Abstract
In addition to its property of enhancing major histocompatibility complex (MHC) class II expression, the class II transactivator (CIITA) was recently demonstrated to be involved in T helper type 1/type 2 (Th1/Th2) differentiation by regulating interleukin-4 (IL-4) gene transcription. There was however, controversy regarding whether CIITA promotes or suppresses IL-4 expression in the experiments with transgenic mice. To clarify the discrepancy by using simpler experimental systems, human Jurkat T cells that express IL-4 but not interferon-γ, even if stimulated with phorbol 12-myristate 13-acetate plus ionomycin, were used for CIITA transfection. Significant suppression of IL-4 gene expression was demonstrated. Simultaneously, histones H3 and H4 in the IL-4 promoter were hypoacetylated. The suppression could be totally reversed by the histone deacetylatase inhibitor trichostatin A. Furthermore, the IL-4 expression was determined in primarily established human Th1/Th2 cells to which CIITA small interference RNA (siRNA) had been introduced. A substantially increased level of IL-4 was recorded in the CIITA siRNA-transfected Th1 cells, which was in parallel with significantly enhanced acetylation in histone H3 of the IL-4 promoter. Chromatin immunoprecipitation analysis indicated that CIITA abrogated the binding of coactivator CBP/p300 and transcription factors STAT6/NFAT1 to IL-4 promoter in the CIITA-transfected cells. In conclusion, CIITA was active in the repression of transcription activation of human IL-4 gene in both the T-cell line and the primary human CD4 T cells by preventing transcription factors from binding to IL-4 promoter through histone hypoacetylation. Our data confirm a potential significant role of CIITA in controlling Th1/Th2 differentiation via modulation of IL-4 gene activation.
Keywords: histone acetylation, interleukin-4, major histocompatibility complex class II transactivator, T helper type1/type 2 subsets
Introduction
Major histocompatibility complex (MHC) class II transactivator (CIITA), a non-DNA-binding protein, is a master regulator in controlling MHC II expression and some genes related to antigen processing.1–4 The transcription activation of the CIITA gene itself is under the control of four independent promoters (P1–PIV).5,6 As a kind of transcription coactivator, CIITA binds to nuclear factors RFX and NF-Y, which are specific for W/S, X, X2 and Y boxes in the class II gene promoter. On the other hand, the N terminus of the CIITA molecule contains domains that either have histone acetyltransferase activity or are capable of interacting with other coactivators, such as cyclic AMP response element binding protein (CREB)-binding protein (CBP)/p300 and pCAF, and chromosome remodelling factor BRG-1, an element of the SW1/SNF complex, to modulate histone acetylation and to recruit RNA polymerase II in controlling class II transcription.7–9
Epigenetic mechanisms for the regulation of gene expression work by modifying the structure of chromatin.10,11 For example, to regulate gene expression, histones may undergo an array of modifications on their tail domains, including acetylation, phosphorylation and methylation.12 A growing body of evidence suggests that diverse modifications of histones may interact with DNA methylation and delicately control gene activation through a complicated programme, termed the ‘histone code’.13 Accumulating data show that high levels of histone acetylation at a particular position correlate well with transcriptional activity, whereas reduced levels result in gene silencing.14,15 The level of histone acetylation is directly modulated by histone acetytransferases and histone deacetylases, which have been closely implicated in transcriptional activation and repression of multiple genes.16 In recent years, much attention has been paid to the role of histone acetylation status in immune gene regulation.17–19
Interleukin-4 (IL-4) is a pleiotropic cytokine with a wide range of biological effects on many haemopoietically and non-haemopoietically derived cells and tissues.20 It also functions as a major determining factor in the differentiation of naive T cells into subsets with T helper type 2 (Th2) phenotype, which facilitates antibody generation and hypersensitivity, as well as protection against worm infection.21 A majority of studies have focused on the molecular mechanisms by which IL-4 expression is regulated in immune responses.22–24 Recently, it has been suggested that the transition of a naive Th cell into a Th2 effector is also under regulation at the level of chromatin. Using a chromatin immunoprecipitation (ChIP) assay, Patrick et al. demonstrated that profound increases in chromatin histone acetylation occur at the interferon-γ (IFN-γ) and IL-4 loci during Th1/Th2 differentiation25 and that GATA-3, the Th2-specific nuclear factor, is able to co-operate with other factors to reinforce IL-4 expression by initiation of chromatin remodelling, especially for the long-range histone hyperacetylation of the Th2-related cytokine genes IL-4, IL-5 and IL-13.26 These data suggest that epigenetic events might be essential in regulation of IL-4 gene expression and T-cell lineage commitment.
Interestingly, CIITA is also active in IL-4 gene transcription and Th1/Th2 differentiation. Reports from Cheong-Hee Chang's laboratory indicated that CIITA-deficient Th1 cells from CIITA–/– xI-E transgenic mice could produce high levels of IL-4 and alter Th1 differentiation.27 The repression of IL-4 expression by CIITA was attributed to its competition with nuclear factor of activated T cells (NFAT) for binding coactivator CBP/p300.28 The opposite conclusion was reached, however, by other investigators who found that CIITA-transgenic CD4 T cells preferentially differentiated into IL-4-secreting Th2 cells upon activation,29,30 implying that overexpressed CIITA enhanced the activation of IL-4. By using CIITA transgenic mice, Park et al. also observed that the enhanced secretion of IL-4 under Th2 conditions was able to repress Th1-dominant experimental autoimmune encephalomyelitis.31 So far, there has been no comprehensive explanation for the discrepancies in previous studies. It is possible that the indirect effects of CIITA on MHC and other related molecules or the complicated conditions for gene co-operation in transgenic mice constructed by different approaches might hinder researchers from reaching a consensus. In this sense, in vitro analysis using cells and cell lines might shed some light on the precise mechanisms underlying the relationship of CIITA and IL-4 gene expression in T cells. Therefore, simplified conditions were adopted in our experiments either using primary human CD4 T cells or Jurkat T cells. Because IL-4 and IL-2, instead of IFN-γ, are constitutively expressed in the Jurkat cell line, these cells are especially suitable for our study to exclude the possible interference of IFN-γ in transcriptions of the IL-4 gene. In addition, little is known about the effects of CIITA on human Th cell differentiation, especially in terms of epigenetic controls.
A clear pattern has been revealed in our experiments that, under the influence of CIITA, the transcription of the IL-4 gene was dramatically repressed in the Jurkat T cells after stimulation by phorbol 12-myristate 13-acetate (PMA) plus ionomycin. In contrast, CIITA small interference RNA (siRNA) was able to enhance the expression of IL-4 in primary human Th1 cells. Mechanism studies further demonstrated that the failure of IL-4 gene transcription resulted from hypoacetylation of histones H3 and H4 in the IL-4 promoter, which in turn caused the failure of CBP/p300 and transcription factors signal transducer and activator of transcription 6 (STAT6)/NFAT1 to bind IL-4 promoter DNA. As far as we know, this is the first time that histone hypoacetylations have been shown to be crucial in down-regulation of IL-4 gene expression by the human CIITA gene. Since IL-4 is involved in Th2 differentiation in relation to a number of diseases, the role of CIITA in epigenetic control of IL-4 expression should not be neglected.
Materials and methods
Cell culture and Th1/Th2 cells
Human T leukaemia cell line Jurkat cells were cultured in RPMI-1640 medium (Gibco Laboratories, Grand Island, NY), supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, 100 μg/ml streptomycin and 2 mm l-glutamine in a humidified atmosphere of 5% CO2 at 37°. In some experiments, the histone deacetylase inhibitor Trichostatin A (TSA) (Sigma-Aldrich, St. Louis, MO) was added for treatment at a concentration of 250 nm for 24 hr. The Th1 and Th2 cells were generated according to Cousins and Lee with minor modifications.32 Human blood was collected from healthy volunteer donors after obtaining informed consent and peripheral blood mononuclear cells were separated by Ficoll–Hypaque gradients separation medium (Lymphoprep, Nycomed, Oslo, Norway). CD4 T cells were isolated using a Dynal CD4 negative isolation kit (Dynal Biotech, Oslo, Norway). Naive CD45RA+ T cells were purified from CD4+ cells by depletion of CD45RO+ cells using anti-human CD45RO antibody (BD Pharmingen, San Diego, CA) and rat anti-mouse immunoglobulin G2a dynabeads (Dynal Biotech, Oslo, Norway) according to the manufacturer's instruction. The purity of CD4+ CD45RA+ T cells was about 98% as determined by fluorescence-activated cell sorter (FACS) FACSCalibur (BD Biosciences, Mountain View, CA). The purified CD4+ CD45RA+ T cells (1·5 × 106/ml) were plated on culture plates and stimulated with immobilized anti-CD3 monoclonal antibodies (mAb) (1 μg/ml), anti-CD28 mAb (2 μg/ml) and recombinant IL-2 (rIL-2; 50 U/ml). For Th1-inducing conditions, IL-12 (2·5 ng/ml) and anti-IL-4 mAb (5 μg/ml) were further added to the cell culture. For Th2-inducing conditions, rIL-4 (12·5 ng/ml), anti-IFN-γ (5 μg/ml) and anti-IL-10 (5 μg/ml) were added. The cytokines and mAbs used were products from R & D Systems (Minneapolis, MN). After 3 days culture, the cells were harvested and restimulated with anti-CD3 (5 μg/ml) and anti-CD28 (2 μg/ml) mAbs for 12 hr.
Plasmid construction and siRNA
Wild type CIITA-containing plasmid pcDNA3.1-FLAG-CIITA was from Dr Ting's laboratory.33 CIITA siRNA were 21-nucleotide long duplexes designed and synthesized by Shanghai GenePharma Co., Ltd (Shanghai, China). The sequence for CIITA suppression was: sense: UCUCCAGUAUAUUCAUCUAdTdT and antisense: UAGAUGAAUAUACUGGAGAdCdC. The specificity of siRNA was examined by aligning against human sequence database.
Transfection
Jurkat cells (1 × 107) negative for human leucocyte antigen (HLA)-DR expression were mixed with 20 μg pcDNA3.1-CIITA. The cells were then electroporated (240 V and 950 μF) in a Gene Pulser (Bio-Rad) for transfection; the efficacy was about 25–30%. Three days later, the HLA-DR-positive cells were enriched by anti-DR microbeads (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany) and stimulated with 12-O-tetradecanoyl-phorbol-13-acetate, PMA (20 ng/ml, Sigma-Aldrich) plus ionomycin (500 ng/ml, Sigma-Aldrich) overnight. Empty vector pcDNA3.1-transfected Jurkat cells were selected by culturing the cells for 3 weeks in the presence of 1 mg/ml G418 (Calbiochem, San Diego, CA) and then maintained at 500 μg/ml G418. An Amaxa Nucleofector System was used for transfection of primary immune cells. Briefly, 5 × 106 CD4+ CD45RA+ T cells were resuspended in 100 μl of the appropriate Amaxa solution and transfected with 100 nm final concentration of siRNA according to manufacturer's protocols U-14. The cells were immediately added to 1 ml RPMI-1640 supplemented with 10% FBS (v/v), 50 IU/ml penicillin, 50 μg/ml streptomycin, 2 mm l-glutamine and 5 × 10−5 m 2-mercaptoethanol and seeded on culture plates as required.
Reverse transcription–polymerase chain reaction (RT-PCR)
Total RNA was prepared with TRIzol reagent (Invitrogen Life Technologies, Grand Island, NY). The RT-PCR were performed with 2 μg RNA and Superscript IITM RNase H-reverse transcriptase (Invitrogen). Hot-start PCR was performed to analyse expression levels with the following primers. IL-4: sense 5′-CTG CAA ATC GAC ACC TAT TA-3′ and antisense, 5′-GAT CGT CTT TAG CCT TTC-3′; IFN-γ: sense 5′-TCG TTT TGG GTT CTC TTG GC-3′ and antisense 5′-GCA GGC AGG ACA ACC ATT AC-3′; IL-2: sense 5′-ACA GCT ACA ACT GGA GCA TT-3′ and antisense 5′-TGC TGT CTC ATC AGC ATA TT-3′; HLA-DR: sense 5′-AAT GGC CAT AAG TGG AGT CC-3′ and antisense 5′-GGA GGT ACA TTG GTG ATC GG-3′; CIITA: sense 5′-CTA CCT GGA GCC TCT TAA CAG CGA T-3′ and antisense 5′-TGG AGA AAG GCA TGG GAA TCT GG-3′; glyceraldehyde-3-phosphate dehydrogenase (GAPDH): sense 5′-CGA CCA CTT TGT CAA GCT CA-3′ and antisense 5′-AGG GGA GAT TCA GTG TGG TG-3′.
Real-time PCR
Total RNA was prepared with TRIzol reagent (Invitrogen Life Technologies). Real-time quantitative PCR was performed by using a cycler (Roche, Hercules, CA) and SYBR green dye. For data analysis, a method designated as 2–ΔΔCT was used to calculate fold change.34 GAPDH expression was set to be unaffected under our treatment conditions and used as reference gene for normalization of threshold cycle (Ct). Triplicates were set for each treatment and error bars represent the range of fold change calculated from the triplicates. The primer sequences for genes GAPDH, IL-2 and IL-4 were the same as given above. The followings are those for CBP and p300. CBP: sense 5′-TGG AAG ACC GAG TGA ACA A-3′ and antisense 5′-GTG GGT GGC AAT GGA AGA-3′; p300: sense 5′-AGG CTG TAT CAG AGC GTA TT-3′ and antisense 5′-ATC CCG ACC ATC CAT CAG-3′.
Experiments were repeated independently at least twice and representative data are displayed in the figures.
Flow cytometric analysis
Suspended in cold phosphate-buffered saline (PBS) supplemented with 1% FBS, the cells were incubated with mAbs for 30–45 min at 2–8° for determining percentages of HLA-DR+ (eBioscience, San Diego, CA) and IL-4 receptor α-positive (IL-4Rα+ R & D Systems) cells, respectively. After removing unreacted antibodies, the cell pellets were resuspended in 400 μl PBS buffer for flow cytometric analysis with FACS Calibur (Becton Dickinson, San Jose, CA).
ChIP assay
ChIP analysis was performed based on the protocol offered by Upstate Biotechnology (Lake Placid, NY). Briefly, cells were fixed with 1% formaldehyde for 10 min at 37°, washed with PBS, and lysed in ChIP lysis buffer (Upstate Biotechnology). Lysate was sonicated three to six times. Each of 100-μg protein/DNA aliquots was separately incubated overnight with following mAbs: anti-acetylated histone H3 and H4 (Upstate Biotechnology), anti-NFAT1 (Upstate Biotechnology), anti-STAT6 (M-20G, Santa Cruz), or anti-CBP/p300 (AC238, abcam, Cambridge, UK) at 5 μg per immunoprecipitation. Protein A–agarose beads (Upstate Biotechnology) were added for 1 hr, then sequentially washed with low-salt buffer, high-salt buffer, LiCl buffer, and TE buffer (with ingredients from Upstate Biotechnology). The beads were eluted with 0·1 m NaHCO3 and 1% sodium dodecyl sulphate (SDS), and the cross-linked protein/DNA was reversed at 65°. DNA from samples without the addition of antibodies was used as negative control and DNA isolated before immunoprecipitation was used as DNA input control. DNA was ethanol-precipitated in the presence of 20 μg glycogen. For PCR, the following primers were used. IL-4 promoter: sense 5′-CCA AGT GAC TGA CAA TCT GGT GTA ACG AA-3′ and antisense 5′-AAT AGG TGT CGA TTT GCA GTG ACA ATG TG −3′; IL-2 promoter: sense 5′-TTT TCT GAG TTA CTT TTG TAT CCC CAC CC-3′ and antisense 5′-TGA AAT CCC TCT TTG TTA CAT TAG CCC AC-3′. The PCR was performed for the chromatin DNA samples at 94° for 4 min, followed by 28 cycles of 94° for 1 min, 60° for 1 min and 72° for 1 min, ending at 72° for 5 min.
Western blotting
Western blotting was performed for whole cell lysate. Aliquots of total protein (50 μg per lane) were electrophoresed on a 12% SDS–polyacrylamide gradient gel and transferred to nitrocellulose membranes (Millipore, Billerica, MA). Washed in rinse buffer at room temperature for 15 min and incubated in blocking buffer (5% fat-free milk in rinse buffer) for 30 min, the membranes were incubated for 2 hr at room temperature with anti CIITA mAb (7-1H, Upstate). Further washed with rinse buffer, the membranes were incubated with 1 : 1000 diluted horseradish peroxidase-conjugated anti-mouse immunoglobulin antibody (Santa Cruz) followed by developing with enhanced chemiluminescence reagents (Amersham, Little Chalfont Buckinghamshire, UK).
Cytokine enzyme-linked immunosorbent assay (ELISA)
Amounts of IL-2, IFN-γ and IL-4 were determined from culture supernatants using the appropriate OptEIA ELISA kit (BD Biosciences) according to the manufacturer's instructions. The coefficient of variation (CV) of interassay and intra-assay for ELISA in our experiments was less than 10%.
Statistical analyses
Results are expressed as means ±SD. Inter-group comparisons were made using one-way analysis of variance (anova). A P-value less than 0·05 was considered statistically significant.
Results
Activity of CIITA in inducing HLA-DR expression in Jurkat cells
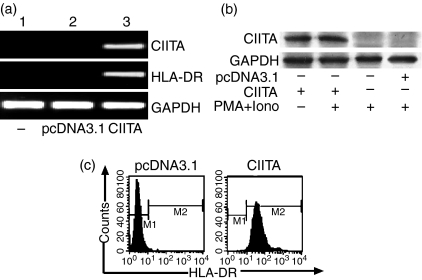
As reported, Jurkat cells usually do not constitutively express MHC class II molecules in the absence of CIITA. Indeed, we did not detect HLA-DR and CIITA mRNA by RT-PCR in the cells (Fig. 1a), even when stimulated with PMA plus ionomycin (PMA + iono) or with IFN-γ (500 U/ml) for 6–24 hr (data not shown). After transfection with CIITA cDNA, however, Jurkat cells expressed CIITA mRNA quite well (Fig. 1a). CIITA protein and HLA-DR molecules could also be detected by Western blotting and flow cytometry, respectively (Fig. 1b,c). Stimulation with PMA plus ionomycin did not change the expression patterns significantly. This indicates that dysfunction of the CIITA gene might account for the HLA-DR gene being silenced in Jurkat cells, and that exogenous CIITA could restore the activation of the class II gene. Accordingly, the CIITA gene we prepared has full native activity for inducing class II gene expression.
Figure 1.
Expressions of CIITA and HLA-DR in Jurkat cells. (a) CIITA and HLA-DR are expressed only in the Jurkat cells transfected with CIITA gene (lane 3) as determined by RT-PCR. Lanes 1 and 2 are negative controls. (b) Similar expression patterns were detectable by Western blotting in the CIITA-transfected Jurkat cells with or without PMA plus ionomycin (Iono) as stimulators. (c) After stimulation with PMA plus ionomycin, HLA-DR expression in CIITA- and empty vector (pcDNA3·1)-transfected Jurkat cells were evaluated by flow cytometric analysis. The DR-positive cells in the CIITA-transfected population have been enriched by anti-HLA-DR microbeads.
Depression of IL-4 expression in the CIITA-transfected Jurkat cells
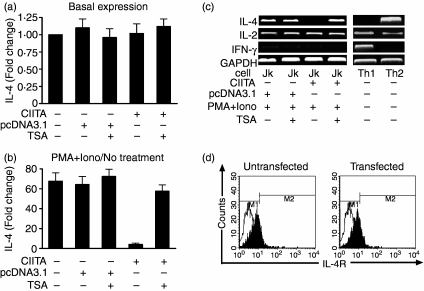
Jurkat cells usually express IL-4 and IL-2 but not IFN-γ after stimulation with PMA + iono.35,36 These cells were thus employed to evaluate the possible role of CIITA in the regulation of IL-4 gene expression. There was no difference in the basal IL-4 expression (without PMA + iono stimulation) in Jurkat cells of different groups as determined by real-time PCR (Fig. 2a). Treatment with PMA + iono for 6 hr could significantly increase IL-4 mRNA transcription in wild-type and pcDNA3.1-transfected Jurkat cells. However, IL-4 expression was dramatically decreased by CIITA transfection (Fig. 2b). The inhibitory effects of CIITA on IL-4 expression were also observed using RT-PCR. As shown in Fig. 2(c) PMA + iono-stimulated IL-4 transcription was almost totally abolished in CIITA-transfected cells. In contrast to IL-4, we found that IL-2 expression was still maintained at high levels in CIITA-transfected cells (Fig. 2c). Because CIITA did not alter the low expression pattern of IFN-γ (Fig. 2c), we excluded the possibility that selective depression of the transcriptional activation of the IL-4 gene was attributable to the expression of IFN-γ, which has been regarded as a counter factor for IL-4 expression via interferon regulatory factors (IRFs) in the IFN-γ receptor-initiated signalling pathway.23,37 In addition, IL-4 was reported to be able to bind to its receptor and enhance its self-expression through a positive feedback in the T cells.38 In our experiments, flow cytometry analysis indicated that the percentages of IL-4R-positive Jurkat cells remained unchanged before and after the cells were transfected with the CIITA gene (Fig. 2d), which also excludes another possibility that CIITA inhibits IL-4 transcription by down-regulation of IL-4R expression.
Figure 2.
Suppressed expression of IL-4 in CIITA-transfected Jurkat cells. (a) There was no difference in basal IL-4 expression in the different groups of Jurkat cells without PMA + Iono stimulation. Fold changes were calculated by comparing the amount of IL-4 mRNA in the different groups with that of wild-type cells followed by normalization to GAPDH levels. The amount of IL-4 mRNA in wild-type cells without TSA treatment was designated as 1·0 (see bar 1). (b) Determination of the suppressed expressions of IL-4 by real-time PCR in the Jurkat cells stimulated with PMA plus ionomycin. Fold changes of transcription in each group were calculated by comparing IL-4 expression in (PMA + Iono)-stimulated cells with that of unstimulated cells (no treatment) followed by normalization to GAPDH levels. The amount of IL-4 mRNA in unstimulated cells of each group was designated as ‘1·0’. Treatment with TSA reversed the CIITA-related suppression of IL-4 expression. (c) The expression of IL-4, instead of IL-2, is almost completely inhibited in the CIITA-transfected Jurkat cells stimulated by PMA plus ionomycin as determined by RT-PCR. The depression can be reversed by TSA, an inhibitor of histone acetyltransferase. Th1 and Th2 cells were used as controls. (d) No change in percentages for IL-4R-positive Jurkat cells (around 20%) before (left) and after (right) transfection with the CIITA gene, suggesting no functional involvement of IL-4R in the CIITA-related suppression of IL-4. Black and white peaks stand for the negative control and the IL-4R-positive cells, respectively.
It is of interest to note, on the other hand, that the CIITA-induced inhibition of the IL-4 gene was reversed by TSA, a histone deacetylatase inhibitor, as determined either by RT-PCR (Fig. 2c) or real-time quantitative PCR (Fig. 2b). It was shown, for example, that CIITA transfection was able to depress IL-4 expression from 64·2 ± 8·22 (cells transfected with pcDNA3.1 as control) to 4·13 ± 1·36 in terms of fold change (Fig. 2b). TSA restored the expression to a level of 57·6 ± 6·32. TSA alone did not induce IL-4 expression after treatment for 6–48 hr in all other experimental groups (data not shown). Taken together, these results strongly suggested that CIITA was able to interfere with histone acetylation, which in turn accounted for the inhibition of the endogenous IL-4 gene by epigenetic mechanisms.
Histone deacetylation at IL-4 promoter
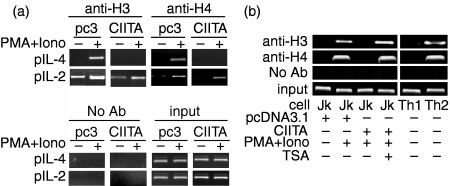
It has been established that histone acetylation initiates a more ‘opened’ chromatin structure to increase the accessibility of transcription complexes to related DNA boxes.16,39 To explore whether the histone deacetylation at the IL-4 gene promoter region was involved in depressing IL-4 gene transcription, both the CIITA-induced HLA-DR+ Jurkat cells, which had been enriched by anti-DR microbeads, and the Jurkat cells transfected with empty vector pcDNA3.1 were activated by PMA + iono for 6 hr. They were then subjected to ChIP analysis using antiacetylated histone H3 and H4 antibody. As shown in Fig. 3(a), histones H3 and H4 of the IL-4 promoter, instead of the IL-2 promoter, retained hypoacetylation status in unstimulated cells. After stimulation with PMA + iono, mock-transfected Jurkat cells retained high levels of histone H3 and H4 acetylation because the IL-4 promoter product was fully detectable using specific primers. In contrast, the DNA–protein complex from the CIITA-transfected cells could not be detected, suggesting that there was a strong abrogation of histone acetylation in the IL-4 promoter region (Fig. 3a). Again, the hypoacetylation could be reversed by TSA (Fig. 3b), which correlated well with the restoration of IL-4 mRNA expression. These results demonstrated that histone hypoacetylation at the promoter region accounted for the inhibitory effects of CIITA on IL-4 gene transcriptional activation.
Figure 3.
Hypoacetylation of histones H3 and H4 at the IL-4 promoter in Jurkat cells transfected with CIITA. (a) Empty vector pcDNA3·1(pc3)- and CIITA-transfected Jurkat cells were activated by PMA plus ionomycin (PMA + Iono). The chromatin harvested from formaldehyde-fixed cells was immunoprecipitated with antiacetylated histone H3 and H4 antibody and amplified by PCR with specific primers to IL-4 or IL-2 promoter. The level of histone acetylation in IL-4 promoter (pIL-4), instead of IL-2 promoter (pIL-2), was depressed in the CIITA-transfected cells as determined by ChIP assay. Results are representative of three independent experiments. (b) ChIP assay indicated that the CIITA-induced H3 and H4 hypoacetylation in the IL-4 promoter of Jurkat cells can be reversed by TSA. Th1 and Th2 cells were used as controls.
Impaired binding of STAT6, NFAT and CBP/p300 to IL-4 promoter
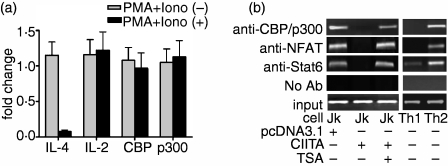
Both CBP and p300 are highly homologous coactivators, which participate in the regulation of gene expressions.40 It was reported that CBP/p300 are required in IL-4-induced gene transcription, and nuclear factor STAT6 is also actively involved through binding to IL-4 promoter.41,42 The intrinsic histone acetyltranferase activity found in CBP/p300 is thought to play a crucial role in transcription by conferring modification of the chromatin structure. To explore the role of the coactivators in the histone hypoacetylation at IL-4 promoter, CBP and p300 expression levels were first examined by RT-PCR in our experiments. The two coactivators were constitutively expressed in Jurkat cells, and stimulation with PMA + iono did not change their expression levels (data not shown). Figure 4(a) shows that CIITA transfection could not affect the expression pattern of CBP and p300, either in unstimulated or in stimulated Jurkat cells. As a control, the PMA + iono-stimulated IL-4 expression in CIITA-transfected Jurkat cells was significantly lower than that in the mock-transfected cells. Real-time PCR also indicated that the expression of the IL-2 gene in CIITA-transfected Jurkat cells was not affected (Fig. 4a). This correlated well with the finding that IL-2 promoter could be immunoprecipitated at the same level both in CIITA-transfected and mock-transfected cells using antiacetylated histone H3 and H4 antibody (Fig. 3a).
Figure 4.
CIITA-related abrogation of STAT6, NF-AT and CBP/p300 binding to IL-4 promoter. (a) The expression patterns of IL-4, IL-2, CBP and p300 in pcDNA3·1- or CIITA-transfected Jurkat cells were examined by real-time PCR. In contrast to depressed expression of IL-4, the relative expression levels of the measured genes between pcDNA3·1- and CIITA-transfected Jurkat cells were basically unaffected (around 1·0). Fold changes were calculated by comparing CIITA-transfected cells with those transfected by pcDNA3·1 (designated as 1·0), followed by normalization to GAPDH levels. (b) The association of CBP/p300 with IL-4 promoter, however, was abrogated in the CIITA-transfected Jurkat cells stimulated with PMA plus ionomycin, as determined by ChIP assay. TSA treatment shows the recovery effect. In the experiment, the chromatin harvested from formaldehyde-fixed cells was immunoprecipitated with anti-CBP/p300 antibody and amplified by PCR with specific primers to IL-4 promoter. The binding of NFAT and STAT6 to IL-4 promoter was also blocked in CIITA-transfected cells.
Although the expression levels of CBP and p300 were not affected, they were not able to bind IL-4 promoter in stimulated Jurkat cells because of CIITA transfection, as determined by ChIP assay with anti-CBP/p300 mAb (Fig. 4b). It has been reported that transcription factors STAT6 and NFAT are also active in IL-4 gene expression. STAT6 regulates the expression of GATA-3, which in turn modulates the production of the Th2-related cytokines and the chromatin remodelling.43,44 NFAT and its cooperating transcription factor AP-1 are implicated in the acute phase of IL-4 gene transcription by differentiated Th2 cells.45,46 NFAT1 is the most abundant NFAT molecule in the T cell.47 To analyse whether the binding of STAT6 and NFAT1 to the IL-4 promoter is involved in CIITA-related suppression of IL-4 transcription, relevant ChIP assays were performed. As expected, formation of a DNA–protein complex was only detected in the control group but not in the CIITA-transfected Jurkat cells (Fig. 4b). Since the accessibility of local chromatin to transcription factors is closely associated with the level of histone acetylation, and thus influences gene expression, our data suggested that the interruption of these proteins binding to the promoter might be one of the critical events for CIITA-induced IL-4 gene suppression. In addition, TSA treatment restored the binding of the three factors to IL-4 promoter (Fig. 4b), indicating the essential role of histone hypoacetylation. This strongly supported the proposal that CIITA is active in down-regulating IL-4 gene expression by epigenetic mechanisms.
CIITA siRNA enhances IL-4 production in human primary Th1 cells
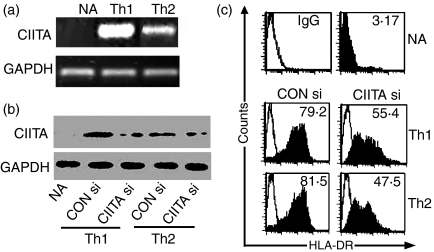
In contrast to rodent T cells, human T cells express substantial amounts of CIITA and MHC class II molecules upon activation. Moreover, we found that differentiated Th1 cells produced higher levels of CIITA mRNA than Th2 cells (Fig. 5a). Flow cytometry analysis indicated that they expressed comparable amounts of HLA-DR (Fig. 5b,c), implying that relatively low levels of CIITA in Th2 cells were sufficient for trans-activating the expression of MHC class II genes. Introduction of CIITA siRNA significantly inhibited CIITA and HLA-DR expression in both Th1 and Th2 cells (P < 0·05). Suppression percentages of DR expression, for example, were 23·8% in Th1 and 34·0% in Th2, respectively (Fig. 5b,c).
Figure 5.
CIITA siRNA inhibited expression of CIITA and HLA-DR in both Th1 and Th2 cells. (a) In contrast to the negative effect of CIITA mRNA in naive Th (NA) cells, a higher transcription level of CIITA mRNA was detected in the in vivo differentiated Th1 cells than in Th2 cells as determined by regular RT-PCR. (b) Unstimulated naive T cells were transfected with control siRNA (CON si) or CIITA siRNA (CIITA si) and then stimulated with anti-CD3 (1 μg/ml) plus anti-CD28 (2 μg/ml) for 72 hr in Th1 or Th2 differential conditions, and the effects of siRNA on CIITA expression in differentiated Th1 or Th2 cells were evaluated by Western blotting. It was shown that siRNA efficiently inhibited CIITA expression in both Th1 and Th2 cells. (c) Flow cytometric analysis indicated that NA cells did not express surface HLA-DR. Th1 and Th2 cells could express comparable levels of HLA-DR molecules, which was significantly inhibited by introduction of CIITA siRNA.
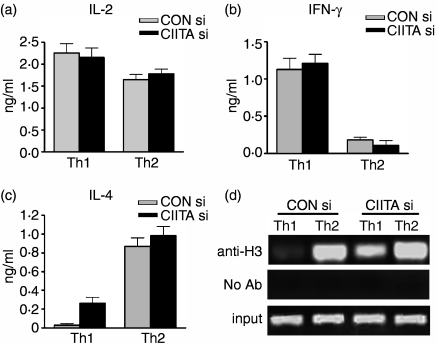
After stimulation with anti-CD3 and anti-CD28 mAbs for 12 hr, the cells were subjected to analysis of the levels of IFN-γ and IL-4 in the supernatants by ELISA. As expected, normal Th1 cells secreted high levels of IFN-γ, whereas IL-4 expression was almost undetectable. Interestingly, CIITA siRNA made these cells produce a greater amount of IL-4 although IFN-γ and IL-2 expressions were not affected (Fig. 6a–c). ChIP assay indicated that CIITA siRNA significantly enhanced the level of histone acetylation of the IL-4 promoter in Th1 cells compared with normal Th1 cells (Fig. 6d). On the other hand, CIITA siRNA did not alter the patterns of low expression of IFN-γ and high expression of IL-4 in Th2 cells.
Figure 6.
CIITA siRNA enhanced IL-4 production in Th1 cells. (a, b, c) Culture supernatants of the Th1 and Th2 cells restimulated with anti-CD3 plus anti-CD28 were collected and the cytokines levels were determined by ELISA. The results show that CIITA siRNA (CIITA si)-transfected Th1 cells produced substantial levels of IL-4 compared to controls, while expression of IFN-γ and IL-2 were basically not changed. On the other hand, decreased levels of CIITA did not significantly alter cytokine expression patterns in Th2 cells. (d) The levels of histone acetylation in the IL-4 promoter of Th1 and Th2 cells were determined by ChIP assay. The acetylated histone H3 of IL-4 promoter was significantly increased in CIITA si-treated Th1 cells, but not in the relevant Th2 cells.
Discussion
CIITA is a master controller for expression of MHC class II genes and thus plays a key role in the immune response. A series of papers have indicated that, in addition to MHC class II genes, CIITA may also be active in regulating other genes within or outside the immune system.27,48–50 To date, the influence of CIITA on Th1/Th2 differentiation remains controversial and the mechanisms by which CIITA influences the expression of type 1 and type 2 cytokines are still far from clear. Th1 cells from CIITA-deficient or CIITA-transgenic mouse models all exhibited enhanced IL-4 expression.27,30 On the other hand, over-expression and transfection assays suggested that CIITA suppressed IL-4 gene transcription.28 The discrepancy might be the result of the different experimental systems and conditions used. Another possibility is the biological difference of CIITA between humans and mice, especially considering that activated murine T cells express undetectable CIITA protein and MHC class II molecules, whereas human T cells express substantial levels of CIITA and MHC class II after activation. In addition, CIITA may influence different target genes by different mechanisms. Sequestration of CBP/p300, for example, was suggested to be involved in the suppression of the activation of collagen I, FasL and IL-4 genes.28,48,50 More recently, an investigation reported that CIITA could be recruited through nuclear factor RFX5 to collagen I gene promoter to suppress its transcription in vivo.51 In the present study, epigenetic mechanisms are demonstrated to be involved in CIITA-mediated human IL-4 gene suppression. Moreover, the effects seem to be cytokine-specific, as IFN-γ and IL-2 expression were not altered in our experiments, either with primary T cells or with the CIITA over-expressed cell line.
It has been proposed that there are three phases or stages in the transcriptional induction of IL-4 expression.52,53 In the initiation phase, naive helper T cells are stimulated by signalling from the ligation of IL-4R and TCR. In the commitment phase, the activation of STAT6 and the induction of GATA-3 transcription initiate remodelling of the IL-4 locus. Last is the acute transcription phase, in which the IL-4 gene is under the influence of lineage-specific and non-specific nuclear factors such as NFAT, AP-1 and c-Maf for final activation. Accordingly, remodelling of the IL-4 locus and epigenetic regulation are key control events. Among them, histone acetylation emerges as a central switch mechanism allowing interconversion between permissive and repressive chromatin domains in terms of transcriptional competence.54 In the case of Th2-biased conditions, histone of the IL-4 locus is acetylated, and chromatin is remodelled into a more permissive status that allows easier accession of NFAT and other transcription factors required for sustained, high-level IL-4 gene expression.55 From the present investigation, we provide evidence that CIITA siRNA can transfer the IL-4 gene from its inactive condition to active expression in human primary Th1 cells. The ChIP assay proved that the expression was associated with increased levels of histone acetylation in the IL-4 promoter. Similarly, in our transfection assay with Jurkat T cells, overexpression of CIITA suppressed endogenous IL-4 transcription, which could be reversed by the histone deacetylatase inhibitor TSA, indicating that CIITA is able to function as a down-regulator in human IL-4 gene expression via epigenetic mechanisms.
In this study, we provide evidence that the binding of transcription factors CBP, STAT6 and NFAT to IL-4 promoter was prevented by CIITA over-expression. Of note, IFN-γ and IL-2 gene transcription were reportedly activated by CBP/p300 and NFAT, respectively,56,57 whereas their expression was not affected in either the CIITA siRNA introduced primary T cells or the CIITA over-expressed cell line in the present study, implying that CIITA is not a global inhibitor of genes that utilize these transcription factors. On the other hand, we suggest that the CIITA-induced histone deacetylation in the IL-4 promoter may play an initial role in IL-4 suppression. First, the expression level of transcription factors CBP/p300, STAT6 and NFAT was not repressed in CIITA overexpressed cells. Second, TSA treatment resulted in the re-binding of these transcription factors to IL-4 promoter and the restoration of IL-4 expression in these cells, which was closely associated with the up-regulation of histone acetylation.
Our data have significant implications for understanding the mechanism of Th1/Th2 differentiation at the level of chromatin. Although the precise mechanisms by which CIITA is simultaneously able to enhance class II expression and inhibit histone acetylation of the IL-4 promoter are not very clear, the results we found may shed some light on developing strategies for interfering with some Th1- and/or Th2-related diseases using epigenetic mechanisms. For example, TSA and other histone deacetylase inhibitors have been extensively tested in antitumour research to induce cancer cell differentiation or apoptosis.58–60 It is worth investigating whether histone deacetylase inhibitors can be used to reverse CIITA-mediated IL-4 suppression and therefore prevent Th1 cell-mediated diseases such as rheumatoid arthritis and autoimmune diabetes. It was also reported that CIITA-deficient Th1 cells can produce IL-4 in the absence of exogenous IL-4, and that the proportions of IL-4+ Th1 cells remained comparable upon multiple restimulations.61 So it is likely that the Th1/Th2 balance can be modulated through CIITA, including using siRNA as proved in our study for human primary T cells. The non-viral gene delivery system62,63 may offer an efficient means to develop relevant T-cell vaccines in vitro. It also provides an opportunity to interfere with Th cell responses in vivo, even in specific cell types, as a benefit of new siRNA technology.64
Acknowledgments
We thank Drs Y. Wang, D-Q. Zhang and W. Sun for their valuable suggestions and discussions. Thanks are also due to Mr T.W. Shen and Y. Ge for their excellent technical assistance. This work was supported by grants from the Natural Science Foundation of China (30530690), the National Key Basic Research Programme of China (2001CB510003), the Shanghai Science-Technology Foundation (05DZ19734), and the Shanghai Leading Academic Discipline Project (T0206).
Abbreviations:
- CIITA
class II transactivator
- CBP
CREB-binding protein
- ChIP
chromatin immunoprecipitation
- TSA
Trichostatin A.
References
- 1.Boss JM, Jensen PE. Transcriptional regulation of the MHC class II antigen presentation pathway. Curr Opin Immunol. 2003;15:105–11. doi: 10.1016/s0952-7915(02)00015-8. [DOI] [PubMed] [Google Scholar]
- 2.Ting JP, Trowsdale J. Genetic control of MHC class II expression. Cell. 2002;109(Suppl.):S21–S33. doi: 10.1016/s0092-8674(02)00696-7. [DOI] [PubMed] [Google Scholar]
- 3.Steimle V, Siegrist CA, Mottet A, Lisowska-Grospierre B, Mach B. Regulation of MHC class II expression by interferon-gamma mediated by the transactivator gene CIITA. Science. 1994;265:106–9. doi: 10.1126/science.8016643. [DOI] [PubMed] [Google Scholar]
- 4.Chang CH, Hong SC, Hughes CC, Janeway CA, Jr, Flavell RA. CIITA activates the expression of MHC class II genes in mouse T cells. Int Immunol. 1995;7:1515–18. doi: 10.1093/intimm/7.9.1515. [DOI] [PubMed] [Google Scholar]
- 5.Holling TMSN, Quinten E, van den Elsen PJ. Activated human T cells accomplish MHC class II expression through T cell-specific occupation of class II transactivator promoter III. J Immunol. 2002;168:763–70. doi: 10.4049/jimmunol.168.2.763. [DOI] [PubMed] [Google Scholar]
- 6.Muhlethaler-Mottet A, Otten LA, Steimle V, Mach B. Expression of MHC class II molecules in different cellular and functional compartments is controlled by differential usage of multiple promoters of the transactivator CIITA. EMBO J. 1997;16:2851–60. doi: 10.1093/emboj/16.10.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fry CJ, Peterson CL. Transcription. Unlocking the gates to gene expression. Science. 2002;295:1847–8. doi: 10.1126/science.1070260. [DOI] [PubMed] [Google Scholar]
- 8.Kretsovali A, Agalioti T, Spilianakis C, Tzortzakaki E, Merika M, Papamatheakis J. Involvement of CREB binding protein in expression of major histocompatibility complex class II genes via interaction with the class II transactivator. Mol Cell Biol. 1998;18:6777–83. doi: 10.1128/mcb.18.11.6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masternak K, Peyraud N, Krawczyk M, Barras E, Reith W. Chromatin remodeling and extragenic transcription at the MHC class II locus control region. Nat Immunol. 2003;4:132–7. doi: 10.1038/ni883. [DOI] [PubMed] [Google Scholar]
- 10.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl.):245–54. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 11.Smale ST, Fisher AG. Chromatin structure and gene regulation in the immune system. Annu Rev Immunol. 2002;20:427–62. doi: 10.1146/annurev.immunol.20.100301.064739. [DOI] [PubMed] [Google Scholar]
- 12.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 13.Cosgrove MS, Wolberger C. How does the histone code work? Biochem Cell Biol. 2005;83:468–76. doi: 10.1139/o05-137. [DOI] [PubMed] [Google Scholar]
- 14.Hebbes TR, Clayton AL, Thorne AW, Crane-Robinson C. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken beta-globin chromosomal domain. EMBO J. 1994;13:1823–30. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Braunstein M, Rose AB, Holmes SG, Allis CD, Broach JR. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 16.Mizzen CA, Allis CD. Linking histone acetylation to transcriptional regulation. Cell Mol Life Sci. 1998;54:6–20. doi: 10.1007/s000180050121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee GR, Kim ST, Spilianakis CG, Fields PE, Flavell RA. T helper cell differentiation. Regulation by cis elements and epigenetics. Immunity. 2006;24:369–79. doi: 10.1016/j.immuni.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 18.Su IH, Tarakhovsky A. Epigenetic control of B cell differentiation. Semin Immunol. 2005;17:167–72. doi: 10.1016/j.smim.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 19.Mostoslavsky R, Alt FW, Bassing CH. Chromatin dynamics and locus accessibility in the immune system. Nat Immunol. 2003;4:603–6. doi: 10.1038/ni0703-603. [DOI] [PubMed] [Google Scholar]
- 20.Paul WE. Interleukin-4: a prototypic immunoregulatory lymphokine. Blood. 1991;77:1859–70. [PubMed] [Google Scholar]
- 21.behsira-Amar O, Gibert M, Joliy M, Theze J, Jankovic DL. IL-4 plays a dominant role in the differential development of Th0 into Th1 and Th2 cells. J Immunol. 1992;148:3820–9. [PubMed] [Google Scholar]
- 22.Kim JI, Ho IC, Grusby MJ, Glimcher LH. The transcription factor c-Maf controls the production of interleukin-4 but not other Th2 cytokines. Immunity. 1999;10:745–51. doi: 10.1016/s1074-7613(00)80073-4. [DOI] [PubMed] [Google Scholar]
- 23.Elser B, Lohoff M, Kock S, et al. IFN-gamma represses IL-4 expression via IRF-1 and IRF-2. Immunity. 2002;17:703–12. doi: 10.1016/s1074-7613(02)00471-5. [DOI] [PubMed] [Google Scholar]
- 24.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–96. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 25.Fields PE, Kim ST, Flavell RA. Cutting edge. Changes in histone acetylation at the IL-4 and IFN-gamma loci accompany Th1/Th2 differentiation. J Immunol. 2002;169:647–50. doi: 10.4049/jimmunol.169.2.647. [DOI] [PubMed] [Google Scholar]
- 26.Yamashita M, Ukai-Tadenuma M, Miyamoto T, et al. Essential role of GATA3 for the maintenance of type 2 helper T (Th2) cytokine production and chromatin remodeling at the Th2 cytokine gene loci. J Biol Chem. 2004;279:26983–90. doi: 10.1074/jbc.M403688200. [DOI] [PubMed] [Google Scholar]
- 27.Gourley T, Roys S, Lukacs NW, Kunkel SL, Flavell RA, Chang CH. A novel role for the major histocompatibility complex class II transactivator CIITA in the repression of IL-4 production. Immunity. 1999;10:377–86. doi: 10.1016/s1074-7613(00)80037-0. [DOI] [PubMed] [Google Scholar]
- 28.Sisk TJ, Gourley T, Roys S, Chang CH. MHC class II transactivator inhibits IL-4 gene transcription by competing with NF-AT to bind the coactivator CREB binding protein (CBP)/p300. J Immunol. 2000;165:2511–17. doi: 10.4049/jimmunol.165.5.2511. [DOI] [PubMed] [Google Scholar]
- 29.Otten LA, Leibundgut-Landmann S, Huarte J, et al. Revisiting the specificity of the MHC class II transactivator CIITA in vivo. Eur J Immunol. 2006;36:1548–58. doi: 10.1002/eji.200535687. [DOI] [PubMed] [Google Scholar]
- 30.Otten LA, Tacchini-Cottier F, Lohoff M, et al. Deregulated MHC class II transactivator expression leads to a strong Th2 bias in CD4+ T lymphocytes. J Immunol. 2003;170:1150–7. doi: 10.4049/jimmunol.170.3.1150. [DOI] [PubMed] [Google Scholar]
- 31.Park WS, Bae Y, Chung DH, et al. T cell expression of CIITA represses Th1 immunity. Int Immunol. 2004;16:1355–64. doi: 10.1093/intimm/dxh132. [DOI] [PubMed] [Google Scholar]
- 32.Cousins DJ, Lee TH, Staynov DZ. Cytokine coexpression during human Th1/Th2 cell differentiation. Direct evidence for coordinated expression of Th2 cytokines. J Immunol. 2002;169:2498–506. doi: 10.4049/jimmunol.169.5.2498. [DOI] [PubMed] [Google Scholar]
- 33.Zika E, Greer SF, Zhu XS, Ting JP. Histone deacetylase 1/mSin3A disrupts gamma interferon-induced CIITA function and major histocompatibility complex class II enhanceosome formation. Mol Cell Biol. 2003;23:3091–102. doi: 10.1128/MCB.23.9.3091-3102.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔC (T) method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Li-Weber M, Giaisi M, Baumann S, Palfi K, Krammer PH. NF-kappa B synergizes with NF-AT and NF-IL6 in activation of the IL-4 gene in T cells. Eur J Immunol. 2004;34:1111–18. doi: 10.1002/eji.200324687. [DOI] [PubMed] [Google Scholar]
- 36.Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–69. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 37.So EY, Park HH, Lee CE. IFN-gamma and IFN-alpha posttranscriptionally down-regulate the IL-4-induced IL-4 receptor gene expression. J Immunol. 2000;165:5472–9. doi: 10.4049/jimmunol.165.10.5472. [DOI] [PubMed] [Google Scholar]
- 38.Nelms K, Keegan AD, Zamorano J, Ryan JJ, Paul WE. The IL-4 receptor: signaling mechanisms and biologic functions. Annu Rev Immunol. 1999;17:701–38. doi: 10.1146/annurev.immunol.17.1.701. [DOI] [PubMed] [Google Scholar]
- 39.Gorisch SM, Wachsmuth M, Toth KF, Lichter P, Rippe K. Histone acetylation increases chromatin accessibility. J Cell Sci. 2005;118:5825–34. doi: 10.1242/jcs.02689. [DOI] [PubMed] [Google Scholar]
- 40.Blobel GA. CBP and p300: versatile coregulators with important roles in hematopoietic gene expression. J Leukoc Biol. 2002;71:545–56. [PubMed] [Google Scholar]
- 41.Gingras S, Simard J, Groner B, Pfitzner E. p300/CBP is required for transcriptional induction by interleukin-4 and interacts with Stat6. Nucl Acids Res. 1999;27:2722–9. doi: 10.1093/nar/27.13.2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McDonald C, Reich NC. Cooperation of the transcriptional coactivators CBP and p300 with Stat6. J Interferon Cytokine Res. 1999;19:711–22. doi: 10.1089/107999099313550. [DOI] [PubMed] [Google Scholar]
- 43.Lee HJ, Takemoto N, Kurata H, et al. GATA-3 induces T helper cell type 2 (Th2) cytokine expression and chromatin remodeling in committed Th1 cells. J Exp Med. 2000;192:105–15. doi: 10.1084/jem.192.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kurata H, Lee HJ, O'Garra A, Arai N. Ectopic expression of activated Stat6 induces the expression of Th2-specific cytokines and transcription factors in developing Th1 cells. Immunity. 1999;11:677–88. doi: 10.1016/s1074-7613(00)80142-9. [DOI] [PubMed] [Google Scholar]
- 45.Burke TF, Casolaro V, Georas SN. Characterization of P5, a novel NFAT/AP-1 site in the human IL-4 promoter. Biochem Biophys Res Commun. 2000;270:1016–23. doi: 10.1006/bbrc.2000.2508. [DOI] [PubMed] [Google Scholar]
- 46.Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol. 1997;15:707–47. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 47.Xanthoudakis S, Viola JP, Shaw KT, et al. An enhanced immune response in mice lacking the transcription factor NFAT1. Science. 1996;272:892–5. doi: 10.1126/science.272.5263.892. [DOI] [PubMed] [Google Scholar]
- 48.Zhu XS, Ting JP. A 36-amino-acid region of CIITA is an effective inhibitor of CBP. novel mechanism of gamma interferon-mediated suppression of collagen alpha(2)(I) and other promoters. Mol Cell Biol. 2001;21:7078–88. doi: 10.1128/MCB.21.20.7078-7088.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wong AW, Brickey WJ, Taxman DJ, et al. CIITA-regulated plexin-A1 affects T-cell–dendritic cell interactions. Nat Immunol. 2003;4:891–8. doi: 10.1038/ni960. [DOI] [PubMed] [Google Scholar]
- 50.Gourley TS, Chang CH. Cutting edge: the class II transactivator prevents activation-induced cell death by inhibiting Fas ligand gene expression. J Immunol. 2001;166:2917–21. doi: 10.4049/jimmunol.166.5.2917. [DOI] [PubMed] [Google Scholar]
- 51.Xu Y, Wang L, Buttice G, Sengupta PK, Smith BD. Major histocompatibility class II transactivator (CIITA) mediates repression of collagen (COL1A2) transcription by interferon gamma (IFN-gamma) J Biol Chem. 2004;279:41319–32. doi: 10.1074/jbc.M404174200. [DOI] [PubMed] [Google Scholar]
- 52.Avni O, Rao A. T cell differentiation: a mechanistic view. Curr Opin Immunol. 2000;12:654–9. doi: 10.1016/s0952-7915(00)00158-8. [DOI] [PubMed] [Google Scholar]
- 53.Murphy KM, Reiner SL. The lineage decisions of helper T cells. Nat Rev Immunol. 2002;2:933–44. doi: 10.1038/nri954. [DOI] [PubMed] [Google Scholar]
- 54.Verdone L, Di Caserta M., ME Role of histone acetylation in the control of gene expression. Biochem Cell Biol. 2005;83:344–53. doi: 10.1139/o05-041. [DOI] [PubMed] [Google Scholar]
- 55.Ansel KM, Lee DU, Rao A. An epigenetic view of helper T cell differentiation. Nat Immunol. 2003;4:616–23. doi: 10.1038/ni0703-616. [DOI] [PubMed] [Google Scholar]
- 56.Nakajima T, Uchida C, Anderson SF, Parvin JD, Montminy M. Analysis of a cAMP-responsive activator reveals a two-component mechanism for transcriptional induction via signal-dependent factors. Genes Dev. 1997;11:738–47. doi: 10.1101/gad.11.6.738. [DOI] [PubMed] [Google Scholar]
- 57.Yea SS, Yang KH, Kaminski NE. Role of nuclear factor of activated T-cells and activator protein-1 in the inhibition of interleukin-2 gene transcription by cannabinol in EL4 T-cells. J Pharmacol Exp Ther. 2000;292:597–605. [PubMed] [Google Scholar]
- 58.Bai J, Demirjian A, Sui J, Marasco W, Callery MP. Histone deacetylase inhibitor trichostatin A and proteasome inhibitor PS-341 synergistically induce apoptosis in pancreatic cancer cells. Biochem Biophys Res Commun. 2006;348:1245–53. doi: 10.1016/j.bbrc.2006.07.185. [DOI] [PubMed] [Google Scholar]
- 59.Zhang X, Yashiro M, Ren J, Hirakawa K. Histone deacetylase inhibitor, trichostatin A, increases the chemosensitivity of anticancer drugs in gastric cancer cell lines. Oncol Rep. 2006;16:563–8. [PubMed] [Google Scholar]
- 60.Alao JP, Stavropoulou AV, Lam EW, Coombes RC, Vigushin DM. Histone deacetylase inhibitor, trichostatin A induces ubiquitin-dependent cyclin D1 degradation in MCF-7 breast cancer cells. Mol Cancer. 2006;5:8. doi: 10.1186/1476-4598-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Patel DR, Li W, Park JS, et al. Constitutive expression of CIITA directs CD4 T cells to produce Th2 cytokines in the thymus. Cell Immunol. 2005;233:30–40. doi: 10.1016/j.cellimm.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 62.Stallwood Y, Briend E, Ray KM, et al. Small interfering RNA-mediated knockdown of notch ligands in primary CD4+ T cells and dendritic cells enhances cytokine production. J Immunol. 2006;177:885–95. doi: 10.4049/jimmunol.177.2.885. [DOI] [PubMed] [Google Scholar]
- 63.Yin J, Ma Z, Selliah N, Shivers DK, Cron RQ, Finkel TH. Effective gene suppression using small interfering RNA in hard-to-transfect human T cells. J Immunol Meth. 2006;312:1–11. doi: 10.1016/j.jim.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 64.Ikeda Y, Taira K. Ligand-targeted delivery of therapeutic siRNA. Pharm Res. 2006;23:1631–40. doi: 10.1007/s11095-006-9001-x. [DOI] [PubMed] [Google Scholar]