Abstract
We and other investigators have demonstrated up-regulation of the expression of the RNA-editing gene 150-kDa adenosine deaminase that acts on RNA (ADAR1) in systemic lupus erythematosus (SLE) T cells and B cells, peripheral blood mononuclear cells (PBMC), natural killer (NK) cells. The presence of a small proportion of activated T cells is the hallmark of SLE. Therefore, it was hypothesized that 150-kDa ADAR1 gene expression is induced by the physiological activation of T cells. To examine this hypothesis, normal T cells were activated by anti-CD3-ε plus anti-CD28 for various time periods from 0 to 48 hr. The expression of 110-kDa and 150-kDa ADAR1, and interleukin (IL)-2 and β-actin gene transcripts was analysed. An approximately fourfold increase in 150-kDa ADAR1 gene expression was observed in activated T cells. ADAR2 gene transcripts are substrates for ADAR1 and ADAR2 enzymes. Therefore, we assessed the role of the 150-kDa ADAR enzyme in editing of ADAR2 gene transcripts. In activated T cells, site-selective editing of the −2 site was observed. Previous studies indicate that this site is predominantly edited by ADAR1. In addition to this, novel editing sites at base positions −56, −48, −45, −28, −19, −15, +46 and +69 were identified in activated T cells. On the basis of these results, it is proposed that 150-kDa ADAR1 gene expression is selectively induced in T cells by anti-CD3-ε and anti-CD28 stimulation and that it may play a role in site-selective editing of gene transcripts and in altering the functions of several gene products of T cells during activation and proliferation.
Keywords: T cells, gene transcripts, RNA editing, mutations
Introduction
Messenger RNA editing was first discovered in the kinetoplast of trypanosomes by Benne et al. in 1986.1 In the intervening 21 years, RNA editing has been described in a diverse spectrum of organisms, ranging from viruses to humans. RNA editing is the co- or post-transcriptional modification of RNA which results in the insertion, deletion or substitution of nucleotides. Two important mechanisms leading to mRNA editing have been identified: (1) guide RNA (gRNA)-directed cleavage of pre-mRNA and subsequent deletion of uridylate (U) residues from, or their insertion into, the mRNA,2,3 and (2) deamination of adenosine and cytosine by specific enzymes.4,5 In mammalian species, adenosine-to-inosine (A→I) editing and cytidine-to-uridine (C→U) editing have been identified as physiological mechanisms.6 Mammalian A→I editing is mediated by adenosine deaminases that act on RNA (ADARs).5 ADAR1 and ADAR2 belong to a family of mammalian RNA-editing enzymes. The human ADAR1 gene transcribes two different sized forms of RNA-specific adenosine deaminases using distinct promoter sites. These are an interferon (IFN)-inducible 150-kDa protein and a constitutively expressed N-terminally truncated 110-kDa protein. The ADAR2 gene transcribes constitutively expressed 80-kDa proteins. These enzymes have a catalytic domain responsible for the conversion of adenosine to inosine and three double-stranded RNA (dsRNA) binding domains.7,8 ADAR family enzymes play an important role in several physiological processes by catalysing the hydrolytic deamination at C6 of the adenosine base in certain mRNAs, which leads to inosine formation.6 Inosines are subsequently recognized as guanosine by the translation machinery. Such editing will result in an adenosine to guanine (A→G) transcript mutation.RNA editing can therefore correct, extend or diversify the information encoded within the corresponding genomic sequence, and frequently alters the function of the affected RNAs. The frequent occurrence of selective ADAR1- and ADAR2-mediated adenosine deamination in mRNA of the genes encoding mammalian glutamate receptors (gluR),9–11 5HT2c serotonin receptors,12 the K(V)1·1 potassium channel13 and ADAR214,15 has been reported. Widespread occurrence of A-to-I editing of Alu-containing and intronic regions of pre-mRNAs in the human transcriptome has been identified recently.16–18 Therefore, RNA editing plays an important role in the regulation of gene expression and the production of phenotypic variability.19,20
T lymphocytes are pivotal cells that regulate the functions of other T cells as well as B cells and antigen-presenting cells. We and other investigators demonstrated up-regulation of 150-kDa ADAR1 expression in systemic lupus erythematosus (SLE) T cells, peripheral blood mononuclear cells (PBMC) and natural killer (NK) cells.21–24 The presence of a small proportion of activated T cells is the hallmark of SLE. Based on this finding, it was hypothesized that 150-kDa ADAR1 gene expression is induced in part by physiological activation of T cells in addition to its up-regulation by type I IFNs.25 The potential significance of ADAR expression in T cells is altered expression of gene(s) by transcript editing and the impact of such editing on T-cell function and on the process of immune regulation. The induction and regulation of ADAR1 during T-cell activation may play a major role in the initiation and propagation of immune functions. The objective of the experiments carried out in this study was to investigate the regulation of ADAR1 during T-cell activation and to determine its role in transcript editing. ADAR2 gene transcripts are substrates for ADAR1 and ADAR2 enzymes, and adenosines selectively edited by these enzymes have previously been identified.14,15 Therefore, the other goal of these experiments was to identify the role of 150-kDa ADAR1 in the editing of ADAR2 gene transcripts of normal and activated T cells.
Materials and methods
T-lymphocyte isolation and phenotypic characterization
Blood samples from normal subjects were obtained at Wake Forest University Baptist Medical Center (WFUBMC) with the approval of the Institute's Review Board (IRB). Only normal subjects who were not on any medication were enrolled in this study. The age of test subjects ranged from 25 to 50 years. T lymphocytes from healthy subjects were purified from PBMC and characterized as previously described.24 Cytofluorographic analysis [fluorescence-activated cell sorting (FACS)] of purified T cells using fluorescein isothiocyanate (FITC)-anti-CD3 (Becton Dickinson, San Jose, CA) demonstrated that 94·5 ± 3·5% of cells were positive for the CD3 membrane complex that delineates T lymphocytes. These purified fresh, quiescent T cells, which had not been exposed to any activators or inhibitors, were utilized in the experiments.
Activation of T lymphocytes
Purified T lymphocytes were activated with CD3-ε and CD28 agonists, as these agonists reproduce physiological conditions of activation by binding to T-cell surface receptors. The T lymphocytes were activated as described previously.26 Briefly, sterile polystyrene culture tubes (12 × 75 mm) were coated with 10 µg/ml of affinity-purified goat anti-mouse immunoglobulin G (IgG) at 4° overnight, washed with phosphate-buffered saline (PBS) (pH 7·4) and then coated with 5 µg/ml of anti-CD3-ε (Becton Dickinson) and incubated at 4° for 2 hr before the addition of T cells. T lymphocytes (6 × 106) were resuspended in RPMI 1640 supplemented with 10 mm HEPES, 200 mm l-glutamine, 10% fetal calf serum (FCS), 50 µg/ml of penicillin and 50 ng/ml of streptomycin. Anti-CD28 (Becton Dickinson) at a concentration of 100 ng/ml was added just before activation. T cells from control and treated series were incubated at 37° in 5% CO2 for time periods from 0 to 48 hr. Activated and non-activated T cells were harvested for further studies.
Isolation of genomic DNA, RNA and cDNA synthesis
Genomic DNA and total cellular RNA were extracted from normal and activated T lymphocytes (6 × 106 in each case) as described previously.24 Single-stranded cDNA (sscDNA) was synthesized from 1 to 2 µg of total RNA using the random primer (Invitrogen, Carlsbad, CA) M-MLV H– reverse transcriptase according to the manufacturer's instructions (Invitrogen).
Oligonucleotide primers for PCR amplification
Oligonucleotide primers for TADAR1 and 150-kDa ADAR1, ADAR2, IL-2 and β-actin were designed based on published sequences using the program oligomer, version 5·0 (Molecular Biology Insights, Cascade, CO) and synthesized by Sigma Genosys (Woodlands, TX).
Amplification of 150-kDa ADAR1, total ADAR1 (TADAR1), ADAR2, IL-2 and β-actin cDNA
The cDNA was amplified by polymerase chain reaction (PCR) using primer sets designed for 110-kDa and 150-kDa ADAR1, IL-2 and β-actin gene transcripts. The following primer sets were used: β-actin forward primer, 5′-TGA CGG GGT CAC CCA CAC TGT GCC CAT CTA-3′ (Accession number M10277; nucleotides 2133–2162); reverse primer, 5′-CTA GGA GCA TTT GCG GTG GAC GAT GGA GGG-3′ (Accession number M10277; nucleotides 2971–3000); IL-2 forward primer, 5′-ATG TAC AGG ATG CAA CTC CTG TCT T-3′ (Accession number U25676; nucleotides 58–72); reverse primer, 5′-GTT AGT GTT GAG ATG ATG CTT TGA C-3′ (Accession number U25676; nucleotides 489–514); 150-kDa ADAR1 forward primer, 5′-CCA CCT CCA GTG CGG AGT AGC G-3′ (exon 1A; nucleotides 71–92); reverse primer, 5′-TGC CCC TTG AGA AAT TCT ATT TGC-3′ (exon 2; nucleotides 289–312); TADAR1 forward primer, 5′-CTG GCA GCC TCC GGG TGT C-3′ (exon 1B; nucleotides 59–77); reverse primer as for 150 kDa ADAR1 (exon 2; nucleotides 289–312); ADAR2 forward primer, 5′-GGA ATT CTA TTA GTC ACT AAG CAA AGT GTC AG-3′; reverse primer, 5′-GCG GTA CCC AGG TGT GCT GCC ATC CTT GG-3′.27 RNA replaced the cDNA in negative control PCR reaction tubes to exclude the possibility of contamination of DNA in cDNA samples, especially in the amplification of the ADAR2 pre-mRNA. The cDNA samples from control and activated groups were amplified as follows. Each reaction mixture consisted of 10% of a single sscDNA reaction, 25 pmol of each primer, 1 × PCR buffer [10 mm Tris-HCl (pH 8·3) and 50 mm KCl], 2·5 mm MgCl2, 200 µm of each dNTP, 1·25 units of Taq polymerase (Perkin-Elmer-Cetus, Emeryville, CA), and double distilled (dd) H2O to a final volume of 50 µl. The reaction mixture was subjected to 30 cycles of denaturation (94°; 1 min) and primer annealing (temperature depending on primer set) for 1 min, and extension for 2 min at 72° plus 2 seconds added for each cycle utilizing a DNA Thermal Cycler (Perkin-Elmer-Cetus). Ten µl of the reaction mixtures was then analysed on a 2% agarose gel in Tris-HCl–acetate–ethylenediaminetetraacetic acid (EDTA) (TAE) buffer. One µg of HaeIII-digested øx174 DNA (Gibco BRL, Gaithersburg, MD) was utilized as molecular weight markers of 1353, 1078, 872, 603, 310, 234, 194, 118 and 72 bp. PCR products were purified using a Qiagen purification system (Qiagen, Valencia, CA). Specific gene amplification was confirmed by the sequencing of the PCR products in an automatic DNA sequencer (ABI Prism 377; Applied Biosystems, Foster City, CA).
Construction of neutral DNA fragments with gene-specific primer ends (MIMICs) for the ADAR1 gene for competitive PCR
We constructed a MIMIC to specifically amplify 150-kDa ADAR1 gene transcripts, as 150-kDa ADAR1 contains a unique sequence at the 5′ end that is not present in 110-kDa ADAR1.7 The 110-kDa ADAR1 transcript does not contain a unique segment which is not a part of 150-kDa ADAR1. It is not possible to generate a MIMIC to specifically quantify the 110-kDa ADAR1 transcript. Therefore, we constructed a MIMIC that can amplify 150-kDa ADAR1 and total ADAR1 (TADAR1) gene transcripts. The concentrations of 110-kDa ADAR1 were obtained by subtracting the 150-kDa ADAR1 values from the TADAR1 values of the respective samples. Neutral DNA of the Lambda ZAP II Vector (Promega, Madison, WI) was used to construct TADAR1 and 150-kDa ADAR1 gene-specific MIMICs as described previously.24 Briefly, composite primers containing TADAR1, 150-kDa ADAR1 gene-specific primer sequences, in addition to 20 nucleotides that hybridize to the neutral DNA fragments, were designed using the program oligomer, version 5·0. Composite primer sequences will be supplied upon request. The above-mentioned TADAR1, 150-kDa ADAR1 gene-specific primers were used in these experiments. The TADAR1, 150-kDa ADAR1 gene-specific MIMICs were generated using composite and TADAR1, 150-kDa ADAR1 gene-specific primers in PCR amplification of neutral DNA fragments. The size of TADAR1, 150-kDa ADAR1 MIMICs was adjusted to distinguish TADAR1 and 150-kDa ADAR1 PCR products. This yielded neutral DNA fragments with TADAR1, 150-kDa ADAR1 gene-specific sequences incorporated at the ends which were designated TADAR1, 150-kDa ADAR1 MIMICs. Following dilution of MIMICs to 100 attomoles/ml, the MIMICs served as stock solutions for internal standards for quantification of transcripts by competitive PCR. The use of MIMICs containing the same primer sequences made it possible to quantify transcripts of TADAR1, 150-kDa ADAR1 because MIMICs and transcripts are amplified with equal efficiencies. In a series of experiments, several concentrations of MIMICs were used as internal standards along with equal quantities of normal cDNA samples to obtain the range of MIMICs to be used in the experimental series.
Quantification of TADAR1, 150-kDa ADAR1 gene transcripts
The twofold dilutions of MIMICs were added to PCR reaction tubes containing equal amounts of cDNA samples. To quantify 150-kDa ADAR1 mRNA, 10% of sscDNA synthesized from 1 µg of total cellular RNA was used. Preliminary studies indicated that the quantity of TADAR1 is about 20-fold higher than that of 150-kDa ADAR1. Therefore, to obtain a better resolution of TADAR1 concentrations, 2% of cDNA synthesized from 1 µg of total RNA isolated from normal or activated T-cells was used along with twofold dilutions of TADAR1 MIMICs as described in the legend to Fig. 1. The sscDNA from control and activated T cells and 25 pmol of each primer, 1 × PCR buffer (10 mm Tris-HCl, pH 8·3, and 50 mm KCl), 200 mm of each dNTP, 25 mm MgCl2, 1·25 units of Taq polymerase and ddH2O to a final volume of 50 µl were subjected to 30 cycles of denaturation at 94° for 1 min, primer annealing (temperature dependent on the primer pair) for 1 min, and extension at 72° for 2 min, plus 2 seconds added for each cycle utilizing a DNA thermal cycler. Because MIMICs and gene-specific PCR products have different base pair lengths, amplified gene transcripts and their respective MIMICs were distinguished by electrophoresis on a 2% agarose gel and staining with ethidium bromide. The amount of gene transcript was estimated by comparison with different concentrations of known standard (MIMIC) and identification of the standard matching the gene product (Fig. 2). The concentrations of 110-kDa ADAR1 were obtained by subtracting the 150-kDa ADAR1 values from TADAR1 values for the respective samples.
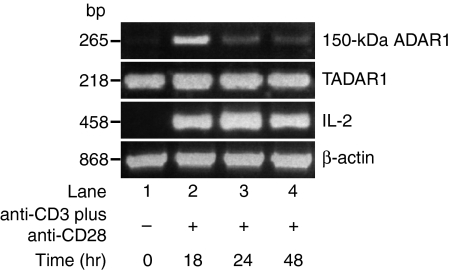
Figure 1.
Induction of 150-kDa adenosine deaminase that acts on RNA 1 (ADAR1) gene expression in T cells by anti-CD3-ε plus anti-CD28 activation. An ethidium bromide-stained agarose gel is shown demonstrating polymerase chain reaction (PCR)-amplified 110-kDa and 150-kDa ADAR1, interleukin (IL)-2 and β-actin gene transcripts at various time-points of T-cell activation by anti-CD3-ε plus anti-CD28. All lanes contain PCR products derived from 10% of cDNA synthesized from 1 µg of total RNA isolated from control and activated T cells at time-points ranging from 0 to 48 hr. The 150-kDa ADAR1 mRNA expression was up-regulated at the 18-hr time-point and had reached control levels by the 48-hr time-point.
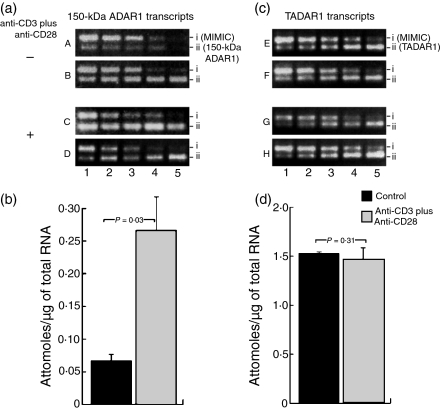
Figure 2.
Quantification of 150-kDa and 110-kDa adenosine deaminase that acts on RNA-1 (ADAR-1) gene transcripts in normal and activated T cells by competitive polymerase chain reaction (PCR). (a) All lanes contain PCR products derived from 10% of cDNA synthesized from 1 µg of total RNA isolated from normal or activated T cells along with twofold dilutions of neutral DNA fragments with gene-specific primer ends (MIMICs) for 150-kDa ADAR1. Amplified MIMICs (row i) and 150-kDa ADAR1 (row ii) transcripts are shown in A–D. A and B are from control T cells; C and D are from T cells activated with anti-CD3 plus anti-CD28. The concentrations of the 150-kDa ADAR1 MIMICs in attomoles in row i are: lane 1, 6·25 × 10−3; lane 2, 3·12 × 10−3; lane 3, 1·56 × 10−3; lane 4, 7·80 × 10−4; lane 5, 3·90 × 10−4 attomoles of 150-kDa ADAR1 MIMICs. (b) Histograms indicating the concentrations of 150-kDa ADAR1 transcripts from normal and activated T cells in attomoles/μg of total RNA. There was a 3·9-fold increase in the amount of 150-kDa ADAR1 gene transcripts (P = 0·03) in activated T cells compared with normal controls. (c) The quantity of total ADAR1 (TADAR1) was about 20-fold higher than that of 150-kDa ADAR1. To obtain better resolution of TADAR1 concentrations, 2% of cDNA synthesized from 1 µg of total RNA isolated from normal or activated T cells along with twofold dilutions of TADAR1 MIMICs was used, as described below. Amplified MIMICs and TADAR1 transcripts are shown in (c). Amplified MIMICs (row i) and TADAR1 (row ii) transcripts are shown in E–H. E and F are from control T cells; G and H are from T cells activated with anti-CD3 plus anti-CD28. The concentrations of the TADAR1 MIMICs in attomoles in row i are: lane 1, 2·5 × 10−2; lane 2, 1·25 × 10−2; lane 3, 6·25 × 10−3; lane 4, 3·12 × 10−3; lane 5, 1·56 × 10−3 attomoles, and each lane contains 2% of cDNA. (d) Mean concentrations, in attomoles/μg of total RNA, ± standard deviation (SD) of 110-kDa ADAR1 transcripts from normal and activated T cells. The concentrations of 110-kDa ADAR1 were obtained by subtracting the 150-kDa ADAR1 values from the TADAR1 values for the respective samples. No significant change (P = 0·30) was observed in the quantities of 110-kDa ADAR1 transcripts from control and activated T cells.
Immunoblotting of 110-kDa and 150-kDa ADAR1 proteins
Cells from the control and experimental series were harvested at various times (0–48 hr) and lysed in 300 µl of lysis buffer [50 mm Tris-HCl (pH 8·0), 150 mm NaCl, 1% Triton X-100, 10 mm sodium pyrophosphate, 50 mm NaF, 1 mm phenylmethylsulphonyl fluoride (PMSF), 0·1 mm DL-dithiothreitol (DTT), and 10 µg/ml each of leupeptin and aprotinin]. Samples were centrifuged at 9300 g at 4° for 5 min in a microcentrifuge to eliminate debris, supernatants were separated and the protein content was quantified. One hundred and fifty micrograms of total protein from each sample was resolved on 10% one-dimensional sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS/PAGE) and the proteins were transferred to immoblion-P (Millipore, Bedford, MA). Membranes were blocked with Blotto (Amersham Biosciences, Corp, Piscataway, NJ) for enhanced chemiluminescence (ECL)-based detection overnight. The membranes were immunoblotted with a 1/1000 dilution of anti-ADAR1 antibody, which detects both 110-kDa and 150-kDa ADAR1 (kindly provided by Dr C. Samuel, Dept. of Molecular, Cellular and Developmental Biology, University of California, Santa Barbara, CA), washed with buffer B [100 mm Tris-HCl (pH 7·5), 500 mm NaCl and 0·1% Tween-20] and probed with a 1/5000 dilution of horseradish peroxidase-labelled sheep anti-mouse IgG or a 1/5000 dilution of peroxidase-conjugated anti-rabbit IgG diluted in Blotto. After washing four times with buffer B, the blots were developed using ECL reagent. The primary and secondary antibodies were stripped from the blots using buffer C [62·5 mm Tris-HCl (pH 6·7), 100 mm 2-mercaptoethanol (2-ME) and 2% SDS] and re-probed with a 1/1000 dilution of rabbit anti-human actin and a 1/5000 dilution of horseradish peroxidase-labelled sheep anti-mouse IgG and ECL. Proteins were quantified by densitometry.
Cloning and sequencing of ADAR2 cDNA and genomic DNA
ADAR2 transcript editing has been used as a specific marker for assessing ADAR enzyme activity.14,15 The ADAR2 transcript has five previously known A to G editing sites (−1, −2, +10, +23 and +24). Both the ADAR1 and ADAR2 enzymes edit ADAR2 gene transcripts. Therefore, in the present study ADAR2 transcript editing was used as a specific marker for assessing ADAR1 and ADAR2 enzyme activity. The amplified products of ADAR2 gene transcripts from activated and nonactivated T lymphocytes of control subjects were cloned and sequenced. These PCR products were t-tailed with Taq polymerase and cloned into pCR2·1-TOPO vectors following the manufacturer's instructions (Invitrogen). Recombinant clones were purified using Qiagen Miniprep kits and sequenced using T7 and M13 primers and an ABI-377 sequencer (Applied Biosystems). Genomic DNA samples from control and activated T cells were amplified using primer sets used for cDNA amplification and sequenced as described previously.24
Data analysis and statistical methods
Data are expressed as mean ± standard deviation (SD) throughout the text unless otherwise stated. Statistical comparisons between control and activated samples were made using Student's t-test and the χ2 test. A value of P < 0·05 was considered to be statistically significant.
Results
AntiCD3-ε and anti-CD28 induce expression of 150-kDa ADAR1 during T-cell activation
T cells were physiologically activated by anti-CD3-ε and anti-CD28 for various time periods from 0 to 48 hr. The cDNA was synthesized from RNA extracted from activated and control T cells. The cDNA was amplified by PCR using primer sets designed for TADAR1, 150-kDa ADAR1, IL-2 and β-actin gene transcripts. The 110-kDa ADAR1 and β-actin gene expressions were used as controls and IL-2 expression as a marker for T-cell activation. The 110-kDa ADAR1 and β-actin transcripts were constitutively expressed at all time-points. Expression of IL-2 from the 18-hr time-point onwards indicates activation of T cells treated with anti-CD3-ε and anti-CD28. TADAR1 and 150-kDa ADAR1 gene expression was analysed by conventional PCR. Expression of the 150-kDa ADAR1 gene was up-regulated at the 18-hr time-point. A decrease was noted at 24 hr and the background level was reached by 48 hr (Fig. 1). Therefore, we analysed the 110-kDa and 150-kDa ADAR1 transcripts from control and activated T cells at the 18-hr time-point by competitive polymerase chain reaction (CPCR).
Quantification of 110-kDa and 150-kDa ADAR1 gene expression in control and activated T cells
The TADAR1 and 150-kDa ADAR1 gene transcripts of three controls and three activated samples at the 18-hr time-point were analysed, as up-regulation of 150-kDa ADAR1 gene expression was observed at this time-point in our preliminary studies. The concentrations of 110-kDa and 150-kDa ADAR1 gene transcripts were calculated in attomoles per µg of total RNA. The concentrations of 110-kDa ADAR1 were obtained by subtracting the 150-kDa ADAR1 values from the TADAR1 values for the respective samples. In normal T cells, the concentration of 150-kDa ADAR1 transcripts ranged between 0·08 and 0·04 attomoles/µg, with a mean value of 0·067 ± 0·13 attomoles/µg of total RNA. In activated T cells, it ranged between 0·32 and 0·16 attomoles/µg with a mean value of 0·266 ± 0·039 attomoles/µg of total RNA (P = 0·03; Table 1). The concentration of 110-kDa ADAR transcripts ranged between 1·52 and 1·56 attomoles/µg, with a mean value of 1·53 ± 0·51 attomoles/µg of total RNA, in control samples and between 1·28 and 1·68 attomoles/µg, with a mean value of 1·47 ± 0·13 attomoles/µg of total RNA, in activated T cells (P = 0·31; Table 1). These results revealed that the ratio of 150-kDa ADAR1 mRNA to 110-kDa ADAR1 mRNA in normal T cells was 1 : 22·8. In activated T cells, we observed an approximately fourfold increase in 150-kDa ADAR1 mRNA content (Fig. 2 and Table 1). This increase in ADAR1 mRNA content in activated T cells resulted in a decrease in the ratio of 150-kDa ADAR1 to 110-kDa ADAR1 mRNA to 1 : 5·5 (Table 1). The increase in ADAR1 gene expression in activated T cells suggests the regulation of ADAR1 during T-cell activation by anti-CD3-ε and anti-CD28 stimulation. No significant change in 110-kDa ADAR gene expression in activated T cells was observed compared with normal controls. These results indicate that 150-kDa ADAR1 gene expression is selectively induced in T cells by anti-CD3-ε and anti-CD28 stimulation and that it may play a role in altering the functions of several gene products of T cells during proliferation and development. The increased expression of 150-kDa ADAR1 gene transcripts in activated T cells suggests that A→I editing of ADAR2 transcripts of activated T cells may be a result of increased RNA-editing enzyme activity.
Table 1.
Concentrations of 150 kDa and 110 kDa ADAR1 transcripts in control and activated T cells
| Sample | 150-kDa ADAR1 (attomoles/µg of total RNA) | 110-kDa ADAR11 (attomoles/µg of total RNA) |
|---|---|---|
| Control 1 | 0·08 | 1·52 |
| Control 2 | 0·08 | 1·52 |
| Control 3 | 0·04 | 1·56 |
| Mean ± SD | 0·067 ± 0·013 | 1·53 ± 0·013 |
| ADAR1 150 kDa:110 kDa | 1 : 22·8 | |
| Activated T cells 1 | 0·32 | 1·68 |
| Activated T cells 2 | 0·32 | 1·28 |
| Activated T cells 3 | 0·16 | 1·44 |
| Mean ± SD | 0·266 ± 0·053 | 1·47 ± 0·116 |
| ADAR1 150 kDa:110 kDa | 1 : 5·5 |
The concentrations of 110-kDa ADAR1 were obtained by subtracting the 150-kDa ADAR1 values from the TADAR1 values for the respective samples.
Expression of 110-kDa and 150-kDa ADAR1 proteins in control and activated T cells
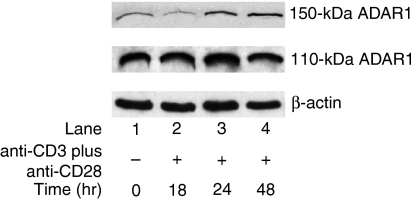
The expression of 150-kDa ADAR1 protein was up-regulated at the 24-hr time-point and thereafter stayed at the same level until the end of the 48-hr period. No significant change was observed in 110-kDa ADAR gene expression at any of the time-points (Fig. 3).
Figure 3.
Induction of 150-kDa adenosine deaminase that acts on RNA 1 (ADAR1) protein expression in T cells by anti-CD3-ε plus anti-CD28 activation. Immunoblots show the 150-kDa and 110-kDa ADAR1 and actin proteins probed with specific antibodies using enhanced chemiluminescence at various time-points of T cell activation by anti-CD3-ε plus anti-CD28. The 150-kDa ADAR1 protein expression was up-regulated approximately threefold at the 24-hr time-point and stayed at the same level until the 48-hr time-point.
Editing of the ADAR2 gene transcripts in activated and non-activated T lymphocytes of control subjects
The cDNAs synthesized from RNA extracted from normal non-activated T cells and from T cells activated with anti-CD3-ε and anti-CD28 for 24 hr were amplified by PCR using specific primer sets designed for ADAR2 mRNA. These transcripts were cloned into pCR2·1-TOPO vectors and analysed for transcript editing. The cDNA was amplified by PCR using specific primers designed for the ADAR2 mRNA region that contains five known A-to-I editing sites, designated −1, 2, +10, +23 and +24. The site co-ordinates are relative to the 3′-splice junction that is known to be created by A-to-I editing.14 Both ADAR1 and ADAR2 enzymes edit ADAR2 gene transcripts at these sites.14,15 A-to-I editing of the −1 site leads to alternative splicing of the ADAR2 pre-mRNA, which in turn results in a non-functional enzyme.14,15 ADAR2 transcript editing has been used as a specific marker for assessing ADAR enzyme activity. The cDNA strands from normal and activated samples were subcloned into TA cloning vectors (pCR2·1) and sequenced using an automated DNA sequencer. A total of 120 recombinant clones from control series and 120 recombinant clones from activated T cells were sequenced and analysed (Table 2). The theoretical background of base discrepancies resulting from errors during reverse transcription and PCR amplification was calculated as described previously.24 The frequencies of guanosine, cytosine and uridine discrepancies in cDNAs from controls and activated samples are within this range, so these discrepancies are probably caused by non-physiological events, such as errors during reverse transcription and PCR. In contrast, adenosine to guanosine was often seen at selective sites, indicating physiological evidence of these base modifications. A-to-I editing was observed in the control and activated T cells at altered frequencies. Multiple sites were edited in some clones compared with other clones. Sequence analysis demonstrated that 10% of the cDNA strands from normal samples showed A-to-I editing (Table 2), whereas such mutations were seen in 20% of cDNA strands from activated T lymphocytes.
Table 2.
Adenosine-to-inosine editing at specific sites in adenosine deaminase that acts on RNA-2 (ADAR-2) pre-mRNA of control and anti-CD3 plus anti-CD28-activated normal T cells
| Number of editing events at sites | ||||||||
|---|---|---|---|---|---|---|---|---|
| Sample | Total number of clones analysed | −2 | −1 | +10 | +23 | +24 | Total | Novel editing sites1 |
| Control 1 | 40 | 1 | 0 | 1 | 1 | 0 | 2 | 1 |
| Control 2 | 40 | 1 | 1 | 1 | 0 | 0 | 2 | 1 |
| Control 3 | 40 | 0 | 2 | 1 | 1 | 1 | 5 | 1 |
| Total | 120 | 2 | 3 | 3 | 2 | 1 | 9 | 3 |
| Activated T cells 1 | 40 | 2 | 0 | 1 | 0 | 1 | 4 | 4 |
| Activated T cells 2 | 40 | 3 | 0 | 1 | 0 | 1 | 5 | 3 |
| Activated T cells 3 | 40 | 2 | 1 | 1 | 0 | 0 | 4 | 4 |
| Total | 120 | 7* | 1 | 3 | 0 | 2 | 13 | 11* |
T cells were activated with anti-CD3 plus anti-CD28 as described in the ‘Materials and methods’.
Novel editing sites were at positions −59, –48, –45, –28, –19, –15, +46 and +69.
P < 0·05 (calculated by the t-test).
Altered editing in ADAR2 gene transcripts of activated T cells
Sequence analysis of ADAR2 transcripts demonstrated the occurrence of A-to-I editing events. All of the edited adenosines are predicted to be located within a base-paired region, and the majority of the 5′-neighbour bases of edited adenosine are either C or A, which is the preferred configuration for ADARs. A-to-G editing at the −2 site was frequently observed in ADAR2 gene transcripts obtained from anti-CD3-ε and anti-CD28 activated T lymphocytes (P < 0·05; Table 2). In addition, heterogeneous A-to-I editing at several other base positions was also identified in the ADAR2 gene transcripts of activated T cells. As far as we know, this is the first identification of the occurrence of ADAR-mediated heterogeneous A-to-I editing in the gene transcripts. These editing events were validated by comparing with known edited sites of ADAR2 gene transcripts. These results demonstrate the occurrence of ADAR1-catalysed A-to-I editing of ADAR2 gene transcripts during antigen-induced T-cell activation in normal subjects.
Novel edited sites identified in ADAR2 gene transcripts of activated T lymphocytes
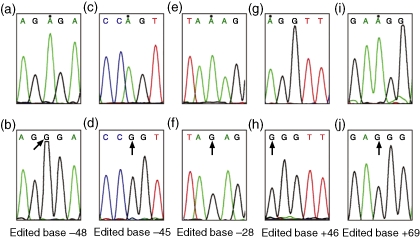
In ADAR2 gene transcripts obtained from control samples, 75% of A-to-G discrepancies observed were clustered in a region that consists of known editing sites (P = 0·001; Table 3), and which is well documented as a substrate for ADARs and as being susceptible to mRNA editing.15 The frequency of such editing in this region in activated T cells was decreased to 53%. This was compensated by an increase in such editing at novel sites located between base positions −56 and −15 and +46 and +69 (Fig. 4 and Table 2; P = 0·015).
Table 3.
Frequency of adenosine-to-inosine editing observed at known and novel sites in adenosine deaminase that acts on RNA 2 (ADAR2) gene transcripts of control and activated T cells
| Known sites | Novel sites | Total | ||||
|---|---|---|---|---|---|---|
| Number1 | % | Number1 | % | Number1 | % | |
| Control | 9 | 75 | 3 | 25 | 12 | 100 |
| Activated | 13 | 53 | 11** | 47 | 24** | 100 |
Total number of editing events observed in all clones analysed in control and activated T cells.
P < 0·001 (calculated by the χ2 test).
Figure 4.
Novel adenosine-to-guanine (A→G) editing observed in adenosine deaminase that acts on RNA 2 (ADAR2) gene transcripts from the control and activated T cells. (a, c, e, g, i) The normal sequence. Normal counterparts of A→G mutated bases at the positions −48, −45, −28, +46 and +69 in different clones are indicated with dots. (b, d, f, h, j) Edited bases at positions −48, −45, −28, +46 and +69, respectively, in different recombinant clones. All edited bases are indicated with arrows. All sequences represent sense strands.
Absence of mutations in ADAR2 genomic DNA
None of the mutations observed in the gene transcripts of ADAR2 from normal and activated T cells was seen in the genomic DNA sequences. The absence of such mutations in the ADAR2 genomic DNA sequences of T cells indicates that their origin is in RNA and that transcript editing is the mechanism involved. In general, the nature of the transcript mutations observed in the present study differs from that of somatic hypermutations.28 The lack of such base changes in genomic DNA of T cells argues against their occurrence as a result of somatic hypermutation and provides plausible evidence of a co- or post-transcriptional modification mechanism affecting ADAR2 gene transcripts in normal and activated T cells.
Discussion
The term ‘RNA editing’ encompasses a wide range of activities that lead to the specific alteration of the primary sequence of an RNA molecule and that play an important role in increasing the flexibility of eukaryotic gene expression.4 Recent studies have shown that the enzymes responsible for site-specific deamination of adenosine to inosine in mRNAs are evolutionarily related to those that carry out this reaction in tRNAs. The ADARs catalyse the covalent modification of dsRNA by hydrolytic C-6 deamination of adenosine to yield inosine.5 Two iosforms of the ADAR gene were identified in humans, but their selective functions are unknown.7,8 However, a selective affinity for editing some adenosines in ADAR2 gene transcripts has been demonstrated in in vitro studies15 and in SLE pathogenesis.24 The inosine generated by the RNA editing process is typically recognized as guanine during translation and cDNA synthesis.5 Messenger RNA editing induced by ADARs and other editing enzymes plays an important role in the regulation of gene expression.9,13 It also plays an important role in pathophysiology.21,23,24,27,29–38 However, the effect of RNA editing and transcript mutations in immune cell regulation has not been explored and is therefore not well understood. Similar mRNA transcript mutations could also occur during other physiological events. Studies investigating this uncharted territory could prove very fruitful and could have a wider importance in improving our understanding the role of RNA editing in gene regulation in physiological events. It is surmised that the abnormal expression of RNA-editing genes such as ADAR1 and cytidine deaminase apolipoprotein B mRNA editing catalytic subunit (APOBEC)-3A might play an important role in immune modulation and defence. Altered expression of RNA-editing genes such as ADAR1 and APOBEC3A has been observed in T cells, PBMC and NK cells of patients with autoimmune diseases in which immune modulations are well documented.22–24,39 The data presented in this paper demonstrate that activation of normal T cells with anti-CD3-ε and anti-CD28 agonists induces the up-regulation of 150-kDa ADAR1. These results also indicate that up-regulated 150-kDa ADAR1 creates novel editing sites and modifies the editing at known sites in ADAR2 gene transcripts.
The results of this study demonstrate that T-cell activation by anti-CD3-ε and anti-CD28 up-regulated ADAR1 expression at the transcript and protein level in addition to its up-regulation via the type I interferon signalling pathway.The mechanism underlying the up-regulation of ADAR1 expression byanti-CD3-ε and anti-CD28 via the T-cell receptor signalling pathway remains to be established. Studies are in progress to identify the downstream molecules involved in 150-kDa ADAR1 up-regulation during anti-CD3-ε and anti-CD28 activation. The increased editing of ADAR2 gene transcripts in activated T-cell samples revealed increased enzymatic activity of 150-kDa ADAR1 by the induction of altered editing in ADAR2 gene transcripts.Although the mechanism underlying the occurrence of editing at novel sites in ADAR2 gene transcripts of activated T cells remains to be established, several factors may be involved. One strong and straightforward possibility is increased enzymatic activity of 150-kDa ADAR1 in T cells. The second possibility is altered expression of ADAR1 and ADAR2 splice variants,40,41 which will be explored in future studies. In addition, alterations of the components of the editing machinery such as editosome, helicases and small nucleolar RNAs (snoRNAs) during T-cell activation might play a role in the regulation of A-to-I editing. Several other cellular factors have recently been implicated in regulating intracellular editing activity by ADARs. For example, it was shown that modification of ADARs by the small ubiquitin modifier (SUMO) reduces their enzyme activity42 and the requirement of inositol hexakisphosphate (IP6) for ADAR2 enzyme activity in general.43 Taken together, these findings suggest possible scenarios explaining how, in activated T cells, editing levels at particular sites in ADAR2 gene transcripts may be modulated. Further studies are warranted to identify the precise mechanism(s) involved in functional variations of these structurally divergent enzymes and their impact on substrate editing to improve our understanding of the role of transcript editing in immune modulations and specifically during T-cell activation and survival.
The other finding of this study is the heterogeneous editing of A to I in the long double-stranded base-paired regions of ADAR2 gene transcripts and the creation of novel editing sites between base positions −56 and −15 and +46 and +69. The occurrence of novel editing at specific sites in ADAR2 gene transcripts of activated T cells may be a result of the up-regulation of ADAR1 and/or formation of ADAR1 and ADAR2 heterodimers, which might have more affinity for the observed novel editing sites.44 On the basis of these results, we suggest a novel concept by which up-regulated ADAR1 can randomly edit adenosines located in double-stranded base-paired coding regions, which are not normally edited by constitutively expressed ADAR1 and ADAR2 isozymes, and cause novel mutations in gene transcripts. Therefore, it is proposed that the up-regulation of 150-kDa ADAR1 by anti-CD3-ε and anti-CD28 activation may tilt the balance of the editing machinery and cause novel mutations in the T-cell transcriptome. Such mutations may contribute to the abnormal function of gene products and to the modulation of gene functions during T-cell activation and proliferation. The precise role of up-regulated 150-kDa ADAR1 in the modulation of editing at several known sites, as well as in the occurrence of editing at new sites, has not yet been identified. Interestingly, a recent study on ADAR2-overproducing mice showed an increase in editing of ADAR2-selective editing targets and a decrease in ADAR1-selective editing at specific base positions.45 It is not known what the impact of intronic editing in ADAR2 pre-mRNA may be on the expression of ADAR2 proteins in vivo. It has recently been demonstrated that hyperedited dsRNAs can be subject to specific cleavage.46 Some of the A-to-G discrepancies in ADAR2 transcripts may induce cleavage of the edited transcripts or influence splicing and their stability.47 On the basis of these findings, it is likely that the transcripts of other genes are also edited by up-regulated ADAR1 in a similar manner, resulting in alterations in the functions of gene products and contributing to the modulation of T-cell functions. Identification of such editing sites in other gene transcripts and their impact on edited gene function will help in assessing the mechanism(s) involved in the regulation of immune functions by mRNA editing.
As indicated in the Introduction, we and other investigators demonstrated up-regulation of 150-kDa ADAR1 expression in SLE T cells, PBMC and NK cells.22–24 The presence of a small proportion of activated T cells is the hallmark of SLE. Therefore, these results are in agreement with previous findings of 150-kDa ADAR1 gene up-regulation in SLE patients.22–24 These studies also indicate that 150-kDa ADAR1 gene expression is induced in part by physiological and/or immune complex-mediated activation of T cells in addition to up-regulation by type I IFNs in SLE pathogenesis. The finding that up-regulated 150-kDa ADAR1 induced novel editing in ADAR2 gene transcripts (Table 2 and Fig. 4) also strengthens our previous finding of novel heterogeneous editing in ADAR2 gene transcripts of SLE T cells.24
In summary, we have identified up-regulation of 150-kDa ADAR1 in T cells physiologically activated with anti-CD3-ε and anti-CD28. Such activation of 150-kDa ADAR1 resulted in selective editing of the −2 site of the ADAR2 transcript. In addition, novel editing sites at base positions −59, −48, −45, −28, −19, −15, +46 and +69 of the ADAR2 transcript were identified in activated T cells. These results demonstrate that 150-kDa ADAR1 gene expression is selectively induced in T cells by anti-CD3-ε and anti-CD28 stimulation and that it may play a role in site-selective editing in ADAR2 gene transcripts during T-cell activation and proliferation. Messenger RNA editing induced by ADARs and other editing enzymes plays an important role in the regulation of gene expression.9,13 The widespread occurrence of A-to-I editing of Alu-containing and intronic regions of pre-mRNAs in the human transcriptome has been identified recently.16–18 The up-regulation of 150-kDa ADAR1 followed by alterations in the editing pattern of ADAR2 gene transcripts observed in the present study indicates the occurrence of such alterations in widespread A-to-I editing in the human transcriptome. Such altered editing could have an enormous impact on gene regulation and function. On the basis of published information and our present findings, we propose that, in addition to specific mutations at the DNA level and defective transcript splicing, transcript editing is a novel unrecognized mechanism that contributes to the modulation and/or regulation of gene expression during T-cell activation and proliferation.
Acknowledgments
We thank our control subjects for their generous contributions of blood samples. We also thank the nursing staff of the General Clinical Research Center of the Wake Forest University School of Medicine for their help in obtaining blood samples, Dr C. Samuel for kindly providing the ADAR antibodies, Dr G. Hawkins for his help with sequencing and Miss Bhargavi Dama for editorial assistance. We thank the anonymous reviewers for their constructive and thoughtful critique of the manuscript. This research was supported by grants from the National Institutes of Health (RO1-AR48628 to DL), the Holcomb Research Fund and the General Clinical Research Center of the Wake Forest University School of Medicine (MO1 RR07122).
Abbreviations:
- ADAR
adenosine deaminase that acts on RNA
- IL
interleukin
- IP6
inositol hexakisphosphate
- MIMICs
neutral DNA fragments with gene-specific primer ends
- snoRNAs
small nucleolar RNAs
- SUMO
small ubiquitin modifier
- TADAR1
total adenosine deaminase that acts on RNA-1.
References
- 1.Benne R, van den Burg J, Brakenhoff JP, Sloof P, Van Boom JH, Tromp MC. Major transcript of the frame shifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell. 1986;46:819–26. doi: 10.1016/0092-8674(86)90063-2. [DOI] [PubMed] [Google Scholar]
- 2.Blum B, Bakalara N, Simpson L. A model for RNA editing in kinetoplastid mitochondria: ‘guide’ RNA molecules transcribed from maxi circle DNA provide the edited information. Cell 1990. 1990;60:189–98. doi: 10.1016/0092-8674(90)90735-w. [DOI] [PubMed] [Google Scholar]
- 3.Kable L, Seiwert SD, Heidmann S, Stuart K. RNA editing: a mechanism for gRNA-specifified uridylate insertion into precursor mRNA. Science. 1996;273:1189–95. doi: 10.1126/science.273.5279.1189. [DOI] [PubMed] [Google Scholar]
- 4.Simpson L, Emeson RB. RNA editing. Annu Rev Neurosci. 1996;19:27–52. doi: 10.1146/annurev.ne.19.030196.000331. [DOI] [PubMed] [Google Scholar]
- 5.Herbert A, Rich A. RNA processing and the evolution of eukaryotes. Nat Genet. 1999;21:265–9. doi: 10.1038/6780. [DOI] [PubMed] [Google Scholar]
- 6.Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–46. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim U, Wang Y, Sanford T, Zeng Y, Nishikura K. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc Natl Acad Sci USA. 1994;91:11457–61. doi: 10.1073/pnas.91.24.11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Melcher T, Maas S, Herb A, Sprengel R, Seeburg PH, Higuchi M. A mammalian RNA editing enzyme. Nature. 1996;379:460–4. doi: 10.1038/379460a0. [DOI] [PubMed] [Google Scholar]
- 9.Sommer B, Kohler M, Sprengel R, Seeburg PH. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991;67:11–9. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- 10.Lomeli H, Mosbacher J, Melcher T, et al. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 1994;266:1709–13. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- 11.O'Connell MA. RNA editing: Rewriting receptors. Curr Biol. 1997;7:R437–9. doi: 10.1016/s0960-9822(06)00212-0. [DOI] [PubMed] [Google Scholar]
- 12.Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, Emeson RB. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–8. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 13.Bhalla T, Rosenthal JJ, Holmgren M, Reenan R. Control of human potassium channel inactivation by editing of a small mRNA hairpin. Nat Struct Mol Biol. 2004;10:950–6. doi: 10.1038/nsmb825. [DOI] [PubMed] [Google Scholar]
- 14.Rueter SM, Dawson TR, Emeson RB. Regulation of alternative splicing by RNA editing. Nature. 1999;399:75–80. doi: 10.1038/19992. [DOI] [PubMed] [Google Scholar]
- 15.Dawson TR, Sansam CL, Emeson RB. Structure and sequence determinants required for the RNA editing of ADAR2 substrates. J Biol Chem. 2004;279:4941–51. doi: 10.1074/jbc.M310068200. [DOI] [PubMed] [Google Scholar]
- 16.Levanon E, Eisenberg E, Yelin R, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol. 2004;22:1001–5. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 17.Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. Plos Biol. 2004;2:1–15. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim DD, Kim TT, Walsh T, Kobayashi Y, Matise TC, Buyske S, Gabriel A. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004;14:1719–25. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maas S, Rich A, Nishikura K. A-to-I RNA editing: recent news and residual mysteries. J Biol Chem. 2003;278:1391–4. doi: 10.1074/jbc.R200025200. [DOI] [PubMed] [Google Scholar]
- 20.Samuel CE. RNA editing minireview series. J. Biol Chem. 2003;278:1389–90. doi: 10.1074/jbc.R200032200. [DOI] [PubMed] [Google Scholar]
- 21.Laxminarayana D, Khan IU, Kammer GM. Transcript mutations of the alpha regulatory subunit of protein kinase A and up-regulation of the RNA editing gene transcripts in lupus T lymphocytes. Lancet. 2002;360:842–9. doi: 10.1016/s0140-6736(02)09966-x. [DOI] [PubMed] [Google Scholar]
- 22.Crow MK, Wohlgemuth J. Microarray analysis of gene expression in lupus. Arthritis Res Ther. 2003;5:279–87. doi: 10.1186/ar1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toyabe S, Kaneko U, Uchiyama M. Decreased DAP12 expression in natural killer lymphocytes from patients with systemic lupus erythematosus is associated with increased transcript mutations. J Autoimmunity. 2004;23:371–8. doi: 10.1016/j.jaut.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 24.Laxminarayana D, O'Rourke KS, Maas S, Olorenshaw I. Altered and novel editing in adenosine deaminase that act on RNA (ADAR) 2 gene transcripts of systemic lupus erythematosus (SLE) T lymphocytes. Immunology. 2007;121:359–69. doi: 10.1111/j.1365-2567.2007.02582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patterson JB, Samuel CE. Expression and regulation by interferon of a double-stranded RNA-specific adenosine deaminase from human cells: Evidence for two forms of the deaminase. Mol Cell Biol. 1995;15:5376–88. doi: 10.1128/mcb.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laxminarayana D, Berrada A, Kammer GM. Early events of human T lymphocyte activation are mediated by type I protein kinase A. J Clin Invest. 1993;92:2207–14. doi: 10.1172/JCI116823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maas S, Patt S, Schrey M, Rich A. Under editing of glutamate receptor GluR-B mRNA in malignant gliomas. Proc Natl Acad Sci USA. 2001;98:14687–92. doi: 10.1073/pnas.251531398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wagner SD, Neuberger MS. Somatic hypermutation of immunoglobulin genes. Annu Rev Immunol. 1996;14:441–57. doi: 10.1146/annurev.immunol.14.1.441. [DOI] [PubMed] [Google Scholar]
- 29.Laxminarayana D, Kammer GM. Messenger RNA mutations of type I protein kinase A regulatory subunit alpha in T lymphocytes of a subject with systemic lupus erythematosus. Int Immunol. 2000;12:1521–9. doi: 10.1093/intimm/12.11.1521. [DOI] [PubMed] [Google Scholar]
- 30.Bourara K, Litvak S, Araya A. Generation of G-to-A and C-to-U changes in HIV-1 transcripts by RNA editing. Science. 2000;289:1564–6. doi: 10.1126/science.289.5484.1564. [DOI] [PubMed] [Google Scholar]
- 31.Vissel B, Royle GA, Christie BR, et al. The role of RNA editing of kainate receptors in synaptic plasticity and seizures. Neuron. 2001;29:217–27. doi: 10.1016/s0896-6273(01)00192-1. [DOI] [PubMed] [Google Scholar]
- 32.Kortenbruck G, Berger E, Speckmann E-J, Musshoff U. RNA editing at the Q/R site for the glutamate receptor subunits GLUR2, GLUR5 and GLUR6 in hippocampus and temporal cortex from epileptic patients. Neurobiol Dis. 2001;8:459–68. doi: 10.1006/nbdi.2001.0394. [DOI] [PubMed] [Google Scholar]
- 33.Sodhi MS, Burnet PW, Makoff AJ, Kerwin RW, Harrison PJ. RNA editing of the 5-HT (2C) receptor is reduced in schizophrenia. Mol Psychiatry. 2001;6:373–9. doi: 10.1038/sj.mp.4000920. [DOI] [PubMed] [Google Scholar]
- 34.Gurevich I, Tamir H, Arango V, Dwork AJ, Mann JJ, Schmauss C. Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron. 2002;34:349–56. doi: 10.1016/s0896-6273(02)00660-8. [DOI] [PubMed] [Google Scholar]
- 35.Kawahara Y, Ito K, Sun H, Aizawa H, Kanazawa I, Kwak S. Glutamate receptors: RNA editing and death of motor neurons. Nature. 2004;427:801. doi: 10.1038/427801a. [DOI] [PubMed] [Google Scholar]
- 36.Lipton SA. Sporadic ALS: blame it on the editor. Nat Med. 2004;10:347. doi: 10.1038/nm0404-347. [DOI] [PubMed] [Google Scholar]
- 37.Kwak S, Kawahara Y. Deficient RNA editing of GluR2 and neuronal death in amyotropic lateral sclerosis. J Mol Med. 2005;83:110–20. doi: 10.1007/s00109-004-0599-z. [DOI] [PubMed] [Google Scholar]
- 38.Kaneko U, Toyabe S, Hara M, Uchiyama M. Increased mutations of CD72 transcript in B-lymphocytes from adolescent patients with systemic lupus erythematosus. Pediatric Allergy Immunol. 2006;17:565–71. doi: 10.1111/j.1399-3038.2006.00466.x. [DOI] [PubMed] [Google Scholar]
- 39.Palucka AK, Blanck JP, Bennett L, Pascual V, Banchereau J. Cross-regulation of TNF and IFN-alpha in autoimmune diseases. Proc Natl Acad Sci USA. 2005;102:3372–7. doi: 10.1073/pnas.0408506102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.George CX, Wagner MV, Samuel CE. Expression of interferon-inducible RNA adenosine deaminase ADAR1 during pathogen infection and mouse embryo development involves tissue-selective promoter utilization and alternative splicing. J Biol Chem. 2005;280:15020–8. doi: 10.1074/jbc.M500476200. [DOI] [PubMed] [Google Scholar]
- 41.Kawahara Y, Ito K, Ito M, Tsuji S, Kwak S. Novel splice variants of human ADAR2 mRNA. Skipping of the exon encoding the dsRNA-binding domains, and multiple C-terminal splice sites. Gene. 2005;363:193–201. doi: 10.1016/j.gene.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 42.Desterro JM, Keegan Jaffray LP, Hay E, O'Connell RT, Carmo-Fonseca M. SUMO-1 modification alters ADAR1 editing activity. Mol Biol Cell. 2005;16:5115–26. doi: 10.1091/mbc.E05-06-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Macbeth MR, Schubert HL, Vandemark AP, Lingam AT, Hill CP, Bass BL. Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science. 2005;309:1534–9. doi: 10.1126/science.1113150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chilibeck KA, Wu T, Liang C, Schellenberg MJ, Gesner EM, Lynch JM, MacMillan AM. FRET analysis of in vivo dimerization by RNA-editing enzymes. J Biol Chem. 2006;281:16530–5. doi: 10.1074/jbc.M511831200. [DOI] [PubMed] [Google Scholar]
- 45.Feng Y, Sansam CL, Singh M, Emeson RB. Altered RNA editing in mice lacking ADAR2 auto regulation. Mol Cell Biol. 2006;26:480–8. doi: 10.1128/MCB.26.2.480-488.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scadden ADJ, Smith CJW. Specific cleavage of hyper-edited dsRNAs. EMBO J. 2001;20:4243–52. doi: 10.1093/emboj/20.15.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schoft VK, Schopoff S, Jantsch MF. Regulation of glutamate receptor B pre-mRNA splicing by RNA editing. Nucl Acids Res. 2007;35:723–32. doi: 10.1093/nar/gkm314. [DOI] [PMC free article] [PubMed] [Google Scholar]