previous subtractive hybridization study. Furthermore, expression of CD94 isoforms varied with promoter usage, with sequence diversity at the exon 4/exon 5 boundary. Our findings may support a role for IFN-γ and the CD94/NKG2A receptor in virus infections and for expression of this receptor on (possibly effector) memory Th1 cells following influenza A virus infection.
Acknowledgments
We would like to thank Anne O'Garra and members of her laboratory for helpful advice on, and reagents for, the real-time quantitative PCR studies.
Abbreviations
- aa
amino acid
- bp
base pair
- HA
haemagglutinin
- IFN-γ
interferon-γ
- interleukin-4
IL-4
- MHC
major histocompatibility complex
- NK
natural killer
- PCR
polymerase chain reaction
- RACE
rapid amplification of cDNA ends
- RT-PCR
reverse transcription PCR
- Th
T helper
References
- 1.Oldstone MBA. Viruses, Plagues and History. Oxford: Oxford University Press; 1998. [Google Scholar]
- 2.Wiley DC, Wilson IA, Skehel JJ. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981;289:373–8. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- 3.Air GM, Els MC, Brown LE, Laver WG, Webster RG. Location of antigenic sites on the three dimensional structure of the influenza N2 virus neuraminidase. Virology. 1985;145:237–48. doi: 10.1016/0042-6822(85)90157-6. [DOI] [PubMed] [Google Scholar]
- 4.Yewdell JW, Hackett C. Specificity and function of T lymphocytes induced by influenza A viruses. In: Krug R, editor. The Influenza Viruses. New York: Plenum Press; 1989. pp. 361–429. [Google Scholar]
- 5.Doherty PC, Christensen JP. Accessing complexity. the dynamics of virus-specific T cell responses. Annu Rev Immunol. 2000;18:561–92. doi: 10.1146/annurev.immunol.18.1.561. [DOI] [PubMed] [Google Scholar]
- 6.Doherty PC, Topham DJ, Tripp RA, Cardin RD, Brooks JW, Stevenson PG. Effector CD4+ and CD8+ T cell mechanisms in the control of respiratory virus infections. Immunol Rev. 1997;59:105–17. doi: 10.1111/j.1600-065x.1997.tb01010.x. [DOI] [PubMed] [Google Scholar]
- 7.Gao XM, Liew FY, Tite JP. A dominant Th epitope in influenza nucleoprotein. Analysis of the fine specificity and functional repertoire of T cell recognising a single determinant. J Immunol. 1990;144:2730–2. [PubMed] [Google Scholar]
- 8.Brown DM, Roman E, Swain SL. CD4 T cell responses to influenza infection. Semin Immunol. 2004;16:171–7. doi: 10.1016/j.smim.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 9.Mills KHG, Skehel JJ, Thomas DB. Extensive diversity in the recognition of influenza virus hemagglutinin by murine T helper clones. J Exp Med. 1986;163:1477–90. doi: 10.1084/jem.163.6.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barnett BC, Graham CM, Burt DS, Skehel JJ, Thomas DB. The immune response of BALB/c mice to influenza hemagglutinin: commonality of the B cell and T cell repertoires and their relevance to antigenic drift. Eur J Immunol. 1989;19:515–21. doi: 10.1002/eji.1830190316. [DOI] [PubMed] [Google Scholar]
- 11.Barnett BC, Burt DS, Graham CM, Warren AP, Skehel JJ, Thomas DB. I-Ad restricted T cell recognition of influenza hemagglutinin. Synthetic peptides identify multiple epitopes corresponding to antibody-binding regions of the HA1 subunit. J Immunol. 1989;143:2663–9. [PubMed] [Google Scholar]
- 12.Burt DS, Mills KHG, Skehel JJ, Thomas DB. Diversity of the class II (I-Ak/I-Ek) -restricted T cell repertoire for influenza hemagglutinin and antigenic drift. Six nonoverlapping epitopes on the HA1 subunit are defined by synthetic peptides. J Exp Med. 1989;170:383–97. doi: 10.1084/jem.170.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith CA, Graham CM, Thomas DB. Immunodominance correlates with T-cell receptor (alpha beta) gene usage in the class II-restricted response to influenza hemagglutinin. Immunology. 1994;82:343–50. [PMC free article] [PubMed] [Google Scholar]
- 14.Graham CM, Smith CA, Thomas DB. Novel diversity in Th1 Th2 type differentiation of hemagglutinin-specific T cell clones elicited by natural influenza virus infection in three major haplotypes (H-2b,d,k) J Immunol. 1998;161:1094–103. [PubMed] [Google Scholar]
- 15.Crowe SR, Miller SC, Brown DM, et al. Uneven distribution of MHC class II epitopes within the influenza virus. Vaccine. 2006;24:457–67. doi: 10.1016/j.vaccine.2005.07.096. [DOI] [PubMed] [Google Scholar]
- 16.Graham CM, Thomas DB. Differential analysis of CD4+ Th memory clones with identical TCRαβ rearrangement (nontransgenic) but distinct lymphokine phenotype reveals diverse and novel gene expression. Immunology. 2004;113:194–202. doi: 10.1111/j.1365-2567.2004.01953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colonna M, Nakajima H, Navarro F, Lopez-Botet M. A novel family of Ig-like receptors for HLA class I molecules that modulate the function of lymphoid and myeloid cells. J Leukoc Biol. 1999;66:375–81. doi: 10.1002/jlb.66.3.375. [DOI] [PubMed] [Google Scholar]
- 18.Natarajan K, Dimasi N, Wang J, Mariuzza DH. Structure and function of natural killer cell receptors: multiple molecular solutions to self-nonself discrimination. Annu Rev Immunol. 2002;20:853–85. doi: 10.1146/annurev.immunol.20.100301.064812. [DOI] [PubMed] [Google Scholar]
- 19.Lohwasser S, Hande P, Mager DL, Takei F. Cloning of murine NKG2A B C: a second family of C-type lectin receptors on murine NK cells. Eur J Immunol. 1999;29:755–61. doi: 10.1002/(SICI)1521-4141(199903)29:03<755::AID-IMMU755>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 20.Vance RE, Tanamachi DM, Hunke T, Raulet DH. Cloning of a mouse homolog of CD94 extends the family of C-type lectins on murine natural killer cells. Eur J Immunol. 1997;27:3236–341. doi: 10.1002/eji.1830271222. [DOI] [PubMed] [Google Scholar]
- 21.Silver ET, Lau JC, Kane KP. Molecular cloning of mouse NKG2A and C. Immunogenetics. 1999;49:727–30. doi: 10.1007/s002510050674. [DOI] [PubMed] [Google Scholar]
- 22.Moretta A, Tambussi G, Bottino C, et al. A novel surface antigen expressed by a subset of human CD3–CD16+ natural killer cells. Role in cell activation and regulation of cytolytic function. J Exp Med. 1990;171:695–714. doi: 10.1084/jem.171.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Litwin V, Gumperz J, Parham P, Phillips JH, Lanier LL. NKB1: a natural killer cell receptor involved in the recognition of polymorphic HLA-B molecules. J Exp Med. 1994;180:537–43. doi: 10.1084/jem.180.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brooks AG, Posch PE, Scorzelli CJ, Borrego F, Coligan JE. NKG2A complexed with CD94 defines a novel inhibitory natural killer receptor. J Exp Med. 1997;185:795–800. doi: 10.1084/jem.185.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gunturi A, Berg RE, Forman J. Preferential survival of CD8 T and NK cells expressing high levels of CD94. J Immunol. 2003;170:1737–145. doi: 10.4049/jimmunol.170.4.1737. [DOI] [PubMed] [Google Scholar]
- 26.Gunturi A, Berg RE, Forman J. The role of CD94/NKG2 in innate and adaptive immunity. Immunol Res. 2004;30:29–34. doi: 10.1385/IR:30:1:029. [DOI] [PubMed] [Google Scholar]
- 27.Vance RE, Kraft JR, Aitman JJ, Jensen PE, Raulet DH. Mouse CD94/NKG2A is a natural killer receptor for the nonclassical major histocompatibility complex (MHC) class I molecule Qa-1b. J Exp Med. 1998;188:1841–6. doi: 10.1084/jem.188.10.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borges L, Hsu ML, Fanger N, Kubin M, Cosman D. A family of human lymphoid and myeloid Ig-like receptors some of which bind to MHC class I molecules. J Immunol. 1997;159:5192–6. [PubMed] [Google Scholar]
- 29.Vance RE, Jamieson AM, Raulet DH. Recognition of the class Ib molecule Qa-1b by putative activating receptors CD94/NKG2C and CD94/NKG2E on mouse natural killer cells. J Exp Med. 1999;190:1801–12. doi: 10.1084/jem.190.12.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bottino C, Castriconi R, Moretta L, Morreta A. Cellular ligands of activating NK receptors. Trends Immunol. 2005;26:221–6. doi: 10.1016/j.it.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 31.Raulet DH. Roles of the NKG2D immunoreceptor and its ligands. Nature Rev Immunol. 2003;3:781–90. doi: 10.1038/nri1199. [DOI] [PubMed] [Google Scholar]
- 32.Bauer SV, Groh J, Wu A, Steinle JH, Phillips LL, Lanier T. Spies activation of NK cells and T cells by NKG2D a receptor for stress-inducible MICA. Science. 1999;285:727–9. doi: 10.1126/science.285.5428.727. [DOI] [PubMed] [Google Scholar]
- 33.Diefenbach A, Jamieson AM, Liu SD, Shastri N, Raulet DH. Ligands for the murine NKG2D receptor: expression by tumor cells and activation of NK cells and macrophages. Nat Immunol. 2000;1:119–26. doi: 10.1038/77793. [DOI] [PubMed] [Google Scholar]
- 34.Haedicke W, Ho FC, Chott A, Moretta L, Rudiger T, Ott G, Muller-Hermelink HK. Expression of CD94/NKG2A and killer immunoglobulin-like receptors in NK cells and a subset of extranodal cytotoxic T-cell lymphomas. Blood. 2000;95:3628–30. [PubMed] [Google Scholar]
- 35.McMahon CW, Raulet DH. Expression and function of NK cell receptors in CD8+ T cells. Curr Opin Immunol. 2001;13:465–70. doi: 10.1016/s0952-7915(00)00242-9. [DOI] [PubMed] [Google Scholar]
- 36.McMahon CW, Zajac AJ, Jamieson AM, Corral L, Hammer GE, Ahmed R, Raulet DH. Viral and bacterial infections induce expression of multiple NK cell receptors in responding CD8+ T cells. J Immunol. 2002;169:1444–52. doi: 10.4049/jimmunol.169.3.1444. [DOI] [PubMed] [Google Scholar]
- 37.Braud VM, Aldemir H, Breart B, Ferlin WG. Expression of CD94–NKG2A inhibitory receptor is restricted to a subset of CD8+ T cells. Trends Immunol. 2003;24:162–4. doi: 10.1016/s1471-4906(03)00064-4. [DOI] [PubMed] [Google Scholar]
- 38.Coles MC, McMahon CW, Takizawa H, Raulet DH. Memory CD8 T lymphocytes express inhibitory MHC-specific Ly49 receptors. Eur J Immunol. 2000;30:236–44. doi: 10.1002/1521-4141(200001)30:1<236::AID-IMMU236>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 39.Beyersdorf NB, Ding X, Karp K, Hanke T. Expression of inhibitory ‘killer cell lectin-like receptor G1’ identifies unique subpopulations of effector and memory CD8 T cells. Eur J Immunol. 2001;31:3443–52. doi: 10.1002/1521-4141(200112)31:12<3443::aid-immu3443>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 40.Robbins SH, Terrizzi SC, Sydora BC, Mikayama T, Brossay L. Differential regulation of killer cell lectin-like receptor G1 expression on T cells. J Immunol. 2003;170:5876–85. doi: 10.4049/jimmunol.170.12.5876. [DOI] [PubMed] [Google Scholar]
- 41.Romero P, Ortega C, Palma A, Molina IJ, Pena J, Santamaria M. Expression of CD94 and NKG2 molecules on human CD4+ T cells in response to CD3-mediated stimulation. J Leukoc Biol. 2001;70:219–24. [PubMed] [Google Scholar]
- 42.Hartt Meyers J, Ryu A, Monney L, Nguyen K, Greenfield EA, Freeman GJ, Kuchroo VK. CD94/NKG2 is expressed on Th1 but not Th2 cells and costimulates Th1 effector functions. J Immunol. 2002;169:5382–6. doi: 10.4049/jimmunol.169.10.5382. [DOI] [PubMed] [Google Scholar]
- 43.Plougastel B, Trowsdale J. Sequence analysis of a 62-kb region overlapping the human KLRC cluster of genes. Genomics. 1998;49:193–9. doi: 10.1006/geno.1997.5197. [DOI] [PubMed] [Google Scholar]
- 44.Lohwasser S, Wilhelm B, Mager DL, Takei F. The genomic organization of the mouse CD94 C-type lectin gene. Eur J Immunogenet. 2000;27:149–51. doi: 10.1046/j.1365-2370.2000.00212.x. [DOI] [PubMed] [Google Scholar]
- 45.Rodriguez A, Carretero M, Glienke J, Bellon T, Ramirez A, Lehrach H, Francis F, Lopez-Botet M. Structure of the human CD94 C-type lectin gene. Immunogenetics. 1998;47:305–9. doi: 10.1007/s002510050362. [DOI] [PubMed] [Google Scholar]
- 46.Lieto LD, Borrego F, You CH, Coligan JE. Human CD94 gene expression: dual promoters differing in responsiveness to IL-2 or IL-15. J Immunol. 2003;171:5277–86. doi: 10.4049/jimmunol.171.10.5277. [DOI] [PubMed] [Google Scholar]
- 47.Wilhelm BT, Landry J-R, Takei F, Mager DL. Transcriptional control of murine CD94 gene: differential usage of dual promoters by lymphoid cell types. J Immunol. 2003;171:4219–26. doi: 10.4049/jimmunol.171.8.4219. [DOI] [PubMed] [Google Scholar]
- 48.Williams NS, Kubota A, Bennett M, Kumar V, Takei F. Clonal analysis of NK cell development from bone marrow progenitors in vitro: orderly acquisition of receptor gene expression. Eur J Immunol. 2000;30:2074–82. doi: 10.1002/1521-4141(200007)30:7<2074::AID-IMMU2074>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 49.Barrat FJ, Cua DJ, Boonstra A, et al. In vitro generation of interleukin 10-producing regulatory CD4+ T cells is induced by immunosuppressive drugs and inhibited by T helper type 1 (Th1)- and Th2-inducing cytokines. J Exp Med. 2002;195:603–16. doi: 10.1084/jem.20011629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vieira PL, Christensen JR, Minaee S, et al. IL-10-secreting T cells do not express Foxp3 but have comparable regulatory function to naturally occurring CD4+CD25+ regulatory T cells. J Immunol. 2004;172:5986–93. doi: 10.4049/jimmunol.172.10.5986. [DOI] [PubMed] [Google Scholar]
- 51.Moser JM, Gibbs J, Jensen PE, Lukacher AE. CD94–NKG2A receptors regulate antiviral CD8+ T cell responses. Nature Immunol. 2002;3:189–95. doi: 10.1038/ni757. [DOI] [PubMed] [Google Scholar]
- 52.Moser JM, Byers AM, Lukacher AE. NK receptors in antiviral immunity. Curr Opin Immunol. 2002;14:509–16. doi: 10.1016/s0952-7915(02)00357-6. [DOI] [PubMed] [Google Scholar]
- 53.Wojtasiak M, Jones CM, Sullivan LC, Winterhalter AC, Carbone FC, Brooks AG. Persistent expression of CD94/NKG2 receptors by virus-specific CD8 T cells is initiated by TCR-mediated signals. Int Immunol. 2004;16:1333–41. doi: 10.1093/intimm/dxh136. [DOI] [PubMed] [Google Scholar]
- 54.Costa P, Rusconi S, Mavilio D, et al. Differential disappearance of inhibitory natural killer cell receptors during HAART and possible impairment of HIV-1-specific CD8 cytotoxic T lymphocytes. AIDS. 2001;15:965–74. doi: 10.1097/00002030-200105250-00004. [DOI] [PubMed] [Google Scholar]
- 55.Galiani MD, Aguado E, Tarazona R, Romero P, Molina I, Santamaria M, Solana R, Pena J. Expression of killer inhibitory receptors on cytotoxic cells from HIV-1-infected individuals. Clin Exp Immunol. 1999;115:472–6. doi: 10.1046/j.1365-2249.1999.00833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Costa P, Rusconi S, Fogli M, et al. Low expression of inhibitory natural killer receptors in CD8 cytotoxic T lymphocytes in long-term nonprogressor HIV-1-infected patients. AIDS. 2003;17:257–60. doi: 10.1097/00002030-200301240-00017. [DOI] [PubMed] [Google Scholar]
- 57.Ortega C, Romero P, Palma A, Orta T, Peña J, García-Vinuesa A, Molina IJ, Santamaría M. Role for NKG2-A and NKG2-C surface receptors in chronic CD4+ T-cell responses. Immunol Cell Biol. 2004;82:587–95. doi: 10.1111/j.0818-9641.2004.01284.x. [DOI] [PubMed] [Google Scholar]
- 58.Hoji A, Rinaldo CR. Human CD8+ T cells specific for influenza A virus M1 display broad expression of maturation-associated phenotypic markers and chemokine receptors. Immunology. 2005;115:239–45. doi: 10.1111/j.1365-2567.2005.02135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saito M, Braud VM, Goon P, et al. Low frequency of CD94/NKG2A+ T lymphocytes in patients with HTLV-1-associated myelopathy/tropical spastic paraparesis but not in asymptomatic carriers. Blood. 2003;102:577–84. doi: 10.1182/blood-2002-09-2855. [DOI] [PubMed] [Google Scholar]
- 60.Peacock CD, Lin MY, Ortaldo JR, Welsh RM. The virus-specific and allospecific cytotoxic T-lymphocyte response to lymphocytic choriomeningitis virus is modified in a subpopulation of CD8+ T cells coexpressing the inhibitory major histocompatibility complex class I receptor Ly49G2. J Virol. 2000;74:7032–8. doi: 10.1128/jvi.74.15.7032-7038.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ugolini S, Vivier E. Regulation of T cell function by NK cell receptors for classical MHC class I molecules. Curr Opin Immunol. 2000;12:295–300. doi: 10.1016/s0952-7915(00)00090-x. [DOI] [PubMed] [Google Scholar]
- 62.Zingoni A, Palmieri G, Morrone S, Carretero M, Lopez-Botel M, Piccoli M, Frati L, Santoni A. CD69-triggered ERK activation and functions are negatively regulated by CD94/NKG2A inhibitory receptor. Eur J Immunol. 2000;30:644–51. doi: 10.1002/1521-4141(200002)30:2<644::AID-IMMU644>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 63.Lu B, Zagouras P, Fischer JE, Lu J, Li B, Flavell RA. Kinetic analysis of genomewide gene expression reveals molecule circuitries that control T cell activation and Th1/2 differentiation. Proc Natl Acad Sci USA. 2004;101:3023–8. doi: 10.1073/pnas.0307743100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mingari MC, Ponte M, Bertone S, Schiavetti F, Vitale C, Bellomo R, Moretta A, Moretta L. HLA class I-specific inhibitory receptors in human T lymphocytes. interleukin 15-induced expression of CD94/NKG2A in superantigen- or alloantigen-activated CD8+ T cells. Proc Natl Acad Sci USA. 1998;95:1172–7. doi: 10.1073/pnas.95.3.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bertone S, Schiavetti F, Bellomo R, Vitale C, Ponte M, Moretta L, Mingari MC. Transforming growth factor-beta-induced expression of CD94/NKG2A inhibitory receptors in human T lymphocytes. Eur J Immunol. 1999;29:23–9. doi: 10.1002/(SICI)1521-4141(199901)29:01<23::AID-IMMU23>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 66.Gunturi A, Berg RE, Crossley E, Murray S, Forman J. The role of TCR stimulation and TGF-beta in controlling the expression of CD94/NKG2A receptors on CD8 T cells. Eur J Immunol. 2005;35:766–75. doi: 10.1002/eji.200425735. [DOI] [PubMed] [Google Scholar]
- 67.Derre L, Corvaisier M, Pandolfino MC, Diez E, Jotereau F, Gervois N. Expression of CD94/NKG2A on human T lymphocytes is induced by IL-12: implications for adoptive immunotherapy. J Immunol. 2002;168:4864–70. doi: 10.4049/jimmunol.168.10.4864. [DOI] [PubMed] [Google Scholar]
- 68.Ota T, Takeda K, Akiba H, et al. IFN-gamma-mediated negative feedback regulation of NKT-cell function by CD94/NKG2. Blood. 2005;106:184–92. doi: 10.1182/blood-2004-11-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bradley LM, Atkins GG, Swain SL. Long-term CD4+ memory T cells from the spleen lack MEL-14, the lymph node homing receptor. J Immunol. 1992;148:324–31. [PubMed] [Google Scholar]
- 70.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–12. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 71.Brown DM, Román E, Swain SL. CD4 T cell responses to influenza infection. Semin Immunol. 2004;16:171–7. doi: 10.1016/j.smim.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 72.Arlettaz L, Villard J, de Rham C, Degermann S, Chapuis B, Huard B, Roosnek E. Activating CD94. NKG2C and inhibitory CD94: NKG2A receptors are expressed by distinct subsets of committed CD8+ TCRαβ lymphocytes. Eur J Immunol. 2004;34:3456–64. doi: 10.1002/eji.200425210. [DOI] [PubMed] [Google Scholar]
- 73.Chen H-W, Liao C-H, Ying C, Chang C-J, Lin C-M. Ex vivo expansion of dendritic-cell-activated antigen-specific CD4+ T cells with anti-CD3/CD28, interleukin-7, and interleukin-15: potential for adoptive T cell immunity. Clin Immunol. 2006;119:21–31. doi: 10.1016/j.clim.2005.11.003. [DOI] [PubMed] [Google Scholar]