Abstract
Coenzyme Q10 (CoQ10) is essential for ATP production in the mitochondria, and is an important antioxidant in every biomembrane and lipoprotein. Due to its hydrophobicity, a binding and transfer protein for CoQ10 is plausible, but none have yet been isolated and characterized. Here we purified a CoQ10-binding protein from human urine and identified it to be saposin B, a housekeeping protein necessary for sphingolipid hydrolysis in lysosomes. We confirmed that cellular saposin B binds CoQ10 in human sperm and the hepatoma cell line HepG2 by using saposin B monoclonal antibody. The molar ratios of CoQ10 to saposin B were estimated to be 0.22 in urine, 0.003 in HepG2, and 0.12 in sperm. We then confirmed that aqueous saposin B extracts CoQ10 from hexane to form a saposin B-CoQ10 complex. Lipid binding affinity to saposin B decreased in the following order: CoQ10>CoQ9>CoQ7>>α-tocopherol>>cholesterol (no binding). The CoQ10-binding affinity to saposin B increased with pH, with maximal binding seen at pH 7.4. On the other hand, the CoQ10-donating activity of the saposin B-CoQ10 complex to erythrocyte ghost membranes increased with decreasing pH. These results suggest that saposin B binds and transports CoQ10 in human cells.
Keywords: coenzyme Q10 binding protein, saposin B, urine, sperm, HepG2
Introduction
Coenzyme Q (CoQ) was first isolated as an essential component of the mitochondrial respiratory chain [1] and is present in all tissues [2, 3]. CoQ is thought to function as a lipid-soluble, front-line antioxidant [2–4], and thus is ubiquitously present in all biomembranes and lipoproteins. CoQ also has many homologues with isoprene chains of various lengths. For example, the isoprene chain is 10 units (CoQ10) in length in humans and guinea pigs, but is 9 units (CoQ9) in length in rats and mice.
CoQ is synthesized in the mitochondria, ER-Golgi system and peroxisomes [2, 3], and therefore, CoQ needs to be transferred intracellularly to other organelles and extracellularly to lipoproteins. Radiolabeled CoQ has been used to confirm this notion. The intravenous injection of 14C-labeled CoQ10 into guinea pigs confirmed uptake by the liver, spleen and adrenal glands, and to a lesser extent, by the kidney and heart [5]. Nakamura et al. intravenously injected 14C-labeled CoQ10 into rats and measured its content in subfractions of the rat heart [6]. At 2 h after administration, radioactivity was found mostly in the cytosolic fraction, followed by the mitochondrial, lysosomal, microsomal and nuclear fractions at 72 h. At two days after intraperitoneal injection of 3H-labeled CoQ10, uptake into various organelles and cytosol of rat liver was confirmed [7]. Oral administration of deuterium-labeled CoQ10 in humans has confirmed uptake into blood circulation [8]. Rosenfeldt et al. measured cardiac mitochondrial CoQ10 levels in patients undergoing cardiac surgery and found that levels were significantly higher in patients orally administrated CoQ10 (300 mg/day for two weeks) than in the placebo control group [9]. These results clearly demonstrate that CoQ10 is transferred intracellularly and extracellularly, resulting in its ubiquitous presence throughout the body.
As CoQ is insoluble in water, the existence of a CoQ-binding and transfer protein has been hypothesized but has yet to be isolated and characterized. We focused on human urine as it contains CoQ10 [10] and is relatively clean and accessible. As expected, CoQ10 was bound to a protein in human urine. We purified this CoQ10-bindng protein using conventional purification methods. The amino acid sequence analysis revealed the binding protein to be saposin B, which is known to be present in all cells and activates lysosomal sphingolipid hydrolase.
In this report, we demonstrate that the saposin B-CoQ10 complex is present in human cells and that saposin B extracts CoQ10 from CoQ10 hexane solution to form the saposin-CoQ10 complex. We also show the evidence that saposin B functions as a CoQ10 transfer protein.
Materials and Methods
Lipid analysis
CoQ homologues and α-tocopherol concentrations were determined using an HPLC-ECD system, as reported previously [11], with minor modification. Briefly, samples were added to a 9-fold volume of HPLC grade 2-propanol (Fisher Chemicals, Fairlawn, NJ), vigorously mixed and centrifuged. Supernatants thus obtained were injected onto the HPLC-ECD system. Mobile phase: 50 mM NaClO4 in methanol/2-propanol (7/3, v/v); flow rate: 1.0 ml/min; analytical column: KANTO RP-18 (L) GP, 5 µm × 150 mm × 4.6 mm (Kanto Chemical, Tokyo, Japan); post-reduction column: RC-10, 15 mm × 4 mm (IRICA, Kyoto, Japan); detector: ECD (600 mV) NANOSPACE SI-1 (Shiseido, Tokyo, Japan).
Purification of urinary CoQ10-binding protein
Urine taken from healthy volunteers was pooled in the presence of sodium azide. Urine (300 ml) was centrifuged at 1,600 × g for 10 min to remove precipitates. Supernatant was applied to a PD-10 column (Amersham Biosciences, Uppsala, Sweden) pre-conditioned with a buffer (20 mM phosphate buffer containing 50 mM NaCl and 0.01% sodium azide, pH 6.0) in order to remove salt and low molecular weight substances. Obtained protein factions were concentrated with an SC250 Express Speed VacR (Thermo Savant, Holbrook, NY) and Centriplus YM-3 membrane (Millipore, Bedford, MA). Samples thus obtained were applied to a DEAE sepharose Fast Flow column (bed volume = 1.0 ml) (Amersham Biosciences) pre-equilibrated with a start buffer (20 mM phosphate buffer containing 50 mM NaCl, pH 6.0). Proteins were eluted at a flow rate of 1.0 ml/min with an elution buffer (1.0 M NaCl in start buffer, pH 6.0) using step-wise gradient of NaCl concentration. CoQ10 concentrations in each fraction were analyzed by HPLC-ECD. CoQ10-rich fractions were concentrated and were subjected to gel filtration (Superdex 200 10/300, bed volume = 24 ml) (Amersham Biosciences). Proteins were eluted with a buffer (50 mM phosphate buffer containing 150 mM NaCl, pH 7.0) at a flow rate of 0.5 ml/min. CoQ10 levels in each fraction were analyzed and CoQ10-rich fractions were pooled. Concentrated fractions from Superdex 200 were then applied to an octyl sepharose column (bed volume = 1 ml) pre-equilibrated with a start buffer (50 mM phosphate buffer, pH 7.0). Proteins were eluted with an elution buffer (50 mM phosphate buffer containing 2% n-octyl-β-D-glucoside) at a flow rate of 1.0 ml/min. CoQ10 concentrations were analyzed and CoQ10-rich fractions were pooled.
Amino acid sequence
The purified protein was subjected to SDS-PAGE, followed by electroblotting onto a PVDF membrane (ATTO, Tokyo, Japan). The blotted membrane was stained with Coomassie brilliant blue. Subsequently, the single stained protein band was excised and analyzed using a gas-phase protein sequencer (PPSQ-21; Shimadzu, Kyoto, Japan).
SDS-PAGE, Silver staining and western blotting
Samples were separated by electrophoresis through an SDS/polyacrylamide gel (15% acrylamide). After electrophoresis, gels were stained with silver staining kits (ATTO). For Western blotting analysis, proteins were transferred to PVDF membranes. The membranes were incubated with mouse anti-saposin B IgG for 1 h at room temperature. Proteins were visualized with HRP-conjugated secondary antibodies (Bio-Rad Japan, Tokyo, Japan).
Preparation of monoclonal antibody against saposin B
Six-week-old female BALB/c mice were immunized subcutaneously with purified human saposin B (50 µg) emulsified in Freund’s complete adjuvant for 7 times. Two weeks later, the mice were injected intravenously with the same antigen (25 µg). Three days later, serum was collected, and spleen cells were fused with P3-X63-Ag8-U1 myeloma cell line. Hybridoma clones were screened by ELISA using purified saposin B. ELISA-positive hybridoma clones were used for preparation of saposin B-specific antibody-producing tumors in pristane (2,6,10,14-tetramethylpentadecane)-primed BALB/c mice. Ascites fluids containing monoclonal antibody were clarified by centrifugation at 2,000 × g for 10 min and stored at −20°C. Immunoglobulin class and subclass were typed with the MonoAB-ID kit (Zymed, San Francisco, CA).
Preparation of HepG2 cells
Cultures of HepG2 cells (Dainippon Sumitomo Pharma, Osaka, Japan) were maintained in Dulbecco’s MEM (Sigma) supplemented with 10% fetal calf serum (HyClone, Logan, UT), 100 units/ml penicillin, 100 µg/ml streptomycin at 37°C in a humidified atmosphere of 5% CO2 and 95% air. At 80–90% confluence, cells were treated with trypsin-EDTA (0.25% trypsin, 0.02% EDTA) at 37°C for 15 min. After addition of culture medium, cells were centrifuged at 1,000 × g for 3 min at 4°C. Cells were washed twice with ice-cold PBS, followed by homogenization in lysis buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.4, 0.1% nonidet P-40 (Nakarai Tesuque, Tokyo, Japan), 0.1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 µg/ml of leupeptin, pepstatin A, N-tosyl-L-phenylalanyl chloromethyl ketone, N-tosyl -L-lysyl chloromethyl ketone) and centrifugation at 10,000 × g for 3 min at 4°C. The supernatant was used for immunoprecipitation.
Preparation of human sperm
Human semen collected from healthy volunteers was centrifuged at 3,000 × g for 10 min at 4°C. Precipitated cells were then washed twice with PBS and homogenized in lysis buffer, as described above.
Immunoprecipitation
Samples placed in protein LoBind tubes (Eppendorf, Hamburg, Germany) were incubated with 5 µg of anti-saposin B IgG or mouse normal IgG for 1 h at room temperature. After incubation, 10 µl of protein G beads (Amersham Biosciences) were added to samples, followed by further incubation for 1 h. Beads were washed three times with PBS. CoQ10 was extracted from beads with 2-propanol and analyzed by HPLC-ECD. A SDS-PAGE sample buffer was mixed with the beads and then applied to a Western blotting analysis of saposin B.
Isoelectric point focusing chromatography
Isoelectric point (pI) focusing chromatography [12] was performed by using the ProteomeLab PF 2D System (Beckman Coulter). Initially the chromatofocusing column was equilibrated for 130 min with start buffer (pH 8.5) at a flow rate of 0.2 ml/min. Then, 0.16 mg of saposin B purified from human urine was injected onto the equilibrated column. After a stable baseline was established, the pH gradient was started by introducing elution buffer (pH 4.0) at a flow rate of 0.2 ml/min. Fractions were collected at 0.3 pH intervals and CoQ10 levels in each fraction were measured. Finally, the column was washed with 1.0 M NaCl. CoQ10-free saposin B and CoQ10-rebound saposin B were analyzed similarly. CoQ10-free saposin B was prepared by washing saposin B solution with 10 volume of hexane and following centrifugation at 1,600 × g for 10 min. CoQ10-rebound saposin B was prepared by mixing CoQ10-free saposin B solution with 10 volume of hexane containing 10 mM CoQ10 and following centrifugation at 1,600 × g for 10 min.
Lipid binding assay
CoQ10-free saposin B (0.5 µM) in 50 mM phosphate buffer (pH 7.4) containing 150 mM NaCl was vigorously mixed for 5 min with hexane containing 10 mM lipids such as CoQ10, CoQ9, CoQ7 and α-tocopherol, followed by centrifugation at 1,600 × g for 10 min. Hexane was removed and the aqueous layer was further centrifuged in Eppendorf-tubes at 15,000 × g for 10 min to separate contaminating hexane. Lipid concentrations in aqueous layer were analyzed by HPLC-ECD. Human serum albumin was used instead of saposin B for comparison. To investigate the pH dependence of CoQ10-binding to saposin B, 150 mM NaCl containing 50 mM acetate buffer (pH 4.5) or 50 mM phosphate buffer (pH 5.4, 6.4 or 8.0) were employed. Saposin B-free buffers were used as blank.
CoQ10 donation activity assay
The crude saposin B fraction was purified on an ion exchange column (DEAE sepharose) followed by a gel filtration column (Superdex 200 10/300). Erythrocyte ghost membranes were prepared by the method of Dodge et al. [13]. Erythrocyte ghost membranes (10 µl), saposin B-containing above buffer (1000 µl) were add to a glass vial and incubated for 60 min with gentle shaking at 37°C. The reaction mixture was transferred to Eppendorf-tubes and centrifuged at 15,000 × g for 3 min at 4°C. The obtained pellets were washed three times with 1.0 ml of buffer. CoQ10 contents in ghost membranes were analyzed by HPLC-ECD. Saposin B-free buffers were used as blank.
Statistical analyses
Data were analyzed by Student t test and values are means ± SD. * and ** indicate significant differences (p<0.05 and 0.01, respectively) between the two groups.
Results and Discussion
Purification and characterization of CoQ10-binding protein
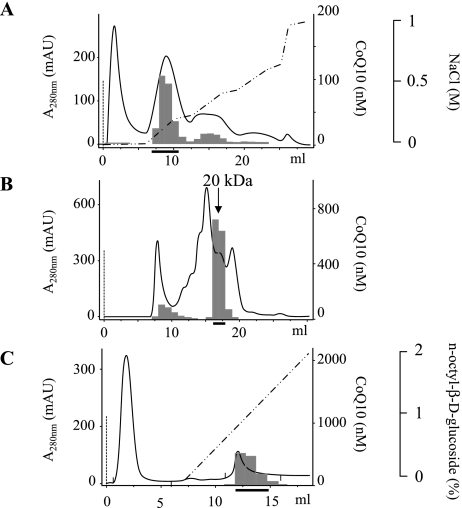
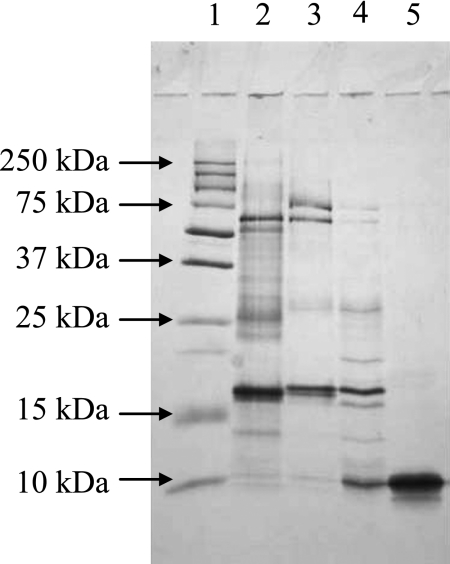
We first confirmed the presence of CoQ10 in human urine from 9 healthy donors. The concentrations ranged from 40–120 nM. Desalted human urine (300 ml) was then loaded onto a DEAE sepharose column. The protein elution profile was determined by monitoring absorbance at 280 nm, as shown in Fig. 1A. The CoQ10 content in each fraction was determined using an HPLC system with an electrochemical detector (ECD) [11] and is shown in the figure. The CoQ10-containing protein fractions indicated by the thick solid line in Fig. 1A were collected and processed by gel filtration. The majority of CoQ10 was eluted with a 20-kDa protein fraction as shown in Fig. 1B. Because the fraction contained several proteins, as determined by SDS polyacrylamide gel electrophoreses (Fig. 2), the CoQ10-binding protein was further purified using an octyl sepharose column (Fig. 1C) and the CoQ10-containing fractions indicated by a thick solid line were isolated. SDS acrylamide gel electrophoreses revealed a single protein band having a molecular weight of 10 kDa, thus suggesting that CoQ10 is bound to a protein homodimer in urine. Table 1 summarizes the purification of the CoQ10-binding protein.
Fig. 1.
Chromatographic profiles of eluates from (A) DEAE sepharose, (B) Superdex 200, and (C) octyl sepharose columns utilized in the purification of saposin B from human urine. Solid bar and dotted line indicate the CoQ10 concentration in the fraction and protein concentration estimated from absorption at 280 nm, respectively. The broken solid line indicates the concentration of NaCl (A) or n-octyl-β-D-glucoside (C) in the eluates. The thick solid line indicates the fractions collected.
Fig. 2.
SDS polyacrylamide gel electrophoresis of the proteins in each collected fraction. Proteins were stained with silver stain kits. Lane 1: Molecular weight markers; Lane 2: Human urine; Lane 3: Fractions obtained from DEAE column chromatography; Lane 4: Fractions obtained from Gel filtration; Lane 5: Fractions obtained from octyl sepharose.
Table 1.
Typical purification of saposin B from 300 ml of human urine.
| Step | Protein (mg) | CoQ10 (nmol) | CoQ10/protein (nmol/mg) | CoQ10 recovery (%) | Purification factor |
|---|---|---|---|---|---|
| Starting material | 17.4 | 7.43 | 0.43 | 100 | 1.0 |
| DEAE Separose | 2.56 | 3.53 | 1.38 | 47 | 3.2 |
| Superdex 200 | 0.51 | 1.35 | 2.65 | 18 | 6.2 |
| Octyl sepharose | 0.084 | 1.00 | 11.9 | 13 | 28 |
We analyzed the N-terminal 20 amino acids of the purified CoQ10-binding protein by Edman sequencing. The amino acid sequence was GDV -QD YIQ MVT DIQ TAV RT, which is identical to human saposin B (GenBank accession number 1N69_A), according to the DNA Data Bank of Japan. Saposin B is a heat-stable glycoprotein comprising 80 amino acids and a carbohydrate component (molecular weights, 9100 and 3000, respectively), having a total molecular weight of 12.1 kDa [14]. As 84 µg of the purified saposin B contained 1.0 nmol CoQ10, the molar ratio of CoQ10 to purified saposin B was calculated: 1.0 nmol/84 µg/12,100 g/mol = 0.14 (Table 1).
Saposin B is known to bind sphingolipids including ganglioside and ceramide [14], as well as glycerophospholipids such as phosphatidylcholine and phosphatidylinositol [15]. This occurs because saposin B is present as a shell-like dimer [16] possessing a hydrophobic cavity for lipid binding. Therefore, it is not surprising that saposin B binds CoQ10. However, CoQ10 binding to saposin B has never been reported.
Saposin B and monoclonal antibody
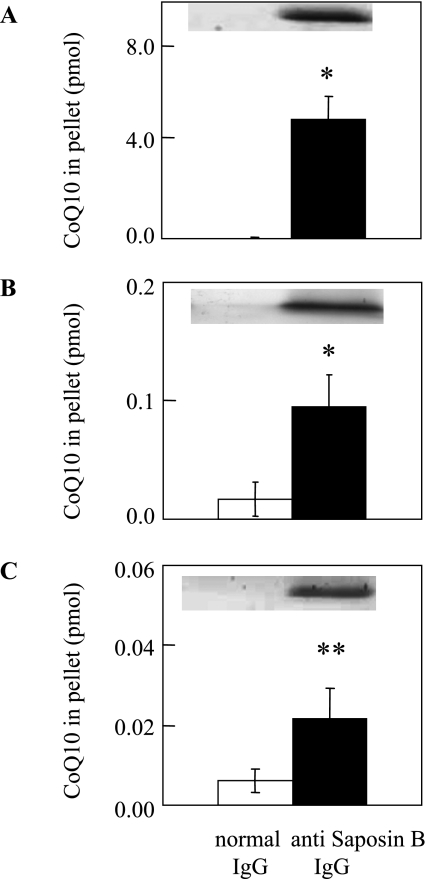
Saposin B is a housekeeping protein necessary for lysosomal sphingolipid hydrolysis [14]. Indeed, it is present in almost every tissue examined [14] as well as blood plasma [17]. CoQ10 is also present in these intra- and extracellular spaces. To confirm the presence of the saposin B-CoQ10 complex in human tissues, we attempted to immunoprecipitate saposin B using a monoclonal antibody and to measure CoQ10 content in the precipitate. For this purpose, we further purified saposin B from human urine and prepared a monoclonal antibody. First, we applied this antibody to desalted human urine passed through a PD-10 column, and found that the precipitate contains both saposin B and CoQ10, as shown in Fig. 3A. On the other hand, urine precipitate using normal IgG contained little saposin B and CoQ10 (Fig. 3A). We also prepared a polyclonal antibody against human saposin B, and immunoprecipitation of human urine with this antibody gave identical results.
Fig. 3.
Presence of saposin B-CoQ10 complex in (A) human urine, (B) HepG2 cells and (C) human sperm. Solid and open bars show the CoQ10 contents in immunoprecipitates obtained with monoclonal anti-saposin B IgG and normal IgG, respectively. Values are means ± SD (n = 3). * and ** indicate significant differences (p<0.05 and 0.01, respectively) between the two groups, as analyzed by the Student t test. Upper panel shows the results of Western blotting with monoclonal anti-saposin B antibody.
Presence of saposin B-CoQ10 complex in human cells
Next, we immunoprecipitated cell lysates obtained from a human hepatoma cell line (HepG2) using a monoclonal antibody against human saposin B in order to confirm the cellular presence of saposin B-CoQ10 complex. As shown in Fig. 3B, anti-saposin B IgG, but not normal IgG, precipitated both saposin B and CoQ10. The saposin B-CoQ10 complex was found in the cytosol of HepG2 cells (data not shown), thus suggesting that saposin B is a CoQ10-transfer protein. It is noteworthy that the majority of exogenous radiolabeled CoQ10 is recovered from the cytosol [6]. Additionally, we detected the saposin B-CoQ10 complex in fresh human sperm (Fig. 3C). Molar ratios of saposin B to CoQ10 were calculated as 0.22 in urine, 0.003 in HepG2 cells, and 0.12 in sperm, thus confirming that considerable amounts of CoQ10 are bound to saposin B in vivo. We also confirmed the presence of the CoQ10-saposin B complex in HepG2 and sperm using a polyclonal antibody against human saposin B.
Saposin B may not be the sole CoQ10 binding protein. In fact, several other proteins are associated with the bindings of CoQ10 (Fig. 1). Saposin B, together with saposins A, C and D, are the proteolysis products of a common precursor protein, prosaposin [14]. All four saposins contain about 80 amino acids and are structurally similar. They have six cysteines and conserved prolines in identical positions. Characterizing the associations between CoQ10 and saposins A, C and D, as well as prosaposin, will require further investigation.
Dissociation and recombination of CoQ10 and saposin B
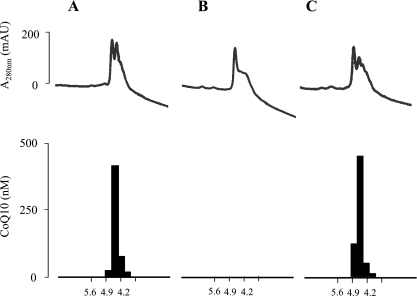
In order to have biological significance, the saposin B-CoQ10 complex must donate CoQ10 to areas of low CoQ10 concentrations and extract CoQ10 from areas of high CoQ10 concentration. We used hexane as a model lipid-carrying space. When purified saposin B from human urine was applied to a chromatofocusing column, saposin B and CoQ10 were coeluted, as shown in Fig. 4A, indicating that saposin B and CoQ10 are present as a complex. The isoelectric point (pI) of the complex is 4.5, which is identical to the reported pI value of saposin B [14]. We then washed an aqueous solution (pH 7.0) of saposin B-CoQ10 complex with 10 volumes of hexane, and injected the aqueous protein phase onto the same column. Figure 4B shows that saposin B was eluted, but CoQ10 had been removed. CoQ10 was detected in the hexane phase, indicating that the saposin B-CoQ10 complex donates CoQ10 to areas of low CoQ10 concentration.
Fig. 4.
Dissociation and rebinding of CoQ10 and saposin B, as analyzed by isoelectric chromatofocusing. The line and bar indicate protein contents estimated from UV absorbance at 280 nm and CoQ concentration in the fractions, respectively. (A) Purified saposin B (0.16 mg) containing CoQ10. (B) Protein solution washed three times with 10 volumes of hexane. (C) Aqueous phase from washed protein solution mixed with 10 volumes of hexane containing 10 mM CoQ10.
We then incubated an aqueous solution (pH 7.0) of CoQ10-free saposin B with 10 volumes of hexane containing 10 mM CoQ10. After vigorous mixing for 10 h, followed by centrifugation, the protein solution was again loaded onto a chromatofocusing column. Fig. 4C shows that saposin B and CoQ10 were coeluted similarly to the original saposin B-CoQ10 complex. The amounts of CoQ10 extracted by saposin B for 5 min increased with CoQ10 concentration in hexane (Fig. 5A). On the other hand, human serum albumin was not able to extract CoQ10 from the hexane solution (Fig. 5A). These data clearly indicate that saposin B extracts CoQ10 from areas of high CoQ10 concentration, while albumin does not.
Fig. 5.
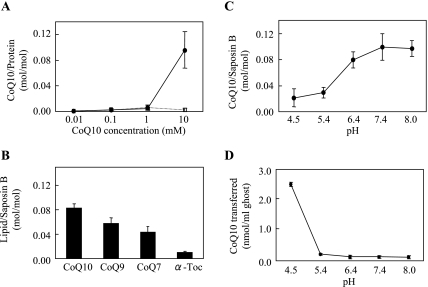
(A) Effects of CoQ10 concentration in hexane on binding to aqueous saposin B. Various concentrations of CoQ10 in hexane were vigorously mixed with 2 ml of 0.5 µM saposin B (solid line) or human serum albumin (dotted line) in 50 mM phosphate buffer containing 150 mM NaCl (pH 7.4) for 5 min. The aqueous phase was separated from hexane by centrifugation. CoQ10 contents in the aqueous layer were plotted. (B) Binding affinity of 10 mM CoQ homologues or α-tocopherol in hexane to aqueous 0.5 µM saposin B (pH = 7.4) for 5 min. (C) Effects of buffer pH on CoQ10-binding to saposin B. (D) Donation of CoQ10 to human erythrocyte ghosts from aqueous saposin B containing 30 pmol CoQ10. All experiments were repeated 3–6 times and mean ± SD are shown.
Binding specificity and pH dependence
The above aqueous CoQ10-free saposin B solution and hexane partition system was useful to examine the substrate specificity and pH dependence of the binding. We selected α-tocopherol and CoQ homologues, such as CoQ9 and CoQ7, as binding substrates and the respective 10 mM hexane solutions were incubated with aqueous CoQ10-free saposin B solution (pH 7.4) for 5 min. The binding affinity was found to decrease in the order of CoQ10>CoQ9>CoQ7>>α-tocopherol (Fig. 5B) while no binding was observed with cholesterol (data not shown). Figure 5C shows that the binding affinity of CoQ10 to saposin B increased with pH, before leveling off at neutral and alkaline pH levels. Peak binding was observed at pH 7.4.
Transfer of CoQ10 from saposin B to erythrocyte ghost membranes
When erythrocyte ghost membranes were incubated with the saposin B-CoQ10 complex, CoQ10 was transferred to the ghost membrane in a time-dependent manner before equilibration at 60 min (data not shown). The amount of CoQ10 transferred in 60 min increased with decreasing pH (Fig. 5D), which was the opposite trend as seen with CoQ10-binding affinity. Under our conditions, about 84% of CoQ10 in saposin B was transferred to the ghost membranes at pH 4.5. The intracellular pH is 8.0 in the mitochondrial matrix and 7.0 in cytosol, while lysosomes are acidic. Therefore, the transfer of CoQ10 from saposin B would favor lysosomes, although even at neutral pH, small but significant amounts of CoQ10 were transferred.
Saposin B and CoQ10 deficiency
The above data suggest that saposin B is a human CoQ10 binding/transfer protein. If this function of saposin B is of physiological importance, saposin B knockout mouse should show symptoms related to CoQ deficiency. Unfortunately, such mice have not yet been established. However, it is also possible to compare reports of human saposin B and CoQ10 deficiencies.
Human saposin B deficiency is a rare disease [18–21]. The onset of the disease was between 6 months and 6 years after birth. Typical symptoms are ataxia and motor deterioration followed by intellectual dysfunction, which resembles metachromatic leukodystrophy. Patients with CoQ10 deficiency are also rare, but show muscle mitochondrial myopathy and ataxia [22–24]. Thus, common features of saposin B and CoQ10 deficiencies are ataxia and motor deterioration. Future analysis of the cellular distribution of CoQ10 in tissues from patients with saposin B deficiency may thus confirm the physiological function of saposin B for CoQ10 transport.
Acknowledgments
We thank Drs. J. Matsuda and K. Suzuki (Tokai University) and Dr. W. Dunlap (Australian Institute of Marine Science) for discussions and comments. This work was supported by the High-Tech Research Center Project for Private Universities from the Ministry of Education, Culture, Sports, Science and Technology (to Y.Y.) and by a grant from Japanese Coenzyme Q Association (to S.Y. and Y.Y.).
References
- 1.Crane F.L., Hatefi Y., Lester R.L., Widmer C. Isolation of a quinone from beef heart mitochondria. Biochim. Biophys. Acta. 1957;25:220–221. doi: 10.1016/0006-3002(57)90457-2. [DOI] [PubMed] [Google Scholar]
- 2.Turunen M., Olsson J., Dallner G. Metabolism and function of coenzyme Q. Biochim. Biophys. Acta. 2004;1660:171–199. doi: 10.1016/j.bbamem.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 3.Dallner G., Stocker R. Coenzyme Q10: Encyclopedia of Dietary Supplements. Marcel Dekker; New York: 2005. pp. 121–131. [Google Scholar]
- 4.Yamamoto Y. Coenzyme Q10 as a front-line antioxidant against oxidative stress. J. Clin. Biochem. Nutr. 2005;36:29–35. [Google Scholar]
- 5.Yuzuriha T., Takada M., Katayama K. Transport of [14C] coenzyme Q10 from the liver to other tissues after intravenous administration to guinea pigs. Biochim. Biophys. Acta. 1983;759:286–291. doi: 10.1016/0304-4165(83)90325-2. [DOI] [PubMed] [Google Scholar]
- 6.Nakamura T., Sanma H., Himeno M., Kato K. In: Transfer of exogenous coenzyme Q10 to the inner membrane of heart mitochondria, in Biomedical and Clinical Aspects of Coenzyme Q, Vol. 2. Yamamura Y., Folkers K., Ito Y., editors. Elsevier; Amsterdam: 1980. pp. 53–64. [Google Scholar]
- 7.Bentinger M., Dallner G., Chojnacki T., Swiezewska E. Distribution and breakdown of labeled coenzyme Q10 in rat. Free Radic. Biol. Med. 2003;34:563–575. doi: 10.1016/s0891-5849(02)01357-6. [DOI] [PubMed] [Google Scholar]
- 8.Tomono Y., Hasegawa J., Seki T., Motegi K., Morishita N. Pharmacokinetic study of deuterium-labeled coenzyme Q10 in man. Int. J. Clin. Pharmacol. Ther. Toxicol. 1986;24:536–541. [PubMed] [Google Scholar]
- 9.Rosenfeldt F., Marasco S., Lyon W., Wowk M., Sheeran F., Bailey M., Esmore D., Pick A., Rabinov M., Smith J., Magley P., Pepe S. Coenzyme Q10 therapy before cardiac surgery improves mitochondrial function and in vitro contractility of myocardial tissue. J. Thorac. Cardiovasc. Surg. 2005;129:25–32. doi: 10.1016/j.jtcvs.2004.03.034. [DOI] [PubMed] [Google Scholar]
- 10.Okamoto T., Fukunaga Y., Ida Y., Kishi T. Determination of reduced and total ubiquinones in biological materials by liquid chromatography with electrochemical detection. J. Chromatogr. 1988;430:11–19. doi: 10.1016/s0378-4347(00)83129-1. [DOI] [PubMed] [Google Scholar]
- 11.Yamashita S., Yamamoto Y. Simultaneous detection of ubiquinol and ubiquinone in human plasma as a marker of oxidative stress. Anal. Biochem. 1997;250:66–73. doi: 10.1006/abio.1997.2187. [DOI] [PubMed] [Google Scholar]
- 12.Soldi M., Sarto C., Valsecchi C., Magni F., Proserpio V., Ticozzi D., Mocarelli P. Proteome profile of human urine with two-dimensional liquid phase fractionation. Proteomics. 2005;5:2641–2647. doi: 10.1002/pmic.200401269. [DOI] [PubMed] [Google Scholar]
- 13.Dodge J.T., Mitchell C., Hanahan D. The preparation and chemical characteristics of hemoglobin-free ghosts of human erythrocytes. Arch. Biochem. Biophys. 1963;100:119–130. doi: 10.1016/0003-9861(63)90042-0. [DOI] [PubMed] [Google Scholar]
- 14.Kishimoto Y., Hiraiwa M., O’Brien J.S. Saposins: structure, function, distribution, and molecular genetics. J. Lipid Res. 1992;33:1255–1267. [PubMed] [Google Scholar]
- 15.Ciaffoni F., Tatti M., Boe A., Salviol i R., Fluharty A., Sonnino S., Vaccaro A.M. Saposin B binds and transfers phospholipids. J. Lipid Res. 2006;47:1045–1053. doi: 10.1194/jlr.M500547-JLR200. [DOI] [PubMed] [Google Scholar]
- 16.Ahn V.E., Faull K.F., Whitelegge J.P., Fluharty A.L., Prive G.G. Crystal structure of saposin B reveals a dimeric shell for lipid binding. Proc. Natl. Acad. Sci. U.S.A. 2003;100:38–43. doi: 10.1073/pnas.0136947100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang M.H., Bindloss C.A., Grabowski G.A., Qi X., Winchester B., Hopwood J.J., Meikle P.J. Saposins A, B, C, and D in plasma of patients with lysosomal storage disorders. Clin. Chem. 2000;46:167–174. [PubMed] [Google Scholar]
- 18.Shapiro L.J., Aleck K.A., Kaback M.M., Itabashi H., Desnick R.J., Brand N., Stephens R.L., Fulhartly A.L., Kihara H. Metachromatic leukodystrophy without arylsulfatase A deficiency. Pediatr. Res. 1979;13:1179–1181. doi: 10.1203/00006450-197910000-00021. [DOI] [PubMed] [Google Scholar]
- 19.Hahn A.F., Gordon B.A., Gilbert J.J., Hinton G.G. The AB-variant of metachromatic leukodystrophy (postulated activator protein deficiency). Light and electron microscopic findings in a sural nerve biopsy. Acta Neuropathol. 1981;55:281–287. doi: 10.1007/BF00690991. [DOI] [PubMed] [Google Scholar]
- 20.Wrobe D., Henseler M., Huettler S., Pascual SIIP., Sandhoff K. A non-glycosylated and functionally deficient mutant (N215H) of the sphingolipid activator protein B (SAP-B) in a novel case of metachromatic leukodystrophy (MLD) J. Inherit. Metab. Dis. 2000;23:63–76. doi: 10.1023/a:1005603014401. [DOI] [PubMed] [Google Scholar]
- 21.Holtschmidt H., Sandholl K., Kwon H.Y., Harzer K., Nakano T., Suzuki K. Sulfatide activator protein. Alternative splicing that generates three mRNAs and a newly found mutation responsible for a clinical disease. J. Biol. Chem. 1991;266:7556–7660. [PubMed] [Google Scholar]
- 22.Ogasahara S., Engel A.G., Frens D., Mack D. Muscle coenzyme Q deficiency in familial mitochondrial encephalomyopathy. Proc. Natl. Acad. Sci. U.S.A. 1989;86:2379–2382. doi: 10.1073/pnas.86.7.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rotig A., Appelkvist E., Geromel V., Chretien D., Kadhom N., Edery P., Lebideau M., Dallner G., Munnich A., Ernster L., Rustin P. Quinone-responsive multiple respiratory-chain dysfunction due to widespread coenzyme Q10 deficiency. Lancet. 2000;356:391–395. doi: 10.1016/S0140-6736(00)02531-9. [DOI] [PubMed] [Google Scholar]
- 24.Musumeci O., Naini A., Slonim A.E., Skavin N., Hadjigeorgiou G.L., Krawiecki N., Weissman B.M., Tsao C.Y., Mendell J.R., Shanske S., De Vivo. D.C., Hirano M., DiMauro S. Familial cerebellar ataxia with muscle coenzyme Q10 deficiency. Neurology. 2001;56:849–855. doi: 10.1212/wnl.56.7.849. [DOI] [PubMed] [Google Scholar]