Abstract
Study Objectives:
Previous genetic investigations of sleep disturbance have shown various measures of sleep quality and sleep pattern to be heritable. But none of these studies have investigated the genetic predisposition to sleep disturbance attributed to caffeine. In this study, the heritability of coffee-attributed sleep disturbance and its relationship with other sleep measures were estimated, and chromosomal regions influencing this trait were identified.
Design:
A classical twin design was used to estimate the heritability of coffee-attributed sleep disturbance and its genetic covariance with other measures of sleep disturbance (e.g., due to anxiety, depression) and sleep quality (e.g., variability in sleep quality). To locate quantitative trait loci influencing coffee-attributed sleep disturbance, a genome-wide linkage screen of 1395 microsatellite markers was performed.
Participants:
The study included 3808 Australian adult twin pairs (n = 1799 monozygous pairs; n = 2009 dizygous pairs). A subsample of 1989 individuals from 1175 families was used for the linkage analysis.
Measurements and Results:
The heritability of coffee-attributed sleep disturbance (measured by self report) was approximately 0.40, with three fourths of this genetic variance explained by genes unrelated to the general sleep disturbance factor. One region of significant linkage to coffee-attributed sleep disturbance was identified on chromosome 2q (LOD score of 2.9).
Conclusions:
Although no candidate genes known to be related to caffeine metabolism or sleep disorder were identified in the significant linkage region, 2 candidates were found under a smaller peak on chromosome 17q.
Citation:
Luciano M; Zhu G; Kirk KM; Gordon SD; Heath AC; Montgomery GW; Martin NG. “No thanks, it keeps me awake”: the genetics of coffee-attributed sleep disturbane. SLEEP 2007;30(10):1378-1386.
Keywords: Coffee-attributed insomnia, sleep disturbance, genome-wide linkage, genes, multivariate analysis, twin study
CAFFEINE IS A WIDELY USED STIMULANT WITH MARKED VARIATIONS IN BEHAVIORAL RESPONSE BETWEEN INDIVIDUALS. POSITIVE BEHAVIORAL responses may include increased alertness and physical activity, although these may represent a withdrawal reversal mechanism operating in habitual caffeine users,1–3 whereas responses such as heightened nervousness and sleep disturbance would be considered negative.4–6 Some people experience no effects whatsoever of caffeine ingestion. Significant individual differences in the extent of wakefulness (i.e., delay of sleep onset) and soundness of sleep due to caffeine have been observed in subjects who were blind to placebo or caffeine administration an hour prior to bedtime.4 In the present study, we investigate the genetic and environmental sources of these individual differences and further analyze the genetic and environmental relationship of caffeine-related sleep disturbance with other types of sleep disturbance and measures of sleep quality.
A number of twin studies have reported substantial heritability (i.e., proportion of variance in a trait due to genes) for amount of coffee and tea consumed.7–9 Kendler et al9 studied female twin pairs and found that twin resemblance for coffee consumption, heavy caffeine use, caffeine intoxication, withdrawal and tolerance could be ascribed solely to genetic factors, with broad heritabilities ranging 0.35 to 0.77. More recently, in our study comprising men and women, overall tea and coffee consumption showed a heritability (h2) of 0.48, although the genetic relationship between tea and coffee consumption differed between sexes, thus highlighting the potential for sex-specific genetic and environmental effects on caffeine related phenotypes.8
Genetic variation in sleep disturbance has also been confirmed in several twin studies.10–12 Partinen and colleagues10 reported a heritability of 0.44 for both duration of sleep and sleep quality in a sample of 2238 monozygotic and 4545 dizygotic Finnish adult twins. In the sample used in the present study, we showed that at least 33% of the variance in sleep disturbance and sleep quality was due to genes, with the remainder accounted for by unique environmental influences, that is, specific to the individual.11 However, we did not investigate the measure of caffeine-related sleep disturbance nor the relationship between the sleep disturbance and quality of sleep measures.
An early exploratory study13 of monozygotic twins showing significant within-pair concordance for insomnia after drinking coffee raises the question of whether sleep disturbance due to caffeine is inherited. We address this question in a large sample of monozygotic and dizygotic twin pairs who have been genotyped for up to 1717 microsatellite markers. First, we investigate whether the etiology of caffeine-induced sleep disturbance differs from that of other sleep disturbance, and then we use genome-wide linkage analysis to see which chromosomal regions (quantitative trait loci) are implicated in coffee-attributed sleep disturbance. We expect to find specific effects of genes on coffee-attributed insomnia, which may relate to genetic variation in caffeine metabolism or sensitivity.14 Significant areas of linkage will be scanned for candidate genes influencing adenosine receptors to which caffeine binds15 and caffeine metabolism.16
METHODS
Between the years 1980 and 1982, a Health and Lifestyle Questionnaire, containing demographic, lifestyle, health, reproductive history (women only), personality, and social attitudes information was mailed to 5867 pairs of twins over the age of 18 who were registered with the Australian Twin Registry. Responses were obtained from 3808 complete twin pairs (1233 monozygotic women, 566 monozygotic men, 746 dizygotic women, 351 dizygotic men, 912 dizygotic opposite sex). At the time of response, the maximum age in the sample was 88 years, and the mean was 34.5 (±14.2) years. Initially, zygosity of twins was mostly determined on the basis of responses to standard questions about physical similarity and the degree to which others confused them with one another. This method has been shown to give at least 95% agreement with the diagnosis based on extensive blood typing.17 Inconsistencies in the twins' responses were followed up by phone and by asking them to send in photos, reducing the misclassification rate still further. More recently, the zygosity of 20.1% of the same-sex pairs has been confirmed through whole genome scans (described below).
Measures of Sleep Disturbance
A number of sleep-disturbance items (initial insomnia, sleep quality, sleep variability) from the sleep questionnaire of Johns18 were included along with items measuring depressed insomnia, anxious insomnia, and sleeplessness from respective Delusions-Symptoms States Inventory depression, Delusions Symptoms States Inventory anxiety, and Eysenck Personality Questionnaire neuroticism scales.19,20 The final item related to sleeplessness induced by coffee consumption.
Measures of subjective sleep quality and sleep disturbance were as follows:
-
Overall quality: “How would you describe the quality of your usual sleep over the last few months?”
(1) very good (2) good (3) fair (4) poor (5) very poor
-
Variability of quality: “How much would you say the quality of your sleep varies from one night to the other?”
(1) not at all (2) slightly (3) moderately (4) very much
-
Initial insomnia: “How often does it take you much longer than usual to get off to sleep?”
(1) less than once a month (2) 1 to 4 times per month (3) more than once a week (4) most nights
-
Anxious insomnia: “Recently, worrying has kept me awake at night.”
(1) not at all (2) a little (3) a lot (4) unbearably
-
Depressed insomnia: “Recently I have been so miserable that I have had difficulty with my sleep.”
(1) not at all (2) a little (3) a lot (4) unbearably
-
Neurotic insomnia: “Do you suffer from sleeplessness?”
(1) no (2) yes
-
Coffee-attributed insomnia: “If you were to drink coffee in the evening, would it stop you from getting to sleep at night?”
(1) never (2) sometimes (3) usually (4) always
Because the data were coded as categorical, they were analyzed in terms of threshold models that assume that underlying each variable is a continuum of liability that is normally distributed in the population and in which thresholds are imposed to define the category boundaries.21
Genetic Linkage Sample
A subsample of 1989 individuals (814 sibling pairs—724 dizygotic, 90 monozygotic; 361 single cases) from 1175 families had been genotyped with 200 to 1395 microsatellite markers (mean of 690 ± 287) spanning the genome. This linkage sample was drawn from a larger Australian sample that had been genotyped in overlapping studies by 6 different facilities—Gemini Genomics, UK; Leiden University Medical Centre, The Netherlands; Mammalian Genotyping Service, Marshfield, Wisconsin, USA; Sequana Therapeutics Inc., San Francisco, CA, USA; Australian Genome Research Facility (AGRF); and The Finnish Genome Centre, University of Helsinki, Finland—as part of various other linkage studies. The scans from 5 of these facilities have been described in detail (see22, but, since then, additional genotyping for 4575 new individuals (from 1204 families) has been performed by the Mammalian Genotyping Service, and new scans are available from AGRF and the Finnish Genome Centre. 10cM genome scans were performed by AGRF and the Finnish Genome Centre using the same standard set of ABI-2 markers (Applied Biosystems, Foster City, CA, USA). The AGRF scan included 1671 individuals from 414 families who had been measured for smoking, anxiety, and alcohol-related traits.23 Those typed by the Finnish Genome Centre were part of a migraine study and comprised 734 individuals from 158 families—the process of the genome-wide screen for this sample has been previously described in detail.24 The additional subjects genotyped through Marshfield were drawn from studies of migraine, asthma, anxiety, and effects of alcohol and were genotyped for a 10cM scan using markers from the Weber screening set 16 (http://research.marshfieldclinic.org/genetics/GeneticResearch/screeningsets.asp). A summary of the genotype datasets from which the sleep sample was drawn is shown in Table 1. This table details individual, family, sibling pairs, and genotype sample sizes and marker information following removal of pedigree errors.
Table 1.
Information for Each of the Genome Scans after Pedigree Errors were Resolved but Before Genotyping Errors were Corrected (Without Marker Cut-off)
| Individual Genotyping Datasets |
|||||||
|---|---|---|---|---|---|---|---|
| Gemini | Leiden | Marshfield | Sequana | Sequana fine-mapping | AGRF | Helsinki | |
| Individuals | 1,144 | 502 | 6,452 | 558 | 2,170 | 1,671 | 734 |
| Families | 387 | 249 | 1,891 | 213 | 607 | 414 | 158 |
| All markers | 384 | 435 | 778 | 499 | 120 | 394 | 383 |
| Autosomal markers | 366 | 416 | 746 | 482 | 120 | 376 | 366 |
| Total number of genotypes | 361,123 | 158,126 | 3,103,329 | 246,966 | 230,542 | 651,433 | 260,755 |
| Genotypes per marker | 987+/−129 | 378+/−105 | 3,989+/−2191 | 511+/−48 | 1,921+/−210 | 1,653+/−45 | 681+/−41 |
| Mean +/− SD (range) | (223–1,124) | (76–493) | (1,659–6,364) | (257–556) | (36–2,115) | (1,053–1,671) | (73–699) |
| Genotypes per individual | 316+/−41 | 315+/−70 | 481+/−165 | 443+/−27 | 106+/−22 | 390+/−12 | 355+/−80 |
| Mean +/− SD (range) | (25–362) | (4–405) | (1–777) | (240–476) | (2–119) | (93–394) | (1–382) |
| Marker heterozygosity % | 78+/−7 | 78+/−9 | 71+/−10 | 74+/−11 | 76+/−10 | 77+/−10 | 77+/−9 |
| Mean +/− SD (range) | (43–94) | (4–98) | (25–91) | (18–100) | (37–93) | (34–93) | (40–94) |
| Inter-marker distance cM | 9.6+/−4.3 | 8.5+/−4.5 | 4.7+/−2.9 | 6.9+/−6.0 | 7.1+/−9.0 | 9.4+/−3.8 | 9.7+/−4.1 |
| Mean +/− SD (range) | (1.4–35.3) | (0.01–22.1) | (0.01–15.9) | (0.001–33.7) | (0.001–82) | (1.4–29.1) | (1.4–29.1) |
| Information content | 0.49+/−0.10 | 0.33+/−0.09 | 0.57+/−0.13 | 0.40+/−0.09 | 0.72+/−0.16 | 0.70+/−0.08 | 0.63+/−0.08 |
| Mean +/− SD (range) | (0.30–0.66) | (0.18–0.48) | (0.01–0.82) | (0.21–0.57) | (0.26–0.88) | (0.34–0.89) | (0.33–0.82) |
Error checking (pedigree errors, Mendelian inconsistencies, map errors) was performed using GRR, RELPAIR, SIBPAIR, MERLIN, GENEHUNTER, and MENDEL programs—this procedure, along with a description of how genome scans were merged, is fully described in Cornes et al.22 Table 2 displays marker and sample sizes of the raw data and progressively cleaned data for the entire genotype data. The cleaned and combined genome scan data included 2161 autosomal markers (of which 763 were duplicates) for 8554 individuals.
Table 2.
Information on Combined Genome Scan Data at 3 Different Stages
| Combined Gemini, Leiden, Marsh.eld, Sequana, AGRF, Helsinki Dataset |
|||
|---|---|---|---|
| Raw | Cleaned | Cleaned + cut-off | |
| Individuals | 9,217 | 9,215 | 8,554 |
| Families | 2,516 | 2,510 | 2,355 |
| Sibpairs | 7,158 | 7,156 | 6,786 |
| All markers | 2,281 | 2,267 | 2,267 |
| Autosomal markers | 2,177 | 2,161 | 2,161 |
| Total number of genotypes | 4,813,091 | 4,760,192 | 4,698,595 |
| Genotyped individuals per marker Mean +/− SD (range) | 2,117+/−1,982 (90–6,364) | 2,100+/−1,964 (90–6,347) | 2,073+/−1,963 (90–6,333) |
| Genotyped markers per individual Mean +/− SD (range) | 522+/−255 (1–1,725) | 517+/−251 (1–1,707) | 549+/−230 (199–1,707) |
| Genotyped sibpairs per marker Mean +/− SD (range) | 1,437+/−1,463 (2–4,419) | 1,421+/−1,420 (1–4,394) | 1,409+/−1,418 (1–4,373) |
| Genotyped markers per sibpair Mean +/− SD (range) | 473+/−207 (1–1,612) | 466+/−203 (1–1,603) | 484+/−190 (1–1,603) |
| Marker heterozygosity % Mean +/− SD (range) | 74+/−11 (4–98) | 74+/−11 (4–98) | 74+/−11 (4–98) |
| Sibpair inter-marker distance cM Mean +/− SD (range) | 8.5+/−3.5 (0.2–122) | 8.5+/−3.5 (0.2–122) | 8.4+/−2.2 (2.3–16) |
| Information content Mean +/− SD (range) | 0.63+/−0.07 (0.27–0.82) | 0.63+/−0.07 (0.27–0.82) | 0.65+/−0.07 (0.27–0.83) |
Note: Raw: data merged but no genotyping errors corrected; Cleaned: genotyping and duplication marker problems resolved; Cleaned and cut off: with individuals genotyped for less than 198 markers omitted.
Statistical Analysis
Multivariate Decomposition of Genetic and Environmental Variance
To estimate the proportions of additive genetic, common environmental, and unique environmental covariance between the measures of sleep disturbance, the classical twin design was used. This design is based on the comparison of monozygotic twin pairs who share 100% of their genes with dizygotic twins who share, on average, 50% of their segregating genes.25 Additive genetic effects (transmissible from parent to child) are considered important if the correlation of monozygotic co-twins on the trait is at least twice that of dizygotic co-twins. Simultaneous equations, established by the known relationship among monozygotic and dizygotic co-twins, were therefore applied to the data [rMZ = A + C; rDZ = ½A + C]. Unique (or nonshared) environmental effects—that include measurement error—are not shared by co-twins and are hence absent from the covariance equations. By using the same logic, the genetic and environmental covariation between traits is based on the comparison of monozygotic and dizygotic cross-trait co-twin correlations. Covariance estimates were derived using a full information maximum likelihood procedure based on analysis of raw categorical data in the statistical package Mx 26, which takes full account of missing data. The fixed effects of age and sex were parameterized in terms of an age regression coefficient and a sex deviation, which were set equal across the 5 zygosity groups (monozygotic women, monozygotic men, dizygotic women, dizygotic men, dizygotic opposite sex).
As the main aim of the multivariate analysis was to establish whether the genetic effects on coffee-attributed insomnia totally overlapped with those influencing other measures of sleep disturbance, an atheoretical model, known as a Cholesky decomposition, was applied to the data. In this decomposition of genetic and environmental covariance, as many factors as there are variables for each source of variance are modeled. The first factor in the model loads on all variables, with each successive factor loading on all variables but the previous one or ones.21 By specifying coffee-attributed insomnia as the last variable in the decomposition, we were able to test whether specific genes (unrelated to the other measures) influence this trait. Reduced models (i.e., with fewer parameters) are favored if the likelihood ratio χ2 comparing the models is less than the critical value (α = 0.05) of the χ2 distribution for the degrees of freedom difference between models. This indicates that there is no significant difference between the saturated model and the reduced model. Initially, a Cholesky decomposition that estimated separate male and female components of variance (additive genetic, common environmental, and unique environmental) was specified, and the equality of these paths was then tested by constraining them equal and evaluating the goodness of model fit.
Genetic Linkage Analysis
Nonparametric linkage analysis of coffee-attributed insomnia was performed in MERLIN27 using a 5cM grid for spacing of markers and including sex and age as covariates. Linkage on the X chromosome was similarly analyzed but using the companion program to MERLIN, MINX.28 Monozygotic twins and single cases, while contributing to trait variance, did not contribute to linkage in the analyses. Because sibling pairs share none, 1, or both their genes at any locus across the genome, these analyses rely on the estimation of probabilities of gene sharing (known as identity by descent [IBD] sharing) between sibling pairs and then the testing of excess IBD sharing among individuals in the same tail of the trait distribution (i.e., affected group). Because there was low endorsement of categories 1 and 2 for this measure (coffee “always” and “usually” keeps me awake), the measure was transformed into a dichotomous scale: categories 2, 3, and 4 versus category 1). Two analyses were performed: one in which the affected group represented those who were kept awake by coffee (always/usually/sometimes), and the other in which those never affected by coffee (i.e., coffee resistant) were treated as the affected group. The significance of the Quantitative Trait Locus (QTL) effect is evaluated by the resulting logarithm-of-odds (LOD) score (based on the comparison of affected pairs' IBD sharing with the null hypothesis of simple Mendelian segregation). Significance levels were derived empirically from 1000 gene-dropping simulations using MERLIN.27 Empirical LOD scores were calculated for suggestive linkage (corresponding to 1 expected false positive per genome scan) and significant linkage (1 expected false positive per 20 genome scans).29,30
Linkage results from the affected sibling-pairs analysis were compared with those from an analysis in Mx that used all data, assuming a threshold model. The unobserved liability of the trait is modeled, and evidence for linkage is obtained if there is a correlation between IBD status and similarity of liability. In this model, information from concordant “coffee-resistant” pairs, concordant “coffee-affected” pairs, and discordant pairs are used. The variance components model in this analysis included additive genetic effects, unique environmental effects, and QTL (Q) effects. The sib-pair covariance included additive genetic effects and Q effects, where Q was conditioned by the estimated proportion of alleles shared (IBD see31,32). This model was fitted to raw ordinal data. Due to the lengthy computational time of this analysis, it was impractical to estimate empirical P values for these results.
RESULTS
Descriptive
The response rates for coffee-attributed sleep disturbance are shown separately for men and women in Table 3, with women reporting higher coffee-attributed insomnia than men. Test-retest data for 87 individuals measured twice over a 3-month interval indicated that the coffee-attributed sleep disturbance item was reliable, demonstrating a polychoric test-retest correlation of 0.80. The frequency distribution of responses for the other sleep-disturbance measures (except neurotic insomnia) and their test-retest reliability have previously been reported by Heath et al.6 For these measures, women reported greater sleep disturbance than men, with the exception of quality of sleep variability. For neurotic insomnia, 21.7% of women reported suffering from sleeplessness, whereas 15.5% of men responded yes to this item. Unlike in the analyses of Heath et al,6 we treated age as a continuous rather than categorical variable and modelled a linear association. We found that older people reported higher rates of neurotic insomnia, variability in sleep quality, initial insomnia, and coffee-attributed insomnia, whereas younger people tended to report more anxious insomnia and depressed insomnia.
Table 3.
Response Rates for Women and Men on the Measure of Coffee-Attributed Sleep Disturbancea
| Always | Usually | Sometimes | Never | No answer | |
|---|---|---|---|---|---|
| Females | |||||
| (N=4870) | 3.7% | 7.0% | 21.4% | 58.7% | 9.2% |
| Males | |||||
| (N=2746) | 2.4% | 5.5% | 18.8% | 64.4% | 8.9% |
Note: Of the 689 individuals who gave no answer, 88% reported that they drank no coffee.
Multivariate Decomposition of Genetic and Environmental Variance
The polychoric co-twin correlations for coffee-attributed insomnia were as follows: 0.39 (monozygotic women; n = 1063 pairs), 0.46 (monozygotic men; n = 500), 0.12 (dizygotic women; n = 619), 0.23 (dizygotic men; n = 292), and 0.22 (dizygotic opposite sex; n = 744). The higher monozygotic than dizygotic correlations indicated that genes influence this trait (h2 approximately 40%), and the observation that the dizygotic opposite-sex correlation was not less than the dizygotic same-sex correlations suggested the absence of sex-limited gene effects. Similarly, for neurotic insomnia, monozygotic correlations were higher than dizygotic correlations, with genetic effects lower for women (h2 = 0.26) than men (h2 = 0.40). Heritabilities of the other traits have previously been shown to range from 0.20 for sleep variability to 0.36 for anxious insomnia.11
Phenotypic correlations between coffee-attributed insomnia and the other sleep-disturbance measures were estimated separately for women and men (see Table 4). These correlations were not as strong as the intercorrelations among the non-coffee sleep-disturbance measures, which ranged between 0.40 (sleep quality - depressed insomnia) and 0.79 (depressed - anxious insomnia) for women and between 0.44 (sleep quality - anxious insomnia) and 0.76 (depressed - anxious insomnia) for men.
Table 4.
Maximum Likelihood Polychoric Phenotypic Correlations of Coffee-Attributed Sleep Disturbance with other Sleep Disturbance Variables, Separately for 4425 Women and 2501 Men
| Neurotic insomnia | Anxious insomnia | Depressed insomnia | Variability of sleep quality | Quality of sleep | Initial insomnia | |
|---|---|---|---|---|---|---|
| Females | 0.31 | 0.23 | 0.26 | 0.24 | 0.24 | 0.37 |
| Males | 0.39 | 0.31 | 0.29 | 0.25 | 0.26 | 0.36 |
All 7 variables were analyzed in a Cholesky decomposition, with separate genetic and environmental parameters estimated for women and men due to their inequality between sexes (Δχ842 = 115.2, P = 0.01). To overcome minimization problems, thresholds for each of the variables were fixed into the model based on their values estimated from the univariate genetic models. Additive genetic and common environmental parameters were tested for significance by first dropping entire factors from the saturated model, and, if these were significant, path coefficients with small values were individually tested for significance. All nonsignificant paths were removed from the model.
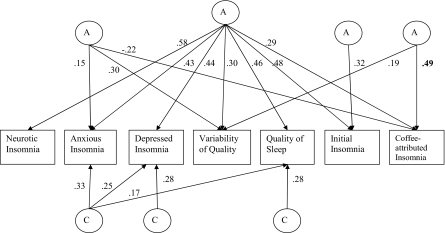
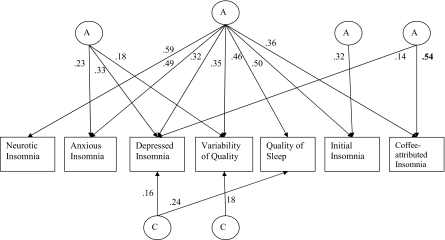
The most parsimonious model showed a nonsignificant χ2 change of 37.97 (for 78 degrees of freedom) from the saturated model. Results of the additive genetic and common environmental factor structure of this model for women and men are presented in Figures 1 and 2, respectively. In these path diagrams, circles represent latent factors (A: additive genes; C: common environment), whereas rectangles represent the measured trait. The arrowed paths leading to the measured traits represent the influence of the latent factor on the trait, with the squared path coefficient an estimate of the percentage of variance explained by the latent factor on the trait. Due to the potential for correlated measurement error between variables, the unique environmental pathways were not reduced and are shown in full in Table 5. The main difference between sexes in the genetic factor structure was that coffee-attributed insomnia was influenced by 3 factors in women and 2 factors in men. The 3 genetic factors in women included a general sleep disturbance factor loading on all variables; a second loading on anxious insomnia, variability of sleep quality, and coffee-attributed insomnia; and a third factor influencing variability of sleep quality and coffee-attributed insomnia. In men, coffee-attributed insomnia was influenced by a general sleep disturbance factor and a second factor, which also influenced depressed insomnia but to a far lesser extent.
Figure 1.
Path diagram representing the additive genetic (A) and common environmental (C) factor structure of the covariance between sleep disturbance measures in women.
Figure 2.
Path diagram representing the additive genetic (A) and common environmental (C) factor structure of the covariance between sleep disturbance measures in men
Table 5.
Parameter Estimates of the Cholesky Decomposition of Unique Environmental Covariance Among Sleep Disturbance Measures in Women and Men (Men in Bold Type)
| Factor 1 |
Factor 2 |
Factor 3 |
Factor 4 |
Factor 5 |
Factor 6 |
Factor 7 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Neurotic insomnia | .81 | .81 | ||||||||||||
| Anxious insomnia | .48 | .35 | .67 | .77 | ||||||||||
| Depressed insomnia | .45 | .46 | .45 | .47 | .51 | .55 | ||||||||
| Variability of sleep quality | .38 | .40 | .10 | .11 | .09 | .07 | .79 | .80 | ||||||
| Quality of sleep | .56 | .49 | .09 | .19 | .07 | .12 | .15 | .18 | .57 | .64 | ||||
| Initial insomnia | .51 | .39 | .12 | .18 | .00 | .13 | .11 | .12 | .09 | .12 | .61 | .65 | ||
| Coffee-attributed insomnia | .25 | −.19 | .05 | .02 | −.01 | −.07 | .03 | .13 | −.04 | .02 | .06 | .18 | .74 | .70 |
Linkage
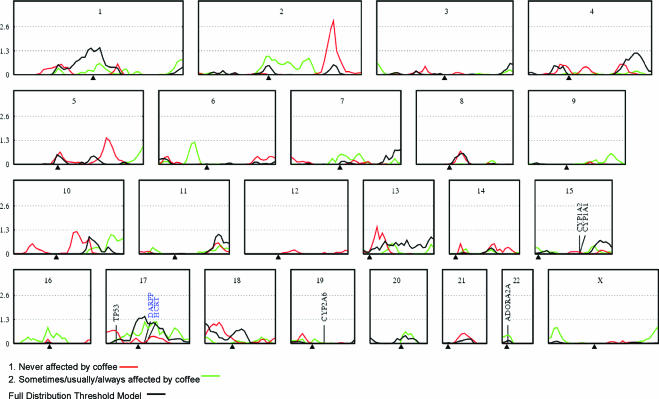
The distribution of coffee-attributed sleep disturbance for this subsample was similar to the full sample, with respective frequencies of 3.6%, 6.6%, 23.5%, and 66.3% for those whose sleep was always, usually, sometimes, and never affected by coffee. Also, the sibling correlation for this trait (r = 0.13) did not differ from the dizygotic twin correlations found in the full sample, further suggesting that there was no sampling bias in this selected genotyping sample. Genome-wide linkage results of the affected-pairs analyses (i.e., where the affected group was treated as either (1) coffee resistant or (2) coffee-affected) and of the full distribution threshold model are shown in Figure 3. These plots are defined by the LOD score on the y-axis and the position of each of the markers along the x-axis. Known human genes involved in caffeine metabolism or central nervous system functioning are indicated on the graph, along with 2 additional candidate genes found under the 17q peak. For the “coffee-resistant” analysis, empirical values were suggestive at a LOD score of 1.3 and significant at a LOD score of 2.6, and, for analysis 2, empirical LOD scores were 1.4 for suggestive and 2.8 for significant linkage. The only significant linkage was observed in the “coffee-resistant” analysis to the 2q region at 220cM (near D2S434; LOD score of 2.92). Suggestive linkage in this analysis was also found on the long arm of chromosome 5, where a LOD score of 1.43 was observed at 150cM (D5S1480), and on 13q, with a LOD of 1.48 at 25cM (D13S1246). Other regions of note in this analysis were 10q22.3, demonstrating a LOD score of 1.21 at 100cM (D10S1696), and 18p (LOD of 1.14 at 25cM; GATA036). In the “coffee-affected” analysis, no linkage peaks exceeded the suggestive criterion, although a peak of 1.18 was observed on the short arm of chromosome 17 at 65cM, and a LOD score of 1.13 was found on 6p at 55cM. The results of the thresholds analysis showed some consistent peaks with those of the affected-pairs analyses; the linkage peaks were lower on chromosomes 2, 10, 13, and 18 but higher on chromosome 17. Linkage peaks were most notably absent on chromosomes 5 and 6, where they had approached suggestive linkage in the respective “coffee-resistant” and “coffee-affected” analyses. Although no significant linkages were observed in the thresholds analysis, suggestive linkage was supported on 1p (LOD score of 1.47 at 145cM; D1S2726) and 17q (LOD score of 1.47 at 65cM; GGAA19G04). Other regions of interest (LOD scores > 1) were found on chromosomes 4q, 11q, and 18p.
Figure 3.
Linkage plot depicting chromosomal regions across the genome that are related to coffee-attributed insomnia. Results are shown for the affected pairs analyses (1. coffee never disturbs sleep; 2. coffee sometimes, usually or always disturbs sleep) and the threshold model analysis. Known genes related to caffeine effects in humans are shown, along with 2 candidate genes (DARPP-32, HCRT) under the 17q peak.
DISCUSSION
Individual differences in coffee-attributed sleep disturbance were shown to have an etiology comprised of additive genetic and unique environmental effects, and these were largely genetically and environmentally distinct from factors influencing general sleep quality and dimensions such as anxiety and depression. The structure of genetic and environmental effects on the covariation between sleep-disturbance measures differed slightly between women and men, but a general additive genetic factor was prominent in both sexes. Importantly, a large specific genetic factor was found for coffee-attributed sleep disturbance in both sexes. A genome-wide screen of markers linked to coffee-attributed insomnia found significant linkage to a chromosomal region on 2q, influencing resilience to caffeine's effects on sleep. Other regions of interest—but falling short of the significance criterion—were on 17q, 5q, 10q, 13q, and 6p.
In our study, less than half of the participants reported disturbed sleep attributed to coffee consumption before bedtime. Women reported a greater disturbance than men, and older people tended to be more sensitive to coffee's stimulant effects on sleep. The heritability of this trait was roughly equal between sexes, estimated at 0.37 in women and 0.42 in men. The remaining variation in this trait was due to the effect of unique environment; such effects are not shared between siblings and may include differences in daily caffeine consumption. We observed only 1% of covariance between caffeine consumption and coffee-attributed sleep disturbance in our sample: those who consumed more coffee were less affected by coffee at bedtime (data not shown). In this analysis, no distinction between regular and decaffeinated coffee was made, since, in the 1980s, decaffeinated products were not in common use in Australia, neither were canned beverages such as Coca-Cola.
Although unique environmental influences were largely specific to coffee-attributed insomnia, a general unique environmental factor was also supported, although its smallest influence was on coffee-attributed insomnia. Other epidemiologic factors shown to influence sleep disturbance and patterns include exercise, education, and marriage,6 all of which may differ between sibling pairs and may contribute to the general effects of unique environment on sleep disturbance. Common environment contributed to variation in some of the sleep-disturbance measures, especially for women, but there was no effect of common environment on coffee-attributed insomnia.
The sleep-disturbance measures were strongly influenced by a general genetic factor, which in both women and men most strongly influenced neurotic insomnia (i.e., suffering from sleeplessness). Although coffee-attributed insomnia was influenced by this factor, so that a person who generally suffered from sleeplessness would also be kept awake by coffee, a greater amount of genetic variance in this measure was explained by nongeneral influences. In women, most genetic variance (24%) was from a factor that also loaded to a lesser extent on variability of sleep quality. In men, this largely specific genetic effect on coffee-attributed insomnia (29%) also influenced depressed insomnia but not variability of sleep quality. In women, there was a further factor that influenced coffee-attributed insomnia. This factor had its largest influence on variability of sleep, and its relationship with coffee-attributed insomnia was in the reverse direction of the phenotypic correlation (although the covariance was smaller than that due to the other factors), so that those kept awake by coffee reported less-variable quality of sleep; they also reported less insomnia due to anxiety. These differences between sexes may partly result from differences in statistical power due to a larger female than male sample. Nonetheless, the results of the multivariate analysis confirmed that the main source of genetic variance for coffee-attributed insomnia was unrelated to a general sleep-disturbance factor.
The genetic linkage results of coffee-attributed insomnia revealed 1 chromosomal region (2q) of major interest, although other peaks falling short of suggestive linkage were also observed. Linkage peaks found in the “coffee-resistant” and “coffee-affected” analyses showed little overlap with each other, even though they represented the extremes of the same trait. However, it should be noted that the sample size of the affected groups in these analyses differed substantially: 340 pairs were “coffee resistant” versus 104 pairs who were “coffee affected.” Consistent with the “coffee-resistant” analysis being more powerful, the results of the threshold model showed a larger number of overlapping linkage peaks with this analysis than that of the “coffee-affected” analysis and suggests that greater confidence should be placed in these results.
A significant region on 2q was observed in the group that was resistant against caffeine's stimulant effect, whereas, in the caffeine-sensitive group, a region of suggestive linkage on 17q was identified. None of the known human genes related to caffeine metabolism or its targets in the central nervous system were located under our linkage peaks (i.e., those with LOD scores greater than 1). This is not entirely surprising since caffeine metabolism per se was not sampled, and the extent to which it plays a role in coffee-attributed sleep disturbance is not known. However, within our chromosome 17q linkage peak lies the DARPP-32 (dopamine- and cyclic AMP-regulated phosphoprotein of relative molecular mass 32,000) gene (located at 68.25cM), the homolog of which, in mice, when knocked out, reduces the stimulatory effect of caffeine on motor activity 33; adenosine (to which caffeine binds) is one class of neurotransmitters that stimulates phosphorylation of DARPP-32.34 Another candidate gene, hypocretin (orexin) neuropeptide precursor (HCRT; located at 70.76cM), lying within this linkage peak has been associated with the regulation of sleep and arousal in humans.35,36 It is possible then that the 17q region is linked to general sleep disturbance rather than specifically to caffeine-related sleep disturbance.
Although our self-report measure of coffee-attributed sleep disturbance showed good reliability over time, the phrasing of the test item was general—not indicating, for example, the quantity of coffee consumed before bedtime and the length of the insomnia—potentially lessening the item's validity. The distribution of our data may have therefore shown less variation than what truly exists in the population, although it was encouraging to observe sex and age effects consistent with other studies for general measures of sleep disturbance.
In summary, we showed that self-reported coffee-attributed insomnia is influenced mostly by genes that do not affect general sleep disturbance. In women, these genes also influence variability in sleep quality, whereas, in men, these genes influence depressive insomnia. A genome-wide linkage analysis of coffee-attributed insomnia revealed 1 significant chromosomal region on 2q in affected sibling pairs who were resilient to caffeine's disruptive effects on sleep. Although no candidate genes fulfilling our requirements of involvement in caffeine metabolism, adenosine-receptor function, or sleep and arousal were found under this peak, 2 candidate genes were identified on 17q, and these may be suitable for future genetic association studies of coffee-attributed sleep disturbance. In the future, we hope to obtain genome-wide association scans for this sample, which will give us much more power to identify the gene variants responsible for coffee-attributed sleep disturbance.
ACKNOWLEDGMENTS
We thank Lien Le for her help with bioinformatics searches, Dr. Peter Visscher for his helpful comments on an earlier version of this manuscript, and the participants for their cooperation. This work was supported by grants from the National Health and Medical Research Council of Australia (951023, 941177, 950998, 981339, 241916 and 941944), and the US National Institute for Alcohol Abuse and Alcoholism (AA007535, AA007728, AA013320, AA013321, AA013326, AA014041, AA10249, AA11998).
Footnotes
Disclosure Statement
This was not an industry supported study. The authors have reported no financial conflicts of interest.
REFERENCES
- 1.James JE, Gregg ME. Effects of dietary caffeine on mood when rested and sleep restricted. Hum Psychopharmacol. 2004;19:333–341. doi: 10.1002/hup.589. [DOI] [PubMed] [Google Scholar]
- 2.Rogers PJ, Martin J, Smith C, Heatherley SV, Smit HJ. Absence of reinforcing, mood and psychomotor performance effects of caffeine in habitual non-consumers of caffeine. Psychopharmacology. 2003;167:54–62. doi: 10.1007/s00213-002-1360-3. [DOI] [PubMed] [Google Scholar]
- 3.Yeomans MR, Ripley R, Davies LH, Rusted JM, Rogers PJ. Effects of caffeine on performance and mood depend on the level of caffeine abstinence. Psychopharmacology. 2002;164:241–249. doi: 10.1007/s00213-002-1204-1. [DOI] [PubMed] [Google Scholar]
- 4.Goldstein A, Warren R, Kaizer S. Psychotropic effects of caffeine in man. I. Individual differences in sensitivity to caffeine-induced wakefulness. J Pharmacol Exp Therap. 1965;149:156–159. [PubMed] [Google Scholar]
- 5.Hughes JR, Higgins ST, Bickel WK, et al. Caffeine self-administration, withdrawal, and adverse effects among coffee drinkers. Arch Gen Psychiatry. 1991;48:611–617. doi: 10.1001/archpsyc.1991.01810310029006. [DOI] [PubMed] [Google Scholar]
- 6.Heath AC, Eaves LJ, Kirk KM, Martin NG. Effects of lifestyle, personality, symptoms of anxiety and depression, and genetic predisposition on subjective sleep disturbance and sleep pattern. Twin Res. 1998;1:176–88. doi: 10.1375/136905298320566140. [DOI] [PubMed] [Google Scholar]
- 7.Hettema JM, Corey LA, Kendler KS. A multivariate genetic analysis of the use of tobacco, alcohol, and caffeine in a population based sample of male and female twins. Drug Alcohol Depend. 1999;57:69–78. doi: 10.1016/s0376-8716(99)00053-8. [DOI] [PubMed] [Google Scholar]
- 8.Luciano M, Kirk KM, H A.C., Martin NG. The genetics of tea and coffee drinking and preference for source of caffeine in a large community sample of Australian twins. Addiction. 2005;100:1510–1517. doi: 10.1111/j.1360-0443.2005.01223.x. [DOI] [PubMed] [Google Scholar]
- 9.Kendler KS, Prescott CA. Caffeine intake, tolerance, and withdrawal in women: a population-based twin study. Am J Psychiatry. 1999;156:223–8. doi: 10.1176/ajp.156.2.223. [DOI] [PubMed] [Google Scholar]
- 10.Partinen M, Kaprio J, Koskenvuo M, Putkonen P, Langinvainio H. Genetic and environmental determination of human sleep. Sleep. 1983;6:179–185. doi: 10.1093/sleep/6.3.179. [DOI] [PubMed] [Google Scholar]
- 11.Heath AC, Kendler KS, Eaves LJ, Martin NG. Evidence for genetic influences on sleep disturbance and sleep pattern in twins. Sleep. 1990;13:318–35. doi: 10.1093/sleep/13.4.318. [DOI] [PubMed] [Google Scholar]
- 12.McCarren M, Goldberg J, Ramakrishnan V, Fabsitz R. Insomnia in Vietnam era veteran twins: influence of genes and combat experience. Sleep. 1994;17:456–61. doi: 10.1093/sleep/17.5.456. [DOI] [PubMed] [Google Scholar]
- 13.Abe K. Reactions to coffee and alcohol in monozygotic twins. J Psychosom Res. 1968;12:199–203. doi: 10.1016/0022-3999(68)90047-0. [DOI] [PubMed] [Google Scholar]
- 14.Levy M, Zylber-Katz E. Caffeine metabolism and coffee-attributed sleep disturbances. Clin Pharmacol Ther. 1983;33:770–5. doi: 10.1038/clpt.1983.105. [DOI] [PubMed] [Google Scholar]
- 15.Retey JV, Adam M, Honegger E, et al. A functional genetic variation of adenosine deaminase affects the duration and intensity of deep sleep in humans. Proc Natl Acad Sci USA. 2005;102:15676–81. doi: 10.1073/pnas.0505414102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cornelis MC, El-Sohemy A, Kabagambe EK, Campos H. Coffee, CYP1A2 genotype, and risk of myocardial infarction. JAMA. 2006;295:1135–41. doi: 10.1001/jama.295.10.1135. [DOI] [PubMed] [Google Scholar]
- 17.Martin NG, Martin PG. The inheritance of scholastric abilities in a sample of twins. I. Ascertainments of the sample and diagnosis of zygosity. Ann Hum Genet. 1975;39:213–8. doi: 10.1111/j.1469-1809.1975.tb00124.x. [DOI] [PubMed] [Google Scholar]
- 18.Johns MW. A new method for measuring daytime sleepiness: the Epworth sleepiness scale. Sleep. 1991;14:540–5. doi: 10.1093/sleep/14.6.540. [DOI] [PubMed] [Google Scholar]
- 19.Bedford A, Foulds GA, Sheffield BF. A new personal disturbance scale (DSSI/sAD) Br J Soc Clin Psychol. 1976;15:387–94. doi: 10.1111/j.2044-8260.1976.tb00050.x. [DOI] [PubMed] [Google Scholar]
- 20.Eysenck HJ, Eysenck SBG. Manual for the Eysenck personality questionnaire. London: Hodder – Stoughton; 1975. [Google Scholar]
- 21.Neale MC, Cardon LR. Netherlands: Kluwer Academic Publishers; 1992. Methodology for Genetic Studies of Twins and Families Dordrecht. [Google Scholar]
- 22.Cornes BK, Medland SE, Ferreira MA, et al. Sex-limited genome-wide linkage scan for body mass index in an unselected sample of 933 Australian twin families. Twin Res Hum Genet. 2005;8:616–32. [PubMed] [Google Scholar]
- 23.Saccone S, Pergadia M, Loukola A, et al. Genetic linkage to chromosome 22q12 for a heavy-smoking quantitative trait in two independent samples. Am J Hum Genet. 2007;80:856–866. doi: 10.1086/513703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wessman M, Kallela M, Kaunisto MA, et al. A susceptibility locus for migraine with aura, on chromosome 4q24. Am J Hum Genet. 2002;70:652–662. doi: 10.1086/339078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Visscher PM, Medland SE, Ferreira MA, et al. Assumption-free estimation of heritability from genome-wide identity-by-descent sharing between full siblings. PLoS Genet. 2006;2:e41. doi: 10.1371/journal.pgen.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neale MC, Boker SM, Xie G, Maes HH. 5th ed. VCU Box 900126, Richmond, VA 23298: Department of Psychiatry; 1999. Mx: Statistical Modeling. [Google Scholar]
- 27.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 28.Abecasis GR. 2004;Vol. 2004 http://www.sph.umich.edu/csg/abecasis/Merlin/reference.html.
- 29.Sawcer S, Jones HB, Judge D, et al. Empirical genomewide significance levels established by whole genome simulations. Genet Epidemiol. 1997;14:223–9. doi: 10.1002/(SICI)1098-2272(1997)14:3<223::AID-GEPI1>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 30.Kruglyak L, Daly MJ. Linkage thresholds for two-stage genome scans. Am J Hum Genet. 1998;62:994–7. doi: 10.1086/301792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fulker DW, Cherney SS, Sham PC, Hewitt JK. Combined linkage and association sib-pair analysis for quantitative traits. Am J Hum Genet. 1999;64:259–267. doi: 10.1086/302193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu G, Duffy DL, Eldridge A, et al. A major quantitative-trait locus for mole density is linked to the familial melanoma gene CDKN2A: A maximum-likelihood combined linkage and association analysis in twins and their sibs. Am J Hum Genet. 1999;65:483–492. doi: 10.1086/302494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindskog M, Svenningsson P, Pozzi L, et al. Involvement of DARPP-32 phosphorylation in the stimulant action of caffeine. Nature. 2002;418:774–8. doi: 10.1038/nature00817. [DOI] [PubMed] [Google Scholar]
- 34.Nishi A, Liu F, Matsuyama S, et al. Metabotropic mGlu5 receptors regulate adenosine A2A receptor signaling. Proc Natl Acad Sci USA. 2003;100:1322–7. doi: 10.1073/pnas.0237126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peyron C, Faraco J, Rogers W, et al. A mutation in a case of early onset narcolepsy and a generalized absence of hypocretin peptides in human narcoleptic brains. Nat Med. 2000;6:991–7. doi: 10.1038/79690. [DOI] [PubMed] [Google Scholar]
- 36.Taheri S, Zeitzer JM, Mignot E. The role of hypocretins (orexins) in sleep regulation and narcolepsy. Annu Rev Neurosci. 2002;25:283–313. doi: 10.1146/annurev.neuro.25.112701.142826. [DOI] [PubMed] [Google Scholar]