Abstract
Retinoid X receptor (RXR) agonists, including the vitamin A metabolite 9-cis retinoic acid, decrease T-lymphocyte apoptosis and promote T helper type 2 (Th2) development ex vivo. To examine the in vivo role of RXR-α in T-lymphocyte development and function, we disrupted the Rxra gene in thymocytes and T lymphocytes using cyclization recombinase (Cre)-loxP-mediated excision of Rxra exon 4. Expression of Cre was targeted to these cells using the Lck promoter. Successful disruption of exon 4 was seen in thymus and T lymphocytes. Mice were healthy and the thymus, spleen and lymph nodes appeared normal. However, knockout mice had a lower percentage of double-positive (CD4+ CD8+) and a higher percentage of double-negative thymocytes than wild-type mice. The percentage of splenic B lymphocytes was lower in unimmunized and ovalbumin-immunized knockout mice and the percentage of T lymphocytes was lower in immunized knockout mice. Ex vivo proliferation was decreased and apoptosis was increased in T lymphocytes from knockout mice. Memory CD4+ T lymphocytes from knockout mice produced more interferon-γ and interleukin-2 (IL-2) and less IL-5 and IL-10 than memory cells from wild-type mice, indicating a Th1 bias in vivo. However, Rxra disruption did not similarly bias ex vivo differentiation of naive CD4+ T lymphocytes, nor did Rxra disruption alter the serum immunoglobulin G1/immunoglobulin G2a response to immunization. In summary, disruption of Rxra altered the percentages of T and B lymphocytes, produced a Th1 bias in vivo, and altered T-lymphocyte proliferation and apoptosis ex vivo. These differences were modest in magnitude and their impact on disease resistance is yet to be examined.
Keywords: knockout mice, retinoid X receptor-α, T helper type 1/T helper type 2, T lymphocyte, vitamin A
Introduction
Prevention of vitamin A deficiency by vitamin A supplements reduces infant and early childhood mortality from infectious diseases.1 As a result, vitamin A supplementation programmes are widely implemented in areas of the world where vitamin A deficiency is common. The programme in Niger, for example, has recently been described.2 Provision of such supplements presumably corrects deficits in immune function caused by vitamin A deficiency and thereby improves recovery from serious infections. A wide range of pathogenic mechanisms are employed by disease-causing micro-organisms and the resulting immune response to these pathogens is quite complex. Given this complexity it is not surprising that vitamin A supplementation does not uniformly improve recovery from all infections, as has recently been discussed for human immunodeficiency virus3 and measles.4 Thus it is important to understand how vitamin A regulates immunity to maximize the benefit of supplementation in subjects at risk of infectious diseases and to make more informed recommendations on vitamin A requirements for healthy individuals.
Only a few mechanistic studies have examined the effect of vitamin A deficiency on immune function in humans, although several groups have pursued experimental studies in rodents.5 From these studies we have learned that T-lymphocyte-mediated antibody responses are impaired by vitamin A deficiency in humans6 and rodents.7–10 In rodents, a T helper type 1 (Th1) bias is produced by vitamin A deficiency11 that involves increased interleukin-12 (IL-12) production by antigen-presenting cells12 and increased interferon-γ (IFN-γ) production by T lymphocytes,13 although contradictory findings have also been reported.14 It is clear, however, that vitamin A can directly affect antigen-presenting cells and T lymphocytes, as well as B lymphocytes.15 Understanding the role of vitamin A in the immune response will therefore require the dissection of its impact on these individual cell types.
The biological activity of vitamin A is mediated by retinaldehyde and retinoic acid. Retinaldehyde is a component of rhodopsin and is required for the visual cycle16 while retinoic acid regulates gene expression by binding to the retinoic acid receptor (RAR) or the retinoid X receptor (RXR). All-trans retinoic acid binds to RAR while 9-cis retinoic acid is a ligand for both receptors.17 RAR forms an obligate heterodimer with RXR to mediate the transcriptional activity of all-trans retinoic acid. This activity does not require the presence of an RXR agonist. However, the RAR–RXR heterodimer can also be activated by RXR agonists as long as an RAR agonist is also bound to RAR. Thus 9-cis retinoic acid can itself activate RAR–RXR heterodimers, while other RXR agonists, such as docosahexaenoic acid18 or pharmacological agents would need to act in concert with an RAR agonist.19 RXR is also a heterodimer partner for many other nuclear receptors, including the vitamin D receptor, the liver X receptor and the peroxisome proliferator-activated receptor (PPAR). RXR agonists do not activate transcription when RXR is a heterodimeric partner with a non-permissive receptor, such as the vitamin D receptor. However, many nuclear receptors are permissive partners, including PPAR and the NR4A family members that do not themselves bind ligands.20 Both PPAR-γ21 and NR4A family members22 are expressed in T lymphocytes. RXR homodimers are also responsive to RXR agonists.23 Thus transcriptional regulation by vitamin A may be mediated via RAR–RXR heterodimers, RXR homodimers, or via the RXR partner of a permissive heterodimer.
In the present study we examined the role of vitamin A in T-lymphocyte development and function by disrupting the Rxra gene in thymocytes and T lymphocytes. We focused on RXR because 9-cis retinoic acid and the RXR agonist AGN19420424 enhance ex vivo Th2 development25 and decrease apoptosis of naive CD4+ T lymphocytes.26Rxra expression was disrupted using cell-specific expression of cyclization recombinase (Cre)27 to excise Rxra exon 4, which was flanked by loxP sites targeted by the Cre enzyme. Exon 4 encodes the majority of the DNA-binding domain of RXR-α.28 Expression of Cre was targeted to thymocytes and T lymphocytes using the Lck promoter to drive Cre expression.29 Germ-line disruption of Rxra cannot be used to examine immune function because such a homozygous mutation is lethal in utero.17 T lymphocytes express both Rxra and Rxrb26 and we plan to disrupt Rxrb in future experiments.
Materials and methods
Mice
Mice with 34-base-pair insertions containing loxP sites27 flanking exon 4 of the Rxra gene (referred to as ‘floxed’) were obtained from Dr Kenneth Chien of the University of California, San Diego.28 The floxed alleles are fully functional.30 These loxP sites allow deletion of the intervening genomic DNA (referred to as ‘floxing out’) in cells expressing the bacteriophage P1 enzyme cyclization recombinase (cre).27 Exon 4 encodes most of the DNA-binding domain of RXR-α. The floxed mice had a mixed C57BL/6 × 129/Sv genetic background. Mice expressing cre under the control of the promoter from the thymocyte and T-lymphocyte-specific protein tyrosine kinase p56 (Lck) gene29 were obtained from Jackson Laboratories (Bar Harbor, ME; strain name, B6.Cg-Tg(Lck-cre)548Jxm/J, stock no. 003802). These mice were crossed to produce mice that were heterozygous (RXR-αko/wt) or homozygous (RXR-αko/ko) for a cell-specific ‘knock-out’ (ko) of the Rxra gene. Mice without such a cell-specific knockout were considered ‘wild-type’ (RXR-αwt/wt). This designation thus included Cre-negative mice with one or two floxed Rxra alleles as well as Cre-positive or Cre-negative mice with no floxed alleles. These ‘wild-type’ genotypes showed similar responses relative to animals with cell-specific disruption of Rxra (data not shown) and were thus grouped together as described.
Cell line
Murine F9 teratocarcinoma cells (CRL-1720) were obtained from the American Type Culture Collection (Manassas, VA).
Reagents
Antibodies and cytokines were purchased from BD-Pharmingen (Mountain View, CA) unless otherwise indicated. The following antibodies were used for flow cytometric analysis or cell sorting: fluorescein isothiocyanate (FITC)-labelled anti-CD3 (#555274, clone 17A2), phycoerytherin (PE)-labelled anti-CD4 (# 553730, clone GK1.5), peridin chlorophyll-a protein (PerCP)-labelled anti-CD8 (# 553036, clone 53–6·7), allophycocyanin-labelled anti-B220 (# 553092, clone RA3-6B2), PE-labelled anti-NK-1.1 (# 557391, clone PK 136) and FITC-labelled anti-T-cell receptor-β (TCR-β; # 553170, clone H57-597). Rabbit anti-RXR-α antibody (catalogue # SC-553) for Western blots was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Reagents used in cell culture included a neutralizing rat monoclonal antibody for murine IL-4 [clone BVD40-1D11, immunoglobulin G2b (IgG2b) isotype], murine IL-4 and murine IL-12 (BD-Pharmingen). Both 9-cis retinoic acid and cell-culture grade dimethylsulphoxide (DMSO) were purchased from Sigma (St Louis, MO). AGN194204 was provided by Dr Rosh Chandraratna of Allergan (Irvine, CA). Russ-10 cell culture medium was prepared as described previously.25
Haematology, flow cytometric analysis and cell-sorting
Heparinized whole blood was analysed using an ABX Pentra 60 automated cell counter (ABX Diagnostics; Montpellier, France) to determine absolute counts and percentages of total white blood cells, lymphocytes, monocytes, neutrophils, basophils and eosinophils, in addition to erythrocyte counts and haemoglobin concentrations. Flow cytometric analysis of whole blood and cell suspensions from tissues was performed on a four-colour, FACSCalibur flow cytometer using CellQuest software (Beckton Dickenson, San Jose, CA). Erythrocytes in whole blood were removed by hypotonic lysis before flow cytometric analysis. Thymocytes were stained with antibodies specific for CD4 and CD8, while whole blood, splenocytes and lymph node cells were stained with antibodies specific for CD3, CD4, CD8 and B220. The percentages of cells in the lymphocyte gate (by forward-scatter, side-scatter analysis) that were positive for each surface antibody stain were determined. The absolute count of lymphocytes from whole blood was used to determine the absolute count of B and T lymphocytes. Subsets of thymocytes and splenocytes were isolated using a MoFlo (Cytomations, Fort Collins, CO) high-speed cell sorter. We routinely obtain populations that were > 96% pure upon re-analysis. T lymphocytes for some analyses were positively selected from splenocytes using anti-Thy-1.2 coated magnetic beads (Dynal-Invitrogen, Carlsbad, CA) according to the manufacturer's instructions.
Polymerase chain reaction analysis of genotype
Genotyping was typically performed on tail DNA from weanling mice. The presence of floxed alleles was identified by polymerase chain reaction (PCR) analysis of genomic DNA using three primers (P1, P2 and P3) that have been described elsewhere.28 P1 and P3 are outside the floxed regions on the 5′ and 3′ sides, respectively. The P1–P3 product from a floxed Rxra allele is approximately 1·5 kilobases (kb) and is 68 base pairs longer than the product from a wild type allele. When the intervening region is floxed-out by the Cre enzyme the P1–P3 product is approximately 0·45 kb. We used primers suggested by Dr Chien's laboratory to identify mice carrying the cre gene (cre-F, 5′-GTTCGCAAGAACCTGATGGACA-3′; cre-R, 5′-CTAGAGCCTGTTTTGCACGTTG-3′). Reaction conditions for the cre primers were as follows: one cycle at 94°, 3 min; 40 cycles at 95° for 30 seconds, 60° for 30 seconds, 72° for 1 min; one cycle at 72° for 8 min. Samples were analysed by agarose gel electrophoresis or using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA) with DNA 1000 or DNA 7500 reagents, and bio sizing software (version A.02.11). The quantity of DNA in each band was assessed by scanning fluorometric analysis using the Bioanalyzer.
Western blot analysis
Cells were washed in cold phosphate-buffered saline (PBS) containing 0·5 mm phenylmethylsulphonyl fluoride (PMSF) before protein extraction. Whole-cell protein extracts were prepared as follows: washed cells were resuspended in lysis buffer (1% nonidet-P40, 30 mm Tris–HCl pH 7·5, 0·5 mm ethylenediaminetetraacetic acid (EDTA) pH 8·0, 150 mm NaCl, 10% glycerol, 0·5 mm PMSF, 1:500 dilution of protease inhibitor solution) and were incubated for 5 min on ice. The protease inhibitor solution consisted of one complete protease inhibitor tablet (Roche Diagnostics, Indianapolis, IN) dissolved in 250 μl water. 5 m NaCl was added to a final concentration of 400 mm and cells were incubated for an additional 10 min on ice. Cells were then passed through a Qiashredder (Qiagen, Valencia, CA) by centrifugation for 10 min at 20 800 g at 4°. Nuclear extracts were prepared as follows: washed cells were resuspended in 100 μl solution A (0·5% nonidet-P40, 10 mm HEPES pH 7·9, 1·5 mm MgCl2, 10 mm KCl) per million cells and incubated for 5 min on ice. Samples were then pelleted by centrifugation at 6800 g at 4° for 5 min. The pellet was resuspended in 25 μl solution B (20 mm HEPES pH 7·9, 25% glycerol, 400 mm NaCl, 1·5 mm MgCl2, 0·5 mm EDTA pH 8, 0·2 mm PMSF, 0·5 mm dithiothreitol, 1:500 dilution of protease inhibitor solution) per million cells. Samples were frozen and thawed three times and passed through a Qiashredder as described above.
Proteins were resolved by sodium dodecyl sulphate–polyacrylamide gel electrophoresis on 4–12% gradient gels (Novex, Carlsbad, CA) and transferred to nitrocellulose (Invitrogen, Carlsbad, CA). Blots were blocked using 10% skimmed milk powder in PBS containing 0·1% Tween (Sigma) (PBST) overnight at 4°. Blots were washed three times with PBST and were blocked again in 5% goat serum (Jackson Immuno Research Laboratories, Inc.; West Grove, PA) in PBST overnight at room temperature. Each blot was then probed using appropriate antibodies diluted at 1:500 at room temperature for 1·5 hr and washed three times with PBST. After washing, the secondary antibody diluted at 1:20 000 was added and incubation lasted another 1·5 hr at room temperature. Finally, the blots were washed three times with PBST and developed using the enhanced chemiluminescence kit (Amersham/Pharmacia Biotech, Piscataway, NJ).
Histopathology
After gross examination the following tissues were collected for microscopic examination: heart, lung, thymus, spleen, liver, intestine with Peyer's patches, lymph nodes (brachial, axillary, inguinal, popliteal, and mesenteric), gastrocnemius muscle with a femur cross-section to examine bone marrow. Tissues were fixed in formalin and the sample containing bone was de-mineralized. Tissues were embedded in paraffin, sectioned, stained with haematoxylin and eosin, and examined microscopically for pathological changes. Tissues were examined first without knowing the genotype and then re-examined with that knowledge.
Immunization
In some experiments mice were immunized with the T-lymphocyte-dependent antigen ovalbumin-dinitrophenol (OVA-DNP) (# D-5051–100 Biosearch Technologies, Inc., Novato, CA). For the first immunization OVA-DNP was diluted in PBS and emulsified with complete Freund's adjuvant (CFA; Sigma). The final injection volume was 0·3 ml per mouse and contained 50 μg OVA-DNP. Mice were injected subcutaneously in both hind footpads (50 μl per site) at the base of the tail (100 μl) and the back of the neck (100 μl). A second immunization was given 2 weeks later using incomplete Freund's adjuvant (IFA). Mice were killed for analysis 1 week after the second immunization.
Cytokine measurements
IL-2, IL-4, IL-5, IL-10, tumour necrosis factor-α (TNF-α) and IFN-γ were measured using Beadlyte Mouse Multi-Cytokine Flex Kit reagents (Upstate Cell Signaling Solutions, Charlottesville, VA) and a Bioplex Suspension Array System (Bio-Rad, Hercules, CA).
Serum IgG enzyme-linked immunosorbent assay (ELISA)
To quantify DNP-specific antibodies, serially diluted serum samples were added to 96-well flat-bottomed plates coated with DNP-conjugated bovine serum albumin (#NC9979122 Fischer Scientific, Pittsburg, PA) in PBS. After washing, bound antibodies were detected using biotin-conjugated goat anti-mouse IgG1 (# M32115), IgG2a (# M32315) and total IgG (# M30115) from Caltag (Burlingame, CA) followed by strepavidin-conjugated horseradish peroxidase (Amersham Biosciences, Piscataway, NJ) and tetramethylbenzidine (TMB) substrate (BD-Pharmingen). Absorbance was measured at 450 nm using an ELX 800 microplate reader (Bio-Tek Instruments, Winooski, VT). A standard curve was created using serial dilutions of a mouse serum with known concentrations of antibody isotypes (MP Biomedicals, Inc., Irvine, CA; # 64901) and wells precoated with goat anti-mouse immunoglobulin (# M30900, Caltag).
CFSE staining to measure cell division
Carboxyfluorescein diacetate, succinimidyl ester (CFSE) was purchased from Molecular Probes (# V12883; Eugene, OR). Cells were adjusted to a concentration of 1 × 106 cells/ml in PBS containing 4% fetal bovine serum and were mixed with an equal volume of 10 μm freshly diluted CFSE in PBS. Cells were then incubated at room temperature on a rocker for 10 min. The CFSE-stained cells were washed with excess Russ-10 medium and were resuspended in Russ-10 at 1 × 106 cells/ml for stimulation. Unstimulated and anti-CD3/anti-CD28-stimulated cultures were evaluated for each mouse at each time-point. The number of cell divisions at each time-point was determined by subtracting the log2 median CFSE intensity for the stimulated culture from the log2 median CFSE intensity for the unstimulated culture.
Measuring proliferation by bromodeoxyuridine incorporation
Splenocytes at 1 × 106/ml in Russ10 medium were stimulated with diluent, concanavalin A at 0·1, 0·3, 1·0 and 3·0 μg/ml, and anti-CD3 plus anti-CD28 antibodies at 1 μg/ml. Bromodeoxyuridine (BrdU) was added for 4 hr on the 3rd day after stimulation and incorporation was measured by ELISA according to the manufacturer's instructions (Roche Cell Proliferation ELISA, Indianapolis, IN).
Annexin V staining to measure apoptosis
Samples of stimulated lymphocyte cultures were stained according to the manufacturer's instructions with PE-Annexin V (# 556421 BD-Pharmingen) to identify apoptotic cells and 7-amino-actinomycin D (7-AAD) (# 559925 BD-Pharmingen) to identify dead cells. In some experiments cells were surface-stained with anti-CD4 and anti-CD8 for 15 min at 4° and washed once with PBS before staining with Annexin V.
Ex vivo Th1/Th2 polarization and intracellular cytokine staining
Naive CD4 T lymphocytes (CD4+ CD62Lhigh) were isolated using the MoFlo cell sorter. Cells were stimulated using plate-bound anti-CD3 (10 μg/ml) and anti-CD28 (10 μg/ml) antibodies under Th2 conditions (10 ng/ml IL-4), Th1 conditions (5 μg/ml anti-IL-4 plus 0·2 ng/ml IL-12), with 100 nm AGN194204 and with diluent only (0·1% DMSO). Cultures were expanded after 3 days by transferring to a fresh well and adding two volumes of Russ-10 medium. On day 7, cells were washed, resuspended at 1 × 106 cells/ml, stimulated for 5 hr with phorbol 12-myristate 13-acetate and ionomycin, treated with brefeldin A for the final 3 hr of culture to block cytokine secretion, and examined for intracellular IL-4 and IFN-γ by flow cytometry on a FACSCalibur as described.14
Experiments
Mice were matched as pairs (homozygous and wild-type) or as groups of three animals (heterozygous, homozygous and wild-type) by litter and gender whenever possible. Mice were aged between 4 and 8 months.
Experiment 1
The goal of this experiment was to demonstrate floxing-out of exon 4 in sorted thymocytes and lymphocytes, to identify histopathological and haematological changes, and to examine thymocyte (CD4/CD8) and lymphocyte (CD3/CD4/CD8/B220) phenotype from the blood and spleen by flow cytometric analysis. This experiment used 15 wild-type mice (eight males; six Cre– RXR-αfloxed/not floxed, two Cre– RXR-αnot floxed/not floxed, six Crenot floxed RXR-αnot floxed/not floxed), 10 heterozygous mice (five males) and 11 homozygous mice (six males).
Experiment 2(a)
The goal of this experiment was to examine the serum antibody and cytokine responses to immunization with the T-lymphocyte-dependent antigen OVA-DNP. Mice were killed for analysis 1 week after the second immunization. Flow cytometry was used to examine thymocyte (CD4/CD8) and lymphocyte (CD3/CD4/CD8/B220; NK1.1/TCR-α) phenotypes. Thy-1.2-selected splenocytes were used for Western blot analysis and to confirm floxing-out of exon 4 of RXR-α. Draining lymph node cells were stimulated with ovalbumin (100 μg/ml) or anti-CD3 plus anti-CD28 antibodies (1 μg/ml of each) to assess cytokine production. Cultures treated with anti-CD3 plus anti-CD28 were also used to assess proliferation (by counting viable cells and with CFSE staining) and apoptosis (by Annexin V staining). This experiment used five wild-type mice (two males; two Cre– RXR-αfl/+, three Cre+ RXR-α+/+), five heterozygous mice (all females) and 11 homozygous mice (two males). Mice were immunized and tissues were processed in groups of three (one of each genotype) that were matched as best as possible by litter and gender. If mice from different litters were used they were matched by age as far as possible. This pairing minimized variation due to gender, age and differences in genetic background because mice had a mixed C57BL/6 × 129/Sv genetic background.
Experiment 2(b)
This experiment used seven wild-type mice (three males; five Cre– RXR-αfl/+, two Cre+ RXR-α+/+) and seven homozygous mice (three males) and involved the same analyses that were performed in Experiment 2, with the exception that BrdU incorporation was used to measure mitogen-stimulated proliferation. Lymphocyte phenotype analysis was also performed on non-draining lymph nodes (mesenteric and Peyer's patch).
Experiment 3
In this experiment five homozygous knockout and five wild-type mice (two male and three female for each genotype; all five wild-types were Cre+ RXR-α+/+) were used to examine the effect of Rxra disruption on proliferation (measured by CFSE staining) and apoptosis (measured by Annexin V staining) on CD4 and CD8 T lymphocytes in anti-CD3/anti-CD28-stimulated splenocyte cultures.
Experiment 4
In this experiment four homozygous knockout and four wild-type mice (three male and one female for each genotype; wild-types included one Cre– RXR-αfl/+, two Cre– RXR-α+/+ and one Cre+ RXR-α+/+) were used to examine the effect of Rxra disruption on Th1/Th2 development. Naive and memory CD4+ T lymphocytes were sorted and stimulated as described above.
Statistical analysis
Statistical analysis was performed using SigmaStat for Windows (version 3·10, Systat Software Inc., Point Richmond, CA). Data are expressed as mean ± standard error (SE) unless otherwise indicated and the P-value for statistical significance was < 0·05 using two-tailed tests. Variables (e.g. flow cytometry data, organ weights) were initially compared for an effect of genotype (heterozygous, homozygous, wild-type) using two-way analysis of variance (anova) to control for gender. If no gender effect was seen, one-way anova was used to compare variables among the three genotypes. Significance tests in anova were made versus the wild-type group using the Holm–Sidak method to control for multiple comparisons. In addition, Student's t-test was used to directly compare homozygous and wild-type animals. In experiments where animals were paired by litter (or age) and gender some comparisons were made using paired t-tests. Two-way anova was also used with Annexin V, CFSE and other data to compare genotypes while controlling for interexperiment variation because data from experiments performed at different times were pooled for analysis. When measurements were made on multiple days statistical comparisons were typically made separately on data from each day. In some cases two-way repeated measures anova was used to compare data from multiple days. Three-way anova was used to compare genotype and a second variable, such as retinoid treatment, while controlling for interexperiment variation.
Results
Generation of mice with floxed-out exon 4 of RXR-α in thymocytes and T lymphocytes
Crosses between lck-cre mice and floxed RXR-α mice were used to produce heterozygous (RXR-αko/wt) and homozygous (RXR-αko/ko) knockout animals, as well as wild-type controls (RXR-αwt/wt). No difficulties in breeding were noted, litter sizes were normal and body weights did not differ by genotype up to 6 months of age (data not shown).
Exon 4 disruption in thymocytes and T lymphocytes
To determine if exon 4 was floxed-out as expected in the thymus, thymocytes from seven homozygous mice, seven heterozygous mice and one wild-type mouse were stained for surface expression of CD4 and CD8 and were sorted into populations of double-positive and single-positive cells. PCR primers flanking the floxed region were used for genomic DNA analysis. In six of seven homozygous mice there was a complete or nearly complete disruption of exon 4 in all double- and single-positive thymocytes (Fig. 1a). That is, the 1·5-kb PCR product representing the floxed allele was not seen (or was barely detectable) but the 0·45-kb product representing the floxed-out allele was seen in six of seven cases. Similarly, in six of seven heterozygous mice the PCR product representing the floxed-out allele was seen in conjunction with the PCR product representing the wild-type allele. Splenocytes from these same 15 mice were stained for surface expression of CD3, CD4, CD8 and B220 to identify CD4+ T lymphocytes (CD3+ CD4+), CD8+ T lymphocytes (CD3+ CD8+) and B lymphocytes (CD3– B220+). The genotyping results for the splenocytes corresponded exactly to the results seen in the thymus, with the same two mice showing anomalous results (i.e. no floxing-out in thymocytes or T lymphocytes). In the remaining 12 of 14 homozygous and heterozygous mice, a complete or nearly complete excision of the floxed allele was seen in both CD4+ and CD8+ T lymphocytes with no detectable excision seen in B lymphocytes. Exon 4 was not deleted in thymocytes or lymphocytes from the wild-type mouse. Genotyping was also performed on unsorted thymocytes and on splenic T lymphocytes selected using magnetic beads labelled with anti-Thy-1.2 antibody. Purity of Thy-1.2-selected T lymphocytes could not be assessed by flow cytometry. These analyses were performed on samples from an additional 23 homozygous or heterozygous mice and the anticipated excision of exon 4 was seen in thymocytes and T lymphocytes in all cases. Thus excision of exon 4 from the floxed allele was observed, as expected, in a total of 35 of the first 37 homozygous or heterozygous mice analysed (95%). The lineage producing anomalous results was not used for further propagation of this line and these anomalies did not recur.
Figure 1.
Decreased Rxra expression in thymocytes and T lymphocytes from cell-specific knockout mice. (a) PCR analysis of genomic DNA confirms floxing-out of exon 4 of the Rxra gene in thymocytes and T lymphocytes but not B lymphocytes from lck-Cre mice with floxed RXR-α allele. Thymocytes from one homozygous (RXR-αko/ko) and one heterozygous (RXR-αwt/ko) mouse were sorted into pools of single-positive (CD4, CD8) and double-positive (DP) cells. Splenocytes from the same mice were sorted into pools of CD4+ T lymphocytes (CD4; CD4+ CD3+), CD8+ T lymphocytes (CD8; CD8+ CD3+) and B lymphocytes (B; B220+ CD3–). As described in the Materials and methods, the floxed (flox), wild-type (wt) and floxed-out (fo) RXR-α alleles were identified with PCR primers (P1 and P3) flanking the floxed region of Rxra. Data are representative of six homozygous and six heterozygous mice sorted in this fashion. Molecular size standards (in base pairs) are shown at the far left. (b) Western blot analysis confirms decreased RXR-α expression in thymocytes and T lymphocytes from knockout mice. Whole-cell protein extracts (10 μg/lane) of total thymocytes (Th), T lymphocytes isolated from splenocytes with Thy-1.2-selective magnetic beads (+), and Thy-1.2-negative splenocytes (–) were analysed by Western blot using an RXR-α-specific antibody. Cell extracts were from one homozygous (RXR-αko/ko) and one wild-type (RXR-αwt/wt) mouse. A nuclear protein extract (10 μg) from F9 (F9) embryocarcinoma cells was included as a positive control for RXR-α, which runs at approximately 54 000 and is indicate by an asterisk (*). Results are representative of blots from six homozygous and five wild-type mice. Molecular size standards are indicated at left. (c) Bar graph showing mean + SE of the intensity of staining for RXR-α by Western blot of Thy-1.2-positive splenic T lymphocytes from six homozygous and five wild-type mice, and from unselected thymocytes of two homozygous and two wild-type mice. The means differed between genotypes at *P =0·013 using paired Student's t-test. Samples of the same cell type from the same blot (which contained the same amount of protein per lane) were paired for analysis. One blot contained T-lymphocyte samples from two homozygous mice and one wild-type mouse and the band intensities for the homozygous knockout mice were averaged for statistical analysis.
RXR-α protein levels in whole thymus and Thy-1.2+ splenocytes
To determine if RXR-α protein production was lower in the thymus and splenic T lymphocytes from knockout mice than in the corresponding cell types from wild-type mice, Western blot analysis was performed on thymus (thymocytes were not separated from stromal cells) and Thy-1.2-selected splenocytes. The RXR-α band (indicated by an asterisk in Fig. 1b) was identified in all T-lymphocyte samples (n = 11) and in four of seven thymus samples (two homozygous knockout and one wild-type sample did not have a detectable band). Production of RXR-α protein was greater in splenic T lymphocytes than in thymus and was often not detected in splenic non-T lymphocytes. In homozygous knockout mice the expression of RXR-α was lower in both T lymphocytes (42% of wild-type) and thymus (54% of wild-type using data from two knockout and two wild-type mice with visible bands) than it was in the corresponding sample from wild-type mice (Fig. 1c). Thus floxing-out exon 4 of RXR-α using the Lck gene promoter to drive cre expression significantly decreased RXR-α protein levels in both thymus and T lymphocytes.
Effect of Rxra disruption on lymphoid tissues of unimmunized mice
Histopathology and haematology
Tissues from 15 wild-type mice (eight male), 10 heterozygous mice (five male) and 11 homozygous mice (six male) were examined microscopically for histopathological changes. The mean age of these 36 mice was 6·0 months (range, 5·0–8·6 months) and did not differ by gender or genotype. The mean body weight (51·5 ± 2·3 g for males and 38·4 ± 2·4 g for females) did not differ by genotype. Spleen and thymus weights and the number of cells per gram of tissue did not differ by genotype (data not shown). Microscopic examination revealed only occasional histopathological changes in thymus, spleen, lymph nodes and other tissues, including variation in secondary lymphoid follicle development, and variation in the splenic white pulp size. However, differences did not correlate with genotypes. Haematological examination did not reveal differences among the genotypes in counts of erythrocytes, total white blood cells, granulocytes, monocytes or lymphocytes, or in haemoglobin concentration or haematocrit (data not shown).
Flow cytometry of thymus, blood and spleen
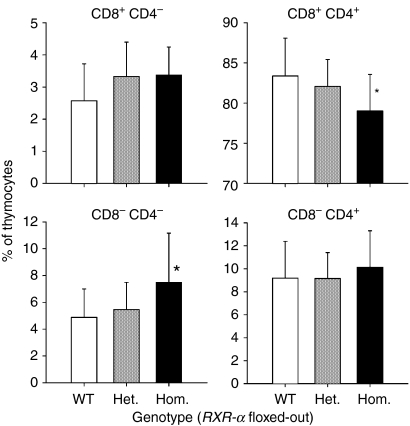
Homozygous knockout mice had a higher percentage of CD4– CD8– double-negative thymocytes (53% higher; P = 0·032 by Student's t-test) and a lower percentage of CD4+ CD8+ double-positive thymocytes (5% lower; P = 0·027 by Student's t-test) than did wild-type mice (Fig. 2). In blood, the number of CD3+ T lymphocytes and B220+ B lymphocytes did not differ by genotype (Fig. 3a). In the spleen the percentage of B220+ B lymphocytes differed among the three genotypes (P = 0·037 by one-way anova), with the homozygous mice having a lower percentage (24% lower; P = 0·024 by one-way anova) than the wild-type mice (Fig. 3a). However, the ratio of B lymphocytes to T lymphocytes in the spleen did not differ by genotype (the mean ± SD for wild-type, heterozygote and homozygote were 1·61 ± 0·66, 1·81 ± 0·85 and 1·32 ± 0·59; P = 0·29 by one-way anova). T-lymphocyte subsets were also examined in blood and the spleen (Fig. 3b). The mean percentage of CD8 T lymphocytes differed among the three genotypes with the percentage being lowest in the homozygous knockout mice, although the mean of this group did not differ significantly from the wild-type mean when compared directly. More double-negative T lymphocytes (CD3+ CD4– CD8–) were seen from homozygous animals in both tissues but the difference was only significant for blood.
Figure 2.
Effect of Rxra disruption on thymocyte CD4/CD8 phenotype. Thymocytes from 15 wild-type mice (WT; RXR-αwt/wt), 10 heterozygous mice (Het.; RXR-αko/wt) and 11 homozygous mice (Hom.; RXR-αko/ko) paired by litter and gender were stained with anti-CD4 and anti-CD8 antibodies and examined by flow cytometry. Bars and error bars indicate means + SD. An asterisk indicates a significant difference from the wild-type by Student's t-test.
Figure 3.
Effect of Rxra disruption on whole blood and splenic lymphocyte populations. Samples of whole blood and spleen from 15 wild-type mice (WT; RXR-αwt/wt), 10 heterozygous mice (Het.; RXR-αko/wt) and 11 homozygous mice (Hom.; RXR-αko/ko) paired by litter and gender. (a) Cells were stained with anti-B220, anti-CD3, anti-CD4 and anti-CD8 antibodies and examined by flow cytometric analysis. The absolute count (blood) and percentage (spleen) of B220+ CD3– B lymphocytes and B220– CD3+ T lymphocytes are shown. (b) The absolute count (blood) and percentage (spleen) of B220– CD3+ T lymphocytes single-positive for CD8, double-negative for CD4 and CD8 and single-positive for CD4 are shown. Bars and error bars indicate means + SD. An asterisk (*) above the bar for a particular genotype indicates a significant difference from the corresponding wild-type mean by one-way anova or t-test. The asterisk in the upper left-hand corner of the graph for splenic CD8+ T lymphocytes indicates a significant difference among the means by one-way anova (P = 0·031) but a significant difference was not seen when individual comparisons were made.
Effect of Rxra disruption on lymphocyte numbers, proliferation and serum antibody response in OVA-DNP-immunized mice
Flow cytometry of spleen and draining lymph node cells
Twelve homozygous knockout mice and 12 wild-type mice were immunized with OVA-DNP using CFA followed by IFA. One week after the second immunization cells from spleen and draining lymph nodes were examined by flow cytometry. The percentages of splenic B and T lymphocytes were both lower in the homozygous mice than in the wild-type mice and the percentage of B lymphocytes was also lower in the draining lymph nodes of knockout mice (Table 1). While no differences in CD4+ and CD8+ T lymphocytes were seen in the spleen, a lower percentage of CD8+ T lymphocytes was seen in the draining lymph nodes of knockout animals. In 14 of these 24 mice (seven homozygous knockouts and seven wild-type) B and T lymphocytes were also counted in non-draining lymph nodes (mesenteric and Peyer's patch) but no differences were seen between the genotypes (data not shown). The percentages of natural killer T cells were examined in the spleens and draining lymph nodes of five homozygous and five wild-type mice but differences were not seen between the genotypes (data not shown).
Table 1.
T-lymphocyte and B-lymphocyte percentages and ratios (mean ± SD) in spleen and draining lymph nodes of 12 mice homozygous for disruption of exon 4 of Rxra (RXR-αko/ko) and 12 wild-type mice (RXR-αwt/wt) following immunization with OVA-DNP
| Tissue and cell type | Wild-type | Homozygous | P-value1 |
|---|---|---|---|
| Spleen | |||
| % B220+ B lymphocytes | 35·9 ± 11·0 | 27·3 ± 10·1 | 0·003 |
| % CD3+ T lymphocytes | 29·5 ± 9·4 | 23·1 ± 8·2 | 0·012 |
| B:T ratio | 1·35 ± 0·64 | 1·31 ± 0·69 | 0·730 |
| % CD4+ T lymphocytes | 56·5 ± 9·7 | 54·8 ± 11·1 | 0·712 |
| % CD8+ T lymphocytes | 27·2 ± 7·7 | 24·0 ± 8·5 | 0·289 |
| CD4:CD8 ratio | 2·27 ± 0·88 | 2·71 ± 1·60 | 0·389 |
| Draining lymph nodes | |||
| % B220+ B lymphocytes | 52·0 ± 14·0 | 46·7 ± 11·1 | 0·098 |
| % CD3+ T lymphocytes | 42·1 ± 13·7 | 46·7 ± 11·7 | 0·117 |
| B:T ratio | 1·41 ± 0·63 | 1·12 ± 0·55 | 0·070 |
| % CD4+ T lymphocytes | 44·7 ± 6·2 | 49·2 ± 9·9 | 0·202 |
| % CD8+ T lymphocytes | 45·5 ± 7·8 | 38·1 ± 9·9 | 0·023 |
| CD4:CD8 ratio | 1·03 ± 0·32 | 1·43 ± 0·66 | 0·016 |
Comparison by paired Student's t-test or Wilcoxon signed-rank test.
Antibody and cytokine response to immunization
The median (25th/75th percentile) serum IgG1 response to OVA-DNP was 211 ng/ml (59/675) in wild-type mice and 108 ng/ml (75/372) in knockout mice (P = 0·38). The median IgG2a response was 12 ng/ml (0·2/98) in wild-type mice compared with 25 ng/ml (6/108) in knockout mice (P = 0·85). The total IgG response was 369 ng/ml (89/2063) in wild-type mice compared with 387 ng/ml (139/712) in knockout mice (P = 0·97). Thus the DNP-specific IgG1, IgG2a and total IgG responses did not differ by genotype. Nor did total IgG1, IgG2a and total IgG concentrations differ by genotype (data not shown). Cytokine concentrations (IL-2, IL-4, IL-5, IL-10, IFN-γ and TNF-α) were low in supernatants from cultures of draining lymph node cells 3 days after stimulation with ovalbumin and did not differ from unstimulated control cultures, nor were differences seen by genotype (data not shown).
Mitogen-stimulated cytokine production
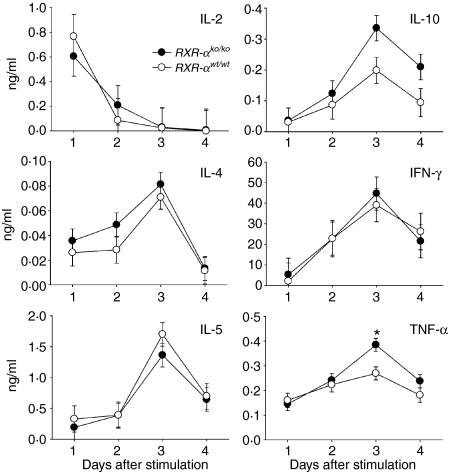
These cytokines were also measured in cultures of the same draining lymph node cells following in vitro stimulation with anti-CD3 plus anti-CD28 antibodies (Fig. 4). The concentrations of TNF-α and IL-10 3 days after stimulation were 43% and 69% greater, respectively, in cultures from knockout mice than in cultures from wild-type mice, although because of the higher variance in the IL-10 data only the difference in TNF-α was statistically significant. The concentrations of the other cytokines did not differ by genotype.
Figure 4.
Effect of Rxra disruption on cytokine production by lymphocyte cultures stimulated with with anti-CD3 (1 μg/ml) plus anti-CD28 (1 μg/ml) antibodies. Data are from seven wild-type mice (RXR-αwt/wt) and seven homozygous knockout mice (RXR-αko/ko) paired by litter and gender, and immunized with ovalbumin-DNP. Cells from draining lymph nodes were cultured for 4 days. The mean ± SE concentrations of cytokines in supernatants are shown. The asterisk indicates significant differences between genotypes by paired Student's t-test (P = 0·014 for TNF-α on day 3).
T-lymphocyte proliferation and apoptosis in response to mitogen stimulation
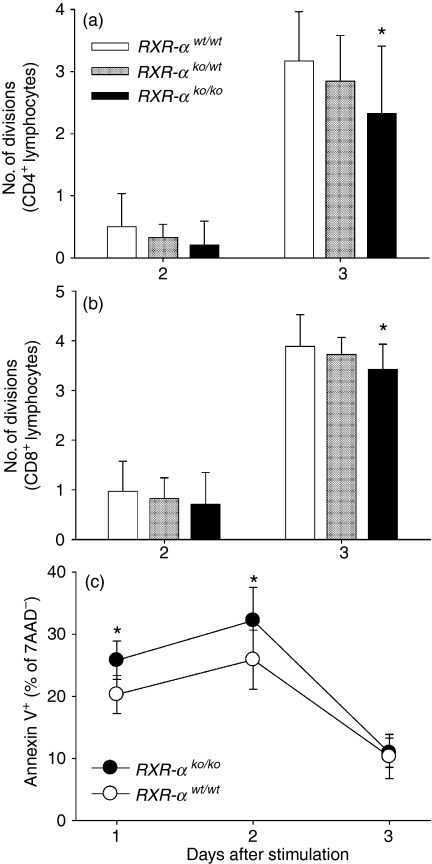
Proliferation was also measured in the draining lymph node cultures stimulated with anti-CD3 plus anti-CD28 antibodies. No differences were seen among the genotypes when total and viable cells were counted using a haemocytometer (data not shown). CFSE staining was also used to characterize proliferation of CD4+ and CD8+ T lymphocytes in these cultures. On day 3 the mean number of cell divisions was significantly lower for both CD4+(Fig. 5a; 27% lower) and CD8+ T lymphocytes (Fig. 5b; 12% lower) from homozygous mice as compared to wild-type mice. The number of cell divisions was intermediate for the heterozygous mice but was not significantly different from the other genotypes.
Figure 5.
Effect of Rxra disruption on lymphocyte division and apoptosis. Data are from five wild-type (RXR-αwt/wt), five heterozygous (RXR-αko/wt) and five homozygous knockout (RXR-αko/ko) mice paired by litter and gender, and immunized with ovalbumin-DNP. Cultures of draining lymph node cells were stimulated with anti-CD3 (1 μg/ml) plus anti-CD28 (1 μg/ml) antibodies added to the supernatant. CD4+ and CD8+ lymphocytes were identified by surface staining of cells in the lymphocyte gate identified by forward-scatter and side-scatter analysis. Cell divisions were counted using CFSE staining. The mean ± SD number of cell divisions 2 and 3 days following stimulation are shown for CD4+ (a) and CD8+ (b) lymphocytes. The asterisks (*) indicate means that are significantly different from the wild-type group (on the same day) by paired Student's t-test (P = 0·028 and 0·021 for CD4+ and CD8+ T lymphocytes, respectively, on day 3). (c) Data are from 10 wild-type (RXR-αwt/wt) and 10 homozygous knockout (RXR-αko/ko) mice immunized with ovalbumin-DNP. Cells from draining lymph nodes were stimulated as described, stained with Annexin V and 7AAD and analysed by flow cytometry. Bars and error bars represent the mean ± SE percentage of 7AAD-negative lymphocytes that were positive for Annexin V. The asterisks indicate significant differences between the groups (on the same day) by paired Student's t-test (P = 0·042 and 0·008 for days 1 and 2, respectively).
Annexin V staining was used as a marker of apoptosis in draining lymph node cultures from homozygous knockout and wild-type mice. On the first and second days after stimulation a significantly greater percentage of viable (judged by 7-AAD staining) lymphocytes from the homozygous knockout mice stained with Annexin V, indicating initiation of apoptosis, as compared to cells from the wild-type mice (Fig. 5c). The percentage of lymphocytes undergoing apoptosis was 27% greater in the homozygous mice than in the wild-type mice on day 1, 24% greater on day 2 and 6% greater on day 3.
In a separate experiment BrdU incorporation was measured to assess DNA synthesis 3 days after mitogen stimulation (using anti-CD3 plus anti-CD28 stimulation as well as concanavalin A) of draining lymph node and splenocyte cultures. No difference was seen between seven homozygous and seven wild-type mice (data not shown).
Effect of Rxra disruption on CD4+ and CD8+ T-lymphocyte proliferation and apoptosis
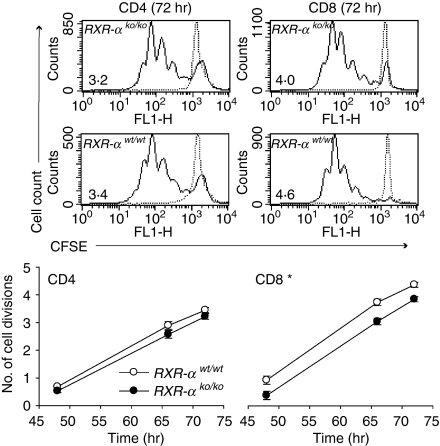
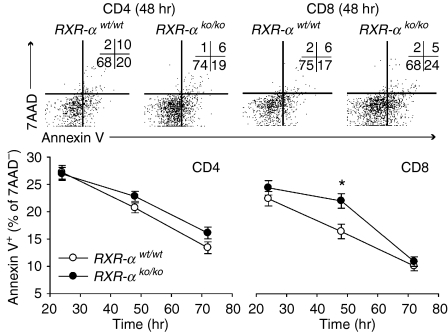
To confirm that Rxra disruption decreases proliferation and increases apoptosis in T lymphocytes from unimmunized mice, cell division was assessed by CFSE staining at 48, 66 and 72 hr and apoptosis was assessed by Annexin V staining at 24, 48 and 72 hr in splenocyte cultures from homozygous knockout and wild-type control mice. In this experiment CD4+ and CD8+ lymphocytes were identified by surface staining. Fewer cell divisions were again seen in both CD4+ and CD8+ T lymphocytes from homozygous knockout mice, although the difference was only statistically significant for CD8+ T lymphocytes (Fig. 6). Similarly a greater percentage of knockout than wild-type CD4+ and CD8+ lymphocytes were Annexin V-positive (Fig. 7), but the difference was again significant only for CD8+ T lymphocytes.
Figure 6.
Effect of Rxra disruption on CD4+ and CD8+ lymphocyte division. Data are from five wild-type (RXR-αwt/wt) and five homozygous knockout (RXR-αko/ko) mice paired by litter and gender. Cultures of splenocytes were stimulated with plate-bound anti-CD3 (10 μg/ml) and anti-CD28 (10 μg/ml) antibodies. CD4+and CD8+ lymphocytes were identified by surface staining of cells in the lymphocyte gate identified by forward-scatter and side-scatter analysis. Cell divisions were counted using CFSE staining. Top panel: CFSE histograms of CD4+ and CD8+ cells from two representative mice 72 hr after stimulation, with the number of cell divisions for each plot shown in the lower left-hand corner. Bottom panel: Mean ± SE number of cell divisions for all five mice of each genotype are shown at 48, 66 and 72 hr. The asterisk indicates that the means for CD8+ cells differed by two-way repeated measures anova (P = 0·022).
Figure 7.
Effect of Rxra disruption on CD4+ and CD8+ lymphocyte apoptosis. Data are from five wild-type (RXR-αwt/wt) and five homozygous knockout (RXR-αko/ko) mice paired by litter and gender. Cultures of splenocytes were stimulated with plate-bound anti-CD3 (10 μg/ml) and anti-CD28 (10 μg/ml). CD4+ and CD8+ lymphocytes were identified by surface staining of cells in the lymphocyte gate identified by forward-scatter and side-scatter analysis. Cells undergoing apoptosis were identified using Annexin V and dead cells were identified using 7AAD. Top panel: Annexin V and CFSE flow analysis plots of CD4+ and CD8+ cells from two representative mice 48 hr after stimulation are shown, with the number of events for each quadrant shown in the upper right-hand corner of each plot. Bottom panel: mean ± SE percentage of viable (7AAD-negative) CD4+ and CD8+ lymphocytes for all five mice of each genotype are shown at 24, 48 and 72 hr. Two-way anova was used to compare genotypes at each time-point (using duplicate measurements for each sample) while controlling for interexperiment variation. The asterisk indicates that the mean for CD8+ cells differed by genotype at 48 hr (P = 0·013).
Effect of Rxra disruption on Th1/Th2 memory phenotype
Cytokine production by memory CD4+ T lymphocytes
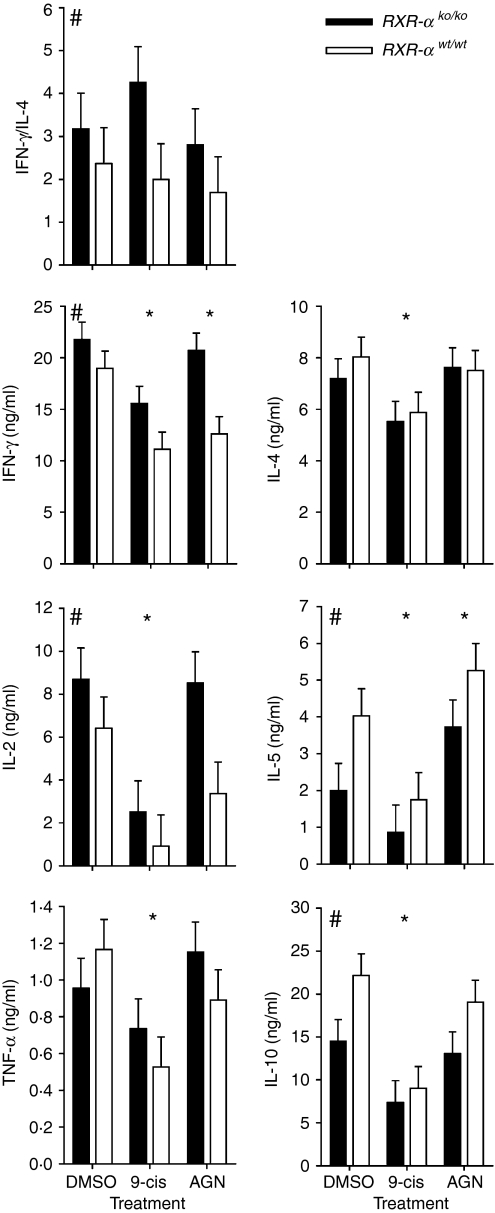
To assess the Th1/Th2 memory phenotype, as indicated by Th1/Th2 cytokine production, naive (CD62Lhigh) and memory (CD62Llow) CD4+ T lymphocytes were sorted and cultured ex vivo for 3 days (memory cells) or 7 days (naive cells) with anti-CD3/anti-CD28 stimulation. Cytokine concentrations were measured in culture supernatants of memory cells, which were also treated with 9-cis retinoic acid, the RXR agonist AGN194204 or vehicle control (Fig. 8). The cytokine profile from cultures of memory CD4+ T lymphocytes showed a bias toward Th1 development in the knockout mice, as the knockout cultures had a significantly higher IFN-γ:IL-4 ratio (3·4 ± 0·5; mean of all treatments) than did cultures from the wild-type mice (2·0 ± 0·5, P = 0·011). Similarly, the concentrations of the Th1 cytokines IFN-γ and IL-2 were significantly greater in supernatants from knockout compared to wild-type cultures (by 36% and 84%, respectively), while the concentrations of the Th2 cytokines IL-5 and IL-10 were both 40% lower in knockout than wild-type cultures. Treatment with the RXR agonist AGN194204 decreased IFN-γ but increased IL-5 concentrations, while 9-cis retinoic acid decreased the concentrations of all cytokines. No statistically significant interactions were seen between genotype and retinoid treatment.
Figure 8.
Effect of Rxra disruption on Th1/Th2 memory cell phenotype. Data are from four wild-type (RXR-αwt/wt) and four homozygous knockout (RXR-αko/ko) mice paired by litter and gender. Memory T-helper lymphocytes (CD3+ CD4+ CD62Llo) were sorted and stimulated with plate-bound anti-CD3 (10 μg/ml) and anti-CD28 (10 μg/ml) antibodies. Cells were also treated with 100 nm 9-cis retinoic acid (9-cis), 100 nm of the RXR agonist AGN194204 (AGN) or vehicle control (DMSO). Cells were cultured for 3 days, then supernatants were collected for measurement of cytokine concentrations. Data were analysed by three-way anova to control for interexperiment variation and to identify differences between genotypes, indicated by the # in the upper left of each graph, and differences between retinoid treatment and the vehicle control are indicated by an asterisk above the appropriate treatment. Bars and error bars represent mean ± SE.
Ex vivo development of Th1/Th2 memory cells
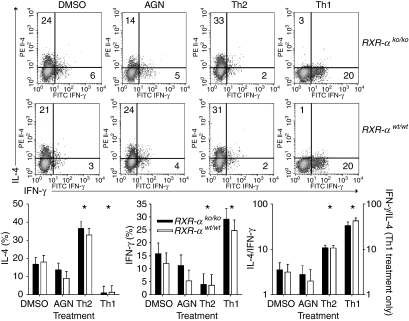
Naive T lymphocytes were treated with anti-CD3 plus anti-CD28 antibodies to stimulate proliferation, and with IL-4 to induce Th2 development, IL-12 plus anti-IL-4 antibody to induce Th1 development, the RXR agonist AGN194204, and vehicle control (Fig. 9). After 7 days in culture the naive Th1/Th2 phenotype was assessed by intracellular cytokine staining. While the Th1- and Th2-promoting treatments both produced the expected polarization, no effect of genotype was seen on Th1/Th2 phenotype development ex vivo. Nor did treatment with the RXR agonist affect Th1/Th2 development.
Figure 9.
Effect of Rxra disruption on ex vivo Th1/Th2 memory cell phenotype development. Data are from four wild-type mice (RXR-αwt/wt) and four homozygous knockout mice (RXR-αko/ko) mice paired by litter and gender. Naive T-helper lymphocytes (CD3+ CD4+ CD62Lhi) were sorted and stimulated with plate-bound anti-CD3 (10 μg/ml) and anti-CD28 (10 μg/ml) antibodies. Cells were also treated with anti-IL-4 (5 μg/ml) plus IL-12 (XX) to promote Th1 development (Th1), IL-4 (XX) to promote Th2 development (Th2), 100 nm of the RXR agonist AGN194204 (AGN) or vehicle control (DMSO). Cells were expanded after 3 days and at 7 days intracellular IL-4 and IFN-γ were measured by flow cytometry following stimulation with phorbol 12-myristate 13-acetate and ionomycin. Top panel: representative scatter plots are shown for two mice, with the percentage of cells positive for each cytokine (and negative for the other) shown in the appropriate quadrant. Bottom panel: bars and error bars represent mean ± SE for the percentage of cells positive for each cytokine, and the ratio of these percentages. Data were analysed by three-way anova to control for interexperiment variation and to identify differences between genotypes, which were not seen, and differences between retinoid treatment and the vehicle control, which are indicated by the asterisk above the appropriate treatment.
Discussion
The floxed RXR-α mouse strain used here was previously used to successfully disrupt RXR-α expression in cardiac myocytes,28 other cardiac cell types31 and hepatocytes30 using Cre. In the present study, using PCR analysis of genomic DNA from FACS-purified thymocytes and T lymphocytes, the extent of Cre-mediated disruption of exon 4 appeared to be complete. Although RXR-α protein expression was significantly reduced in the thymus and in splenic T lymphocytes, higher than expected residual protein expression was detected, suggesting that the gene knockout was incomplete. Alternatively, residual RXR-α protein synthesized before Rxra gene disruption in thymocytes, and/or RXR-α produced by thymic stromal cells not targeted by the Cre excision, may have contributed, in some degree, to the persistent background levels of RXR-α protein in the thymus. These two factors were presumably less important in Western blot analysis of the more mature and more highly purified splenic T lymphocytes. The range of RXR-α protein expression in T lymphocytes varied from 8% to 74% of matched controls. Cell-to-cell variability in disruption of lck-Cre-targeted genes as well as enhanced survival of T lymphocytes that ‘escape’ gene disruption may have contributed to our observations. Earlier studies of lck-Cre-targeted genes have reported similar findings in peripheral T lymphocytes, including a 25% residual presence for the intact integrin-like kinase gene,32 7–34% for the intact N-acetylgalactosaminyl transferase gene29 and 59–86% for the intact core binding factor beta gene.33 Future studies would be needed to evaluate this hypothesis for RXR-α. In any case, our present model represents a significant, but not complete, disruption of Rxra gene expression in T lymphocytes.
Thymus size and cell number did not differ between homozygous knockout and wild-type mice, indicating that Rxra disruption, which would be initiated in double-negative thymocytes using the lck gene promoter,34 did not substantially reduce thymocyte survival. However, an increase in double-negative and a decrease in double-positive thymocyte percentages suggest that some aspect of thymocyte development has been altered. Decreased RXR-α expression may delay maturation from the double-negative stage to the double-positive stage. However, such a disruption might also result in lower percentages of single-positive cells, which was not observed. Future studies will be needed to clarify the impact of Rxra disruption on thymocyte proliferation and survival.
The percentages of T and B lymphocytes in spleen and lymph nodes were altered by Rxra disruption, although the total number of lymphocytes was not affected. These data indicate that T-lymphocyte survival or distribution is altered by Rxra disruption. The decrease in T lymphocytes tended to be greater in the immunized than unimmunized mice, raising the possibility that the effect of Rxra disruption may be more pronounced in proliferating cells, or in cells exposed to paracrine signalling molecules in lymphoid tissue during an active immune response. The lower percentages of B lymphocytes suggest that some aspect of T-lymphocyte help for B-lymphocyte growth and development may be altered. A switch toward a Th1 phenotype could diminish cytokine help for B-lymphocyte development, which is provided by Th2 cytokines, particularly IL-4.35,36 Thus a negative effect of Rxra disruption in T lymphocytes on B-lymphocyte numbers is not unexpected.
Previous studies have shown that RXR agonists modulate important lymphocyte signalling pathways37 and increase survival of T lymphocytes.38,39 In the present study Rxra disruption decreased T-lymphocyte proliferation and increased apoptosis. CFSE analysis revealed decreased proliferation of both CD4+ and CD8+ lymphocytes, although the difference was seen more consistently in CD8+ cells. In addition, a greater percentage of proliferating T lymphocytes from knockout mice were apoptotic as compared to wild-type controls. These data suggest that increased apoptosis in the knockout mice may be responsible, at least in part, for the decreased proliferation also seen in these cultures. We have previously shown that Bcl2a1, an anti-apoptotic gene found in T lymphocytes, is regulated by RXR agonists.26 It is thus possible that decreased Bcl2a1 expression in knockout mice may be responsible for the increased apoptosis seen in these studies. Future studies will examine this hypothesis.
Previous work has shown that ex vivo treatment of purified, naive T lymphocytes with RXR agonists promotes Th2 development25 and that treatment with an RXR antagonist does not block Th2 development under permissive conditions (i.e. the presence of IL-4).40 A recent study used mice in which a point mutation in Rxra results in a 90% decrease in RXR-α transcriptional activating activity.41 This mutation did not block Th2 development of purified, naive T lymphocytes cultured under Th2-permissive conditions but did promote Th1 development under Th1-skewing or mixed Th1/Th2 conditions. These studies indicate that stimulation of RXR in T lymphocytes promotes Th2 development under permissive conditions but disrupting RXR signalling (although not completely eliminating it) does not block Th2 development under the same conditions.
Based on these data we predicted that Rxra disruption in T lymphocytes would impair Th2 development and promote Th1 development, although this effect might be minimized by Th2-promoting conditions. Results from these experiments support a modest effect of Rxra disruption on blocking Th2 development in vivo. The principal evidence for this comes from two observations. First, mitogen-stimulated production of TNF-α, a cytokine produced by Th1 memory cells, was higher in splenocyte cultures from knockout mice than from wild-type mice. Second, cytokine production by purified memory CD4+ T lymphocytes from knockout mice had a marked Th1 bias (i.e. increased IFN-γ and IL-2, decreased IL-5 and IL-10) relative to those from wild-type mice. On the other hand, no skewing of the IgG1/IgG2a antibody response was seen. In addition, ex vivo development of naive into memory Th1/Th2 cells was not altered by Rxra disruption. This may have resulted from residual expression of RXR-α protein, as well as expression of RXR-β, which is also found in T lymphocytes.26 In summary, these findings indicate that disruption of the Rxra gene in T lymphocytes produces a modest bias toward development of a Th1 memory phenotype in vivo.
In conclusion, we have demonstrated that disruption of Rxra in T lymphocytes modestly promoted Th1 development in vivo, presumably by decreasing the expression of one or a few Th2-promoting genes regulated by RXR homo- or heterodimers. We have also shown that T-lymphocyte proliferation and survival ex vivo following TCR stimulation is diminished by Rxra disruption. This decreased survival may result from altered transcriptional regulation of an apoptosis-related gene. One candidate is the anti-apoptotic gene Bcl2a1, which is up-regulated by RXR agonists in T lymphocytes.26 This study represents the first attempt to examine the cell-specific role of RXR in cells of the immune system using in vivo methods. Examining the role of RXR in this manner may reveal novel approaches to therapeutically modulate the immune response to Th1/Th2-mediated infectious or chronic inflammatory diseases. Such an approach might use pharmacological RXR agonists or antagonists or, perhaps, may involve one of the many nutrients (or their metabolites) that act as agonists for this pathway.
Acknowledgments
The authors thank Xiaowen Jiang and Alina Wettstein for their technical expertise and diligence in performing many of these experiments, and Paula Do for her assistance in genotyping the mice used in these studies. This work was supported by NIH grant no. R01 AI 50863, USDA CRIS Project no. 5306-51530-006-00D, and USDA co-operative agreement no. 585306-0-233. The U.C. Davis Mouse Biology Program assisted with the development of transgenic mice. Fluorescence-activated cell sorting was conducted in a facility constructed with support from Research Facilities Improvement Program Grant no. C06 RR-12088–01 from the National Center for Research Resources, NIH.
Abbreviations
- 7-AAD
7-amino-actinomycin D
- anova
analysis of variance
- BrdU
bromodeoxyuridine
- CFA
complete Freund's adjuvant
- CFSE
carboxyfluorescein diacetate, succinimidyl ester
- DMSO
dimethylsulphoxide
- EDTA
ethylenediaminetetraacetic acid
- ELISA
enzyme-linked immunosorbent assay
- FITC
fluorescein isothiocyanate
- IFA
incomplete Freund's adjuvant
- IFN
interferon
- IL
interleukin
- OVA-DNP
ovalbumin–dinitrophenol
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- PE
phycoerytherin
- PMSF
phenylmethylsulphonyl fluoride
- PPAR
peroxisome proliferator-activated receptor
- RAR
retinoic acid receptor
- RXR
retinoid X receptor
- Th1
T helper 1
- Th2
T helper 2
- TNF-α
tumour necrosis factor-α
- Tween
polyoxyethylenesorbitan monolaurate.
References
- 1.Villamor E, Fawzi WW. Vitamin A supplementation: implications for morbidity and mortality in children. J Infect Dis. 2000;182(Suppl. 1):S122–33. doi: 10.1086/315921. [DOI] [PubMed] [Google Scholar]
- 2.Aguayo VM, Baker SK, Crespin X, Hamani H, Mamadou I, Taibou A. Maintaining high vitamin A supplementation coverage in children: lessons from Niger. Food Nutr Bull. 2005;26:26–31. doi: 10.1177/156482650502600103. [DOI] [PubMed] [Google Scholar]
- 3.Wiysonge CS, Shey MS, Sterne JA, Brocklehurst P. Vitamin A supplementation for reducing the risk of mother-to-child transmission of HIV infection. Cochrane Database Syst Rev. 2005 doi: 10.1002/14651858.CD003648.pub2. CD003648. [DOI] [PubMed] [Google Scholar]
- 4.Huiming Y, Chaomin W, Meng M. Vitamin A for treating measles in children. Cochrane Database Syst Rev. 2005 doi: 10.1002/14651858.CD001479.pub3. CD001479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stephensen CB. Vitamin A, infection, and immune function. Annu Rev Nutr. 2001;21:167–92. doi: 10.1146/annurev.nutr.21.1.167. [DOI] [PubMed] [Google Scholar]
- 6.Semba RD, Muhilal Scott AL, et al. Depressed immune response to tetanus in children with vitamin A deficiency. J Nutr. 1992;122:101–7. doi: 10.1093/jn/122.1.101. [DOI] [PubMed] [Google Scholar]
- 7.Stephensen CB, Moldoveanu Z, Gangopadhyay NN. Vitamin A deficiency diminishes the salivary immunoglobulin A response and enhances the serum immunoglobulin G response to influenza A virus infection in BALB/c mice. J Nutr. 1996;126:94–102. doi: 10.1093/jn/126.1.94. [DOI] [PubMed] [Google Scholar]
- 8.Wiedermann U, Hanson LA, Kahu H, Dahlgren UI. Aberrant T-cell function in vitro and impaired T-cell dependent antibody response in vivo in vitamin A-deficient rats. Immunology. 1993;80:581–6. [PMC free article] [PubMed] [Google Scholar]
- 9.Smith SM, Hayes CE. Contrasting impairments in IgM and IgG responses of vitamin A-deficient mice. Proc Natl Acad Sci USA. 1987;84:5878–82. doi: 10.1073/pnas.84.16.5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pasatiempo AM, Kinoshita M, Taylor CE, Ross AC. Antibody production in vitamin A-depleted rats is impaired after immunization with bacterial polysaccharide or protein antigens. FASEB J. 1990;4:2518–27. doi: 10.1096/fasebj.4.8.2110538. [DOI] [PubMed] [Google Scholar]
- 11.Cantorna MT, Nashold FE, Hayes CE. In vitamin A deficiency multiple mechanisms establish a regulatory T helper cell imbalance with excess Th1 and insufficient Th2 function. J Immunol. 1994;152:1515–22. [PubMed] [Google Scholar]
- 12.Cantorna MT, Nashold FE, Hayes CE. Vitamin A deficiency results in a priming environment conducive for Th1 cell development. Eur J Immunol. 1995;25:1673–9. doi: 10.1002/eji.1830250629. [DOI] [PubMed] [Google Scholar]
- 13.Carman JA, Hayes CE. Abnormal regulation of IFN-gamma secretion in vitamin A deficiency. J Immunol. 1991;147:1247–52. [PubMed] [Google Scholar]
- 14.Stephensen CB, Jiang X, Freytag T. Vitamin A deficiency increases the in vivo development of IL-10-positive Th2 cells and decreases development of Th1 cells in mice. J Nutr. 2004;134:2660–6. doi: 10.1093/jn/134.10.2660. [DOI] [PubMed] [Google Scholar]
- 15.Chen Q, Ross AC. Inaugural article: vitamin A and immune function. retinoic acid modulates population dynamics in antigen receptor and CD38-stimulated splenic B cells. Proc Natl Acad Sci USA. 2005;102:14142–9. doi: 10.1073/pnas.0505018102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saari JC. Retinoids in photosensitive systems. In: Sporn MB, Roberts AB, Goodman DS, editors. The Retinoids: Biology, Chemistry and Medicine. New York: Raven Press, Ltd; 1994. pp. 351–86. [Google Scholar]
- 17.Mark M, Ghyselinck NB, Chambon P. Function of retinoid nuclear receptors: lessons from genetic and pharmacological dissections of the retinoic acid signaling pathway during mouse embryogenesis. Annu Rev Pharmacol Toxicol. 2006;46:451–80. doi: 10.1146/annurev.pharmtox.46.120604.141156. [DOI] [PubMed] [Google Scholar]
- 18.de Urquiza AM, Liu S, Sjoberg M, et al. Docosahexaenoic acid, a ligand for the retinoid X receptor in mouse brain. Science. 2000;290:2140–4. doi: 10.1126/science.290.5499.2140. [DOI] [PubMed] [Google Scholar]
- 19.Gronemeyer H, Gustafsson JA, Laudet V. Principles for modulation of the nuclear receptor superfamily. Nat Rev Drug Discov. 2004;3:950–64. doi: 10.1038/nrd1551. [DOI] [PubMed] [Google Scholar]
- 20.Maxwell MA, Muscat GE. The NR4A subgroup: immediate early response genes with pleiotropic physiological roles. Nucl Recept Signal. 2006;4:e002. doi: 10.1621/nrs.04002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clark RB, Bishop-Bailey D, Estrada-Hernandez T, Hla T, Puddington L, Padula SJ. The nuclear receptor PPAR gamma and immunoregulation: PPAR gamma mediates inhibition of helper T cell responses. J Immunol. 2000;164:1364–71. doi: 10.4049/jimmunol.164.3.1364. [DOI] [PubMed] [Google Scholar]
- 22.Winoto A, Littman DR. Nuclear hormone receptors in T lymphocytes. Cell. 2002;109(Suppl.):S57–66. doi: 10.1016/s0092-8674(02)00710-9. [DOI] [PubMed] [Google Scholar]
- 23.Szanto A, Narkar V, Shen Q, Uray IP, Davies PJ, Nagy L. Retinoid X receptors: X-ploring their (patho)physiological functions. Cell Death Differ. 2004;11(Suppl. 2):S126–43. doi: 10.1038/sj.cdd.4401533. [DOI] [PubMed] [Google Scholar]
- 24.Vuligonda V, Thacher SM, Chandraratna RA. Enantioselective syntheses of potent retinoid X receptor ligands: differential biological activities of individual antipodes. J Med Chem. 2001;44:2298–303. doi: 10.1021/jm0100584. [DOI] [PubMed] [Google Scholar]
- 25.Stephensen CB, Rasooly R, Jiang X, et al. Vitamin A enhances in vitro Th2 development via retinoid X receptor pathway. J Immunol. 2002;168:4495–503. doi: 10.4049/jimmunol.168.9.4495. [DOI] [PubMed] [Google Scholar]
- 26.Rasooly R, Schuster GU, Gregg JP, Xiao JH, Chandraratna RA, Stephensen CB. Retinoid x receptor agonists increase bcl2a1 expression and decrease apoptosis of naive T lymphocytes. J Immunol. 2005;175:7916–29. doi: 10.4049/jimmunol.175.12.7916. [DOI] [PubMed] [Google Scholar]
- 27.Sauer B. Inducible gene targeting in mice using the Cre/lox system. Methods. 1998;14:381–92. doi: 10.1006/meth.1998.0593. [DOI] [PubMed] [Google Scholar]
- 28.Chen J, Kubalak SW, Chien KR. Ventricular muscle-restricted targeting of the RXRalpha gene reveals a non-cell-autonomous requirement in cardiac chamber morphogenesis. Development. 1998;125:1943–9. doi: 10.1242/dev.125.10.1943. [DOI] [PubMed] [Google Scholar]
- 29.Hennet T, Hagen FK, Tabak LA, Marth JD. T-cell-specific deletion of a polypeptide N-acetylgalactosaminyl-transferase gene by site-directed recombination. Proc Natl Acad Sci USA. 1995;92:12070–4. doi: 10.1073/pnas.92.26.12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wan YJ, An D, Cai Y, et al. Hepatocyte-specific mutation establishes retinoid X receptor alpha as a heterodimeric integrator of multiple physiological processes in the liver. Mol Cell Biol. 2000;20:4436–44. doi: 10.1128/mcb.20.12.4436-4444.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merki E, Zamora M, Raya A, et al. Epicardial retinoid X receptor alpha is required for myocardial growth and coronary artery formation. Proc Natl Acad Sci USA. 2005;102:18455–60. doi: 10.1073/pnas.0504343102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu E, Sinha S, Williams C, et al. Targeted deletion of integrin-linked kinase reveals a role in T-cell chemotaxis and survival. Mol Cell Biol. 2005;25:11145–55. doi: 10.1128/MCB.25.24.11145-11155.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao L, Cannons JL, Anderson S, et al. CBFB-MYH11 hinders early T cell development and induces massive cell death in the thymus. Blood. 2006 doi: 10.1182/blood-2006-10-051508. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zamoyska R, Basson A, Filby A, Legname G, Lovatt M, Seddon B. The influence of the src-family kinases, Lck and Fyn, on T cell differentiation, survival and activation. Immunol Rev. 2003;191:107–18. doi: 10.1034/j.1600-065x.2003.00015.x. [DOI] [PubMed] [Google Scholar]
- 35.Mosmann TR, Coffman RL. TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu Rev Immunol. 1989;7:145–73. doi: 10.1146/annurev.iy.07.040189.001045. [DOI] [PubMed] [Google Scholar]
- 36.Mackay CR. Follicular homing T helper (Th) cells and the Th1/Th2 paradigm. J Exp Med. 2000;192:F31–4. doi: 10.1084/jem.192.11.f31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ishaq M, Fan M, Wigmore K, Gaddam A, Natarajan V. Regulation of retinoid X receptor responsive element-dependent transcription in T lymphocytes by Ser/Thr phosphatases. functional divergence of protein kinase C (PKC) theta; and PKC alpha in mediating calcineurin-induced transactivation. J Immunol. 2002;169:732–8. doi: 10.4049/jimmunol.169.2.732. [DOI] [PubMed] [Google Scholar]
- 38.Bissonnette RP, Brunner T, Lazarchik SB, et al. 9-cis retinoic acid inhibition of activation-induced apoptosis is mediated via regulation of fas ligand and requires retinoic acid receptor and retinoid X receptor activation. Mol Cell Biol. 1995;15:5576–85. doi: 10.1128/mcb.15.10.5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szondy Z, Reichert U, Fesus L. Retinoic acids regulate apoptosis of T lymphocytes through an interplay between RAR and RXR receptors. Cell Death Differ. 1998;5:4–10. doi: 10.1038/sj.cdd.4400313. [DOI] [PubMed] [Google Scholar]
- 40.Grenningloh R, Gho A, di Lucia P, et al. Cutting edge: inhibition of the retinoid X receptor (RXR) blocks T helper 2 differentiation and prevents allergic lung inflammation. J Immunol. 2006;176:5161–6. doi: 10.4049/jimmunol.176.9.5161. [DOI] [PubMed] [Google Scholar]
- 41.Du X, Tabeta K, Mann N, Crozat K, Mudd S, Beutler B. An essential role for Rxralpha in the development of Th2 responses. Eur J Immunol. 2005;35:3414–23. doi: 10.1002/eji.200535366. [DOI] [PubMed] [Google Scholar]