Abstract
Inactivation of the genes involved in DNA mismatch repair is associated with microsatellite instability (MSI) in colorectal cancer. We report that hypermethylation of the 5′ CpG island of hMLH1 is found in the majority of sporadic primary colorectal cancers with MSI, and that this methylation was often, but not invariably, associated with loss of hMLH1 protein expression. Such methylation also occurred, but was less common, in MSI− tumors, as well as in MSI+ tumors with known mutations of a mismatch repair gene (MMR). No hypermethylation of hMSH2 was found. Hypermethylation of colorectal cancer cell lines with MSI also was frequently observed, and in such cases, reversal of the methylation with 5-aza-2′-deoxycytidine not only resulted in reexpression of hMLH1 protein, but also in restoration of the MMR capacity in MMR-deficient cell lines. Our results suggest that microsatellite instability in sporadic colorectal cancer often results from epigenetic inactivation of hMLH1 in association with DNA methylation.
Mismatch repair is required for the cell to accurately copy its genome during cellular proliferation. Deficiencies of this system result in mutation rates 100-fold greater than those observed in normal cells (1, 2). These mutations are particularly evident in microsatellite sequences, consisting of repeats of 1–4 bp. Microsatellite instability (MSI) is thereby a hallmark of mismatch repair gene (MMR)-deficient cancers. MSI has been observed in approximately 13% of sporadic colorectal cancers (CRC) and in virtually all CRC arising in patients with hereditary nonpolyposis colorectal cancer (HNPCC) (3, 4). HNPCC generally is associated with germ-line mutations in one of two MMR genes, hMLH1 and hMSH2, with mutations of other MMR genes being rare (5, 6). In MSI+ cancers from patients without HNPCC, these same genes often are mutationally inactivated. However, in a significant subset of sporadic tumors with MSI+, no mutations of MMR genes could be identified (7–11) and it was speculated that nonmutational mechanisms or novel genes were responsible for the defect (10, 11).
Alternative modes of inactivation of genes during the development of cancer include an epigenetic process marked by promoter region hypermethylation associated with transcriptional loss, as demonstrated for several tumor suppressor genes (12–14). Interestingly, two lines of experimentation have suggested an intimate relationship between MMR and altered DNA methylation in human cells. First, exogenous and endogenous sequences appear to be methylated at much higher levels in MMR-deficient colorectal tumors than in their MMR-proficient counterparts (15, 16). Second, a subset of MSI+ sporadic colorectal tumors and MSI+ tumor cell lines derived from a variety of tumor types lack hMLH1 protein without apparent structural alterations of this gene (7, 17), and the promoter of the hMLH1 gene has been shown to be methylated in four primary colorectal tumors and tumor cell lines (18). These results raise a variety of questions about the causal relationship between MMR deficiency and DNA methylation. To address these questions, we have analyzed hMLH1 promoter methylation in several subtypes of CRC, including those with known mutations of MMR genes. We have matched these data to patterns of hMLH1 expression and tested the functional consequences of promoter region methylation of this gene in MMR-deficient cell lines.
MATERIALS AND METHODS
Tissue Samples and Cell Cultures.
Colorectal mucosa and primary sporadic colorectal specimens were obtained as described (16). The HNPCC kindreds from which tumors were studied have been described (19, 20). Colorectal cancer cell lines (21) used in this study have been characterized previously for their MSI status (22), mutations of MMR genes in the case of MSI+ tumors (10), and, in some cases, their ability to perform DNA mismatch repair in vitro (17, 23). Cell lines were maintained in appropriate media and were treated with 1 μM 5-aza-2′-deoxycytidine for 5 days (RKO and SW48 cells) or with 5 μM 5′-azacytidine for 1 or 3 days (AN3CA).
Methylation-Specific PCR (MSP).
DNA methylation patterns in the CpG islands of hMLH1 and hMSH2 genes were determined by chemical treatment with sodium bisulfite and subsequent MSP as described (24). Primer sequences of hMLH1 for unmethylated reaction were 5′-TTTTGATGTAGATGTTTTATTAGGGTTGT-3′ (sense) and 5′-ACCACCTCATCATAACTACCCACA-3′ (antisense), and for methylated reaction were 5′-ACGTAGACGTTTTATTAGGGTCGC-3′ (sense) and 5′-CCTCATCGT AAC-TACCCGCG-3′ (antisense). Primer sequences of hMSH2 for unmethylated reaction were 5′-GGTTGTTGTGGTTGGATGTTGTTT-3′ (sense) and 5′-CAACTACAACATCTCCTTCAACTACACCA-3′ (antisense) and for methylated reaction were 5′-TCGTGGTCGGACGTCGTTC-3′ (sense) and CAACGTCTCCTTCGACTACACCG-3′ (antisense). Paraffin-embedded samples first were amplified with flanking PCR primers that amplify bisulfite-modified DNA but that would not preferentially amplify methylated or unmethylated DNA. The primers used were 5′-GAGTAGTTTTTTTTTTAGGAGTGAAG-3′(sense) and 5′-AAAAACTATAAAACCCTATACCTAATCTA-3′ (antisense). All PCRs were performed with positive controls for both unmethylated and methylated alleles, and no DNA control. Human placental DNA treated in vitro with excess SssI methyltransferase (New England Biolabs), generating DNA completely methylated at all CpG sites, served as the positive control for methylated hMSH2.
Western Analysis.
Cells (≈1 × 105) were lysed in SDS sample buffer (2% SDS/60 mM Tris, pH 6.8/10% glycerol/0.1 M DTT) and resolved by electrophoresis on a 4–20% SDS-polyacrylamide gradient gel (NOVEX, San Diego), transferred to Immobilon P membranes (Millipore), and probed with anti-human MLH1 mAb (Oncogene Science, Ab-1) at 1 μg/ml concentration. After incubation with horseradish peroxidase-coupled secondary antibody (Pierce), reactive proteins were visualized with enhanced chemiluminescence (Amersham).
Immunohistochemistry of hMLH1.
Sections (6 μm) of formalin-fixed, paraffin-embedded tissue were deparaffinized with xylenes for 30 min and dehydrated by using graded ethanols. Antigen retrieval was performed by using a heat-induced epitope retrieval method (25). Immunoperoxidase staining using diaminobenzidine as chromogen was performed with the TechMate 1000 automatic staining system (Ventana, BioTek Solutions, Tucson, AZ). Mouse mAb to hMLH1 gene product (PharMingen) was used at 1:300 dilution. Staining of tumor nuclei was evaluated as present or absent in coded slides by one author (S.R.H.) who had no knowledge of the results of the molecular analyses.
Mismatch Repair Assay.
Preparation of cell-free extracts and mismatched substrates, and procedures for measuring mismatch repair activity, have been described (26). DNA mismatch repair reactions (25 μl) contained 30 mM 4-(2-hydroxyethyl)-1-piperazine-ethanesulfonic acid (pH 7.8); 7 mM MgCl2; 200 μM each CTP, GTP, UTP; 4 mM ATP; 100 μM each dCTP, dATP, dGTP, dTTP; 40 mM creatine phosphate; 100 mg/ml creatine phosphokinase; 15 mM sodium phosphate (pH 7.5); 1 fmol of indicated DNA substrate; and 50 μg of extract proteins. After incubating at 37°C for either 15 or 30 min, samples were processed and introduced into E. coli NR9162 (mutS) via electroporation. Cells were plated, M13 mp2 plaque colors were scored, and repair efficiencies (in %) were calculated as described (26).
RESULTS
Methylation Status of hMLH1 in Normal Cells and Cultured Tumors.
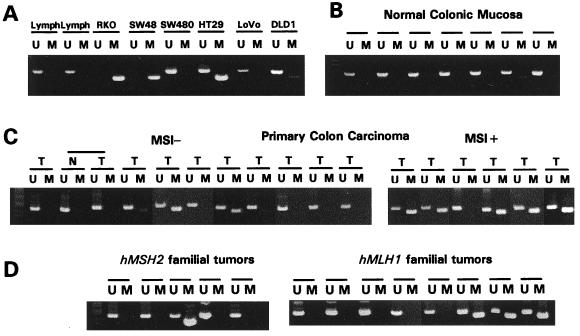
To examine promoter region methylation of hMLH1 and hMSH2, we adapted MSP for the 5′ CpG islands present in both genes (24). The region chosen for hMLH1 spans the area of greatest CpG density immediately 5′ to the transcription start site, in an area previously studied for methylation changes (18). In colorectal mucosa samples from 10 patients without cancer (Fig. 1B) and normal lymphocytes (Fig. 1A), only unmethylated hMLH1 genes were present, as would be expected for the 5′ CpG island of this and other nonimprinted genes in normal tissues (27). In the nonexpressing cell line SW48 (10, 17), found previously by another PCR assay to have hypermethylation of the 5′ hMLH1 CpG island (18), we found only methylated hMLH1 (Fig. 1A).
Figure 1.
Methylation of hMLH1 promoter region CpG island in cell lines and primary human samples. The presence of a visible PCR product in those lanes marked U indicates the presence of unmethylated genes of hMLH1; the presence of product in those lanes marked M indicates the presence of methylated genes. (A) Normal lymphocytes and colorectal cell lines. Normal lymphocytes and the MSI− colorectal cell line SW480 contain only unmethylated hMLH1. MSI+ cell lines RKO and SW48 contain only methylated hMLH1. MSI+ cell lines Lovo and DLD1, both with mutations in MMR genes, are unmethylated at hMLH1. HT29 contains both unmethylated and methylated hMLH1 genes. (B) Normal colonic mucosa samples, all unmethylated at hMLH1. (C) Primary sporadic colon carcinomas (T), with the MSI phenotype shown above. All primary tumors include amplification with the U primer set, a result of the presence of normal contaminating tissue. Included is one MSI− tumor with adjacent normal mucosa, labeled N. (D) Primary colon carcinomas from patients with either inherited hMSH2 mutations (Left) or hMLH1 mutations (Right).
We examined hypermethylation of hMLH1 in 37 CRC cell lines (examples in Fig. 1A). The MSI+ cell lines RKO, VACO5, and VACO6, previously characterized as lacking mutations in any mismatch repair gene (10), also were completely methylated at the hMLH1 locus. VACO5 and VACO6 previously have been shown to lack expression of hMLH1 mRNA (10). We next examined four MSI+ CRC cell lines in which hMSH2, hMLH1, hPMS2, and hPMS1 were all expressed as determined by reverse transcription–PCR (RT-PCR) analysis and in which the entire coding sequences were wild type. In three of these lines, the hMLH1 genes were methylated (VACO481, VACO444, and x587), whereas the other (x543) contained only unmethylated genes. Thus, seven of eight cell lines with MSI+ phenotype and no known MMR gene mutation have a methylated hMLH1. In four MSI+ lines with known mutations of a MMR gene, one (Cx2), with a deletion of the first six exons of hMSH2, was partially methylated whereas the other three (LoVo, x595, and DLD-1), with inactivating mutations of hMSH2, hMLH1, and hMSH6, respectively, exhibited no hMLH1 promoter methylation. We also examined 25 CRC cell lines without MSI and, of these, one, HT29 (Fig. lA), had partial methylation of the hMLH1 gene, whereas two were fully methylated. Finally, we found no hMLH1 promoter methylation in 29 cancer cell lines derived from organs other than the colon (data not shown), including the hMLH1 mutant prostate cancer cell line DU145. This suggests that methylation of hMLH1 most often was found in cell lines with the MSI+ phenotype and without mutational inactivation of a MMR gene.
hMLH1 and hMSH2 Methylation Status in Primary Colorectal Cancer.
Primary cancers were analyzed by using MSP to determine the prevalence of hMLH1 promoter methylation in CRC in their natural environment. Eleven of 13 (84%) MSI+ cancers (16) exhibited prominent methylation, compared with only 2 of 21 MSI-primary cancers (Fig. 1C, Fisher’s exact P < 0.0001). Unlike the situation with the cell lines, however, the primary MSI+ cancers always had both methylated and nonmethylated hMLH1 genes present (compare Fig. 1 A with C). It is likely that a significant fraction of the unmethylated genes was derived from the non-neoplastic cells (stromal, inflammatory, vascular, etc.), which invariably are present within primary tumors but are not found in cultured cell lines.
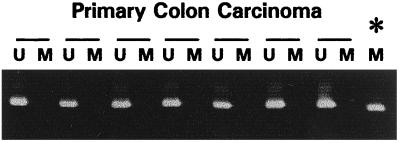
Because germ-line mutations occur as frequently in hMSH2 as in hMLH1 in HNPCC kindreds (5, 6), we examined these sporadic colorectal tumors for hypermethylation of hMSH2. hMSH2 also contains a CpG island in the 5′ promoter region of the gene. As expected for a 5′ CpG island, normal lymphocytes are unmethylated at this locus (not shown). In contrast to the results for hMLH1, we found that 0 of 34 sporadic colorectal tumors, including the 13 MSI+ cases, harbored hypermethylation in the hMSH2 5′ CpG island (Fig. 2).
Figure 2.
Methylation of hMSH2 in primary sporadic colorectal cancer. The presence of a visible PCR product in those lanes marked U indicates the presence of unmethylated genes of hMSH2; the presence of product in those lanes marked M indicates the presence of methylated genes, seen only in the in vitro methylated control DNA (∗). All primary colorectal tumors contain only unmethylated hMSH2 genes.
One would expect that if hMLH1 methylation was the cause of the MSI+ phenotype, tumors with classical mutations in a mismatch repair gene should not exhibit methylation of hMLH1. However, it is difficult to determine the mutational status of MMR genes in primary cancers for several reasons. In addition to the fact that many different genetic alterations can cause the MSI+ phenotype, the presence of non-neoplastic cells within the primary tumors greatly complicates the ability to reliably detect mutations. Therefore we turned to primary cancers from HNPCC patients. These patients have germ-line mutations of one allele of a MMR gene, and the tumors that develop frequently contain inactivating mutations or losses of the normal allele inherited from the unaffected parent. Thus, such tumors provide an opportunity to study the relationship between hMLH1 methylation and MSI in primary tumors with well characterized genetic defects in MMR genes. Four of 18 such tumors (22%) were found to contain methylated hMLH1 genes (Fig. 1D). Three of these four tumors occurred in families with germ-line mutations of hMLH1, whereas the fourth occurred in a patient with a germ-line mutation of hMSH2. The frequency of hMLH1 methylation in these tumors was significantly reduced relative to sporadic MIN tumors (hMLH1 families vs. sporadic MSI+, P < 0.01; hMSH2 families vs. sporadic MSI+, P < 0.002; combined families vs. sporadic MSI+, P < 0.001).
Expression of hMLH1 Protein in Primary Colorectal Cancers.
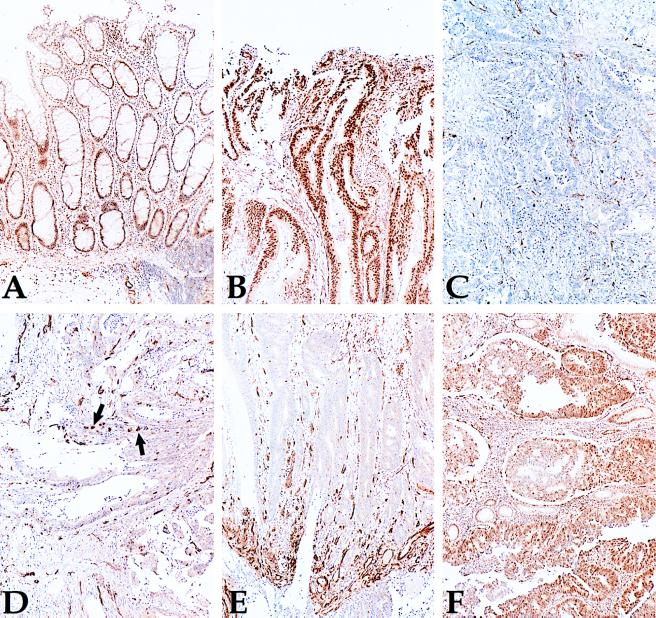
Five of the primary MSI+ CRC with hMLH1 promoter region methylation were examined immunohistochemically with a mAb to hMLH1 to determine the relationship between hMLH1 expression and methylation. Four of the five tumors had no detectable hMLH1 expression in neoplastic cells (Fig. 3C), whereas one had a heterogeneous staining pattern (Fig. 3 D and E). In all cases, the positive staining of non-neoplastic cells provided an internal control for the integrity of the immunohistochemical procedures. In contrast, six MSI− tumors lacking hMLH1 promoter methylation each exhibited uniform staining of neoplastic cells with the same antibody (example Fig. 3B). In two MSI− cancers with methylated hMLH1 genes, heterogeneous staining by the anti-MLH1 antibody was observed, with most cancer cells expressing hMLH1 (Fig. 3F).
Figure 3.
Immunohistochemistry of hMLH1 in primary colon cancer. A is normal colon adjacent to the MSI+ carcinoma shown in C, which is methylated at hMLH1 and does not express any protein within the cancer cells. B is a MSI− tumor that is unmethylated at hMLH1 and expresses the protein. D and E are from a MSI+ tumor with hypermethylation of hMLH1 that expresses hMLH1 in only some of the cancer cells, which are shown near arrows. In E, control vascular structures at the bottom stain for hMLH1, whereas the carcinoma nuclei do not. F is a MSI− tumor that has hypermethylation of hMLH1 and expresses hMLH1 in most cells.
Functional Consequences of hMLH1 Methylation in Colorectal Cancer.
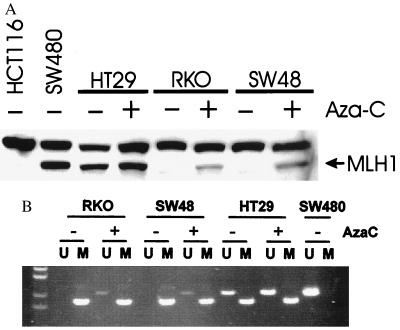
The above results were consistent with the idea that the methylation of hMLH1 is associated with its decreased expression in CRC, contributing to the MSI+ phenotype. We therefore examined the expression of hMLH1 in colorectal cell lines and correlated this expression to the status of MMR genes, MIN phenotype, and hMLH1 methylation. The anti-hMLH1 antibody is directed at the C terminus of the protein and, therefore, no hMLH1 protein was detected in HCT116 cells, which have a truncating mutation of hMLH1 (Fig. 4A). We also found that the SW48 and RKO cell lines, which are hypermethylated at hMLH1, contained no hMLH1 protein, whereas the MMR-proficient cell line SW480 and MSI− cell line HT29 exhibited a protein of the expected size (Fig. 4A). We also examined expression at the level of RT-PCR in selected MSI− colorectal cell lines. We found that all MSI− cell lines expressed hMLH1 mRNA by this sensitive assay, including one displaying methylated hMLH1 genes (data not shown).
Figure 4.
(A) Western blot analysis of hMLH1 in colorectal cell lines. Note detectable protein in SW480 and HT29 before drug treatment (AzaC), but in RKO and SW48 only after drug treatment. (B) Demethylation analysis of cell lines after azacytidine treatment. The presence of U product in RKO and SW48 after 5-aza-2′-deoxycytidine indicates the presence of demethylation of the hMLH1 promoter in these cell lines.
To more directly address whether the promoter region methylation was itself inhibiting the expression of hMLH1, we treated cell lines with 5-aza-2′-deoxycytidine, an agent that results in the demethylation of DNA. After a 5-day treatment with the demethylating agent 5-aza-2′-deoxycytidine, expression of hMLH1 protein was restored substantially in SW48 and RKO cells, whereas this drug minimally increased the expression of hMLH1 in HT29 cells. This reactivation was associated with the presence of unmethylated hMLH1 alleles in both SW48 and RKO, which could not be detected before drug treatment (Fig. 4B).
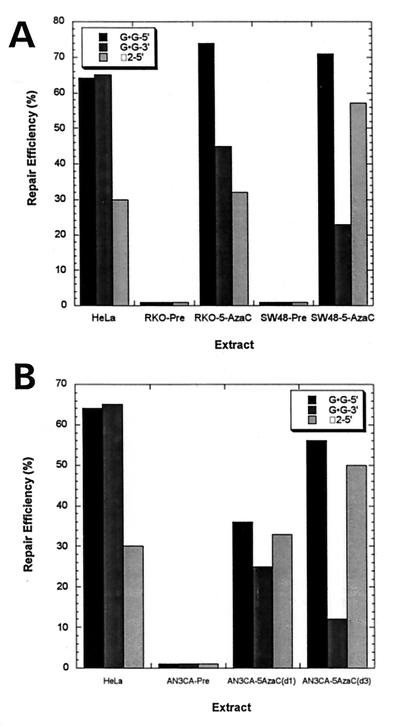
To determine whether the methylation of hMLH1 plays a direct role in mediating the MSI+ phenotype, extracts from untreated and 5-aza-2′-deoxycytidine-treated SW48 and RKO cells were tested for ability to repair base⋅base and insertion/deletion mismatches. Extracts of untreated SW48 or RKO cells that were not expressing hMLH1 failed to repair a G⋅G mismatch with a nick either 5′ or 3′ to mismatch or a substrate containing two extra bases and a nick 5′ to the unpaired bases (Fig. 5A). However, after treatment with 5-deoxy-2′-azacytidine for 5 days, these cells not only expressed hMLH1 protein, but also performed strand-specific mismatch repair of both substrates (Fig. 5A). A separate set of experiments also was performed with the endometrial carcinoma cell line AN3CA. This cell line exhibits MSI and lacks mismatch repair activity (23), lacks hMLH1 mRNA expression (17), and has a methylated hMLH1 promoter (18). Treatment of AN3CA cells with 5-azacytidine for 1 or 3 days led to demethylation of the hMLH1 promoter (not shown), restored expression of hMLH1 message (data not shown) as determined by RT-PCR (17), and restored the ability of extracts to perform strand-specific repair of substrates containing either a G⋅G mismatch with a nick either 5′ or 3′ to mismatch or a substrate containing two extra bases and a nick 5′ to unpaired bases (Fig. 5B).
Figure 5.
(A) Mismatch repair activity in extracts of tumor cell lines treated with 5′-aza 2′-deoxycytidine. Repair reactions were incubated for 30 min (except G⋅G-3′ for 15 min), and the products were analyzed as described in Materials and Methods. Results are for the mismatched substrates G⋅G-5′/3′ and 2 unpaired bases with a nick 5′ to unpaired bases. DNA substrates contained a nick in the minus strand at either position −264 (for the 3′ nicked substrate) or at position +276 (for the 5′ nicked substrate), where position +1 is the first transcribed base of the lacZα-complementation gene. The G⋅G mismatch is at position 88, and α2 is at 90, 91 of the lacZα gene. HeLa, RKO-pretreatment (RKO-Pre), and SW48-pretreatment (SW48-Pre) are compared with cell extracts of RKO (RKO-5-AzaC) and SW48 (SW48-5-AzaC) made after 5 days of treatment with 5-aza-2′-deoxycytidine. (B) Mismatch repair activity in extracts of the AN3CA tumor cell line either untreated or treated with 5′ azacytidine. Above described substrates are tested in AN3CA cell extracts either pre, 1 day, or 3 days posttreatment with 5′ azacytidine.
DISCUSSION
Our findings demonstrate several points about the relationship between hMLH1 promoter methylation and MMR deficiency. First, methylation of the hMLH1 promoter occurs commonly in both cell lines and primary cancers with MMR deficiency. Second, such methylation is correlated with decreased expression of the hMLH1 gene, both at the RNA and protein levels. Third, and most important, demethylation of the hMLH1 promoter results in reexpression of hMLH1 in each of three cell lines tested. Not only was protein expressed, but MMR activity was restored, formally excluding the possibility that functionally important mutational defects in the coding regions of any MMR gene (missense or nonsense) were primarily responsible for the absence of MMR activity in these lines. Although multiple other silenced and hypermethylated genes in tumor cells can be reexpressed after demethylation (12–14), this is the clearest example of the restoration of a key normal function previously lost in the neoplastic cells.
In the present study, we found hypermethylation of hMLH1 in the majority (84%) of MSI+ sporadic colorectal cancers. Does this reflect the true incidence of this change in the subset of CRC with MMR deficiency and that an epigenetic mechanism is responsible for this defect in the majority of such tumors? The answer to this question must await even larger studies that include both methylation analyses and the actual prevalence of MMR gene mutations in these tumors. It is possible that some of the 11 sporadic MSI+ cancers we found to have hMLH1 hypermethylation may have structural alterations of a MMR gene. As mentioned previously, evaluation of MMR gene mutations in primary cancers can be difficult because of the large number of genes that can cause the phenotype and the masking of mutations by non-neoplastic cells present within the tumors. One way to estimate the proportion of sporadic MIN cancers in which methylation of the hMLH1 promoter plays a role is to consider the proportion of cases, analyzed in detail, where structural mutations of a MMR gene have not been identifiable. Our analysis of the relevant literature on this point (7–11) suggests that such mutations are identifiable in at least 26% of cancers, leaving the remaining 100 − 26 (74%) as possibly attributable to methylation of the hMLH1 promoter. This estimate can also be reached by subtracting from 84% the “background” methylation of hMLH1 (2 of 21 sporadic MSI− primary tumors, 4 of 18 HNPCC primary tumors, 1 of 5 cell lines with MMR gene mutation, and 3 of 25 MSI− colorectal cell lines). Combining these groups give a background rate of 15%. If 85% of hMLH1 methylation is “specific” and 84% of MSI+ CRC is methylated, then in 71% (0.85 × 0.84) methylation is functional and leads to inactivation of MMR. Thus, even by these conservative estimates, and judging by our functional analyses in cell culture, hypermethylation-associated silencing of hMLH1 results in MMR deficiency in a high number of sporadic CRC.
Although, the bulk of our data suggest that methylation of the hMLHI promoter is an epigenetic event that plays a causal role in the MMR defect in many MSI+ cancers, we report several observations that complicate this interpretation. First, as noted above, methylation of the hMLH1 promoter is not totally confined to MSI+ tumors, because it occurs in a small subset of MSI− cancers. Second, methylation of the hMLH1 promoter occurred in several tumors with coding region mutations of either hMLH1 or another MMR gene. In such cases, the methylation of this promoter may not be the primary cause of the MMR deficiency. There are several reasons that aberrant methylation might be seen in tumors where it may not be of functional significance. First, the sensitivity of our assay may detect a level of allelic silencing that does not yet produce a MSI+ phenotype, as the MSI− cell line HT29 demonstrates. Such partial methylation may also explain the heterogeneous staining pattern for hMLH1 protein in two of our MSI− primary colorectal tumors. However, the two MSI− colorectal cell lines with only methylated hMLH1 genes raise another interesting possibility. Although hypermethylation of hMLH1 was associated with mRNA still detectable at the RT-PCR level in one of these cell lines, normal levels of hMLH1 may not be expressed, and the MMR proficiency of these cell lines has not been determined. Diminished hMLH1 expression may lead to MMR deficiency without MSI in cell lines tolerating alkylating DNA damage (28).
A second explanation for our findings of inherited colorectal tumors with hMLH1 methylation concerns the frequency of LOH. LOH generally is found in familial cancers associated with mutated tumor-suppressor genes. For hMLH1, even though such loss always involves the wild-type allele, LOH was reported in only 44% of these tumors (20). Therefore, some tumors from families with germ-line hMLH1 mutations may have hypermethylation, rather than LOH, of the wild-type allele. In fact, one of the three HNPCC tumors we studied with hMLH1 hypermethylation did not have LOH of 3p. Such hypermethylation of the wild-type allele has been observed for the von Hippel-Lindau gene (VHL) in 6 of 18 tumors from patients with inherited mutations of VHL without LOH (29), and for Rb in a tumor from a patient with a germ-line mutation of this tumor-suppressor gene (30).
Our present study highlights recently observed correlations between the MSI+ phenotype and DNA methylation. Two previous studies have suggested that alterations in the mismatch repair pathways correlate with hypermethylation of both exogenous and endogenous DNA sequences (15, 16). For example, of the 13 primary MSI+ cancers described in this study, a striking methylation of several different genes was observed previously (16). Exogenously added sequences also become methylated to a much higher degree in MSI+ than in MSI− cell lines, regardless of the defective MMR gene involved (15). Our data suggest that for sporadic CRC, by targeting the hMLH1 promoter region, a propensity for methylation of endogenous genes in colon cancers is the cause, and not the consequence of, microsatellite instability. Further support for this sequence of events is suggested by the frequency of p16 hypermethylation in the samples from the present study, an event correlated with the MSI+ phenotype in sporadic colon cancer (16). In tumors from patients with HNPCC, p16 hypermethylation was present in 5 of 23 (22%) of these inherited MSI+ tumors (data not shown). This is a much lower frequency of p16 methylation than reported previously in MSI+ sporadic tumors (9 of 15 = 60%, P < 0.02), and similar to that observed in sporadic MSI− tumors (22%) (16). Thus, the MSI+ phenotype produced by genetic inactivation of the MMR genes is not associated with an increased frequency of p16 hypermethylation, whereas epigenetic inactivation of hMLH1 through hypermethylation often is associated with p16 hypermethylation.
Our results suggest that DNA methylation associated with transcriptional silencing of hMLH1 is the underlying cause of MMR defects in most sporadic colorectal cancers having a MSI+ phenotype. The resulting mutator phenotype is associated with mutation of functionally important genes such as the transforming growth factor type β II receptor (22) and BAX (31). Thus, hypermethylation of hMLH1 and the associated MSI+ phenotype in sporadic colon cancers may represent an unusual setting in which an epigenetic event may lead to multiple genetic alterations in tumor cells.
Acknowledgments
We thank Drs. Albert de la Chapelle, Lauri Aaltonin, and Paivi Peltomaki for tumors from HNPCC families. This research was supported by grants from the National Institutes of Health (CA43318, CA54396, CA44704, and GM50006) and a Gastrointestinal Cancer SPORE Grant (CA-62924). J.G.H. is a V Foundation Scholar. J.G.H. and S.B.B. receive research funding and are entitled to sales royalties from ONCOR, which is developing products related to research described in this paper. The terms of this arrangement have been reviewed and approved by The Johns Hopkins University in accordance with its conflict of interest policies.
ABBREVIATIONS
- MSI
microsatellite instability
- MMR
mismatch repair
- CRC
colorectal carcinomas
- HNPCC
hereditary nonpolyposis colorectal cancer
- MSP
methylation-specific PCR
- RT-PCR
reverse transcription–PCR
- LOH
loss of heterozygosity
References
- 1.Thomas D C, Umar A, Kunkel T A. Mutat Res. 1996;350:201–205. doi: 10.1016/0027-5107(95)00112-3. [DOI] [PubMed] [Google Scholar]
- 2.Modrich P, Lahue R. Annu Rev Biochem. 1996;65:101–33. doi: 10.1146/annurev.bi.65.070196.000533. , 101–133. [DOI] [PubMed] [Google Scholar]
- 3.Aaltonen L A, Peltomaki P, Leach F S, Sistonen P, Pylkkanen L, Mecklin J P, Jarvinen H, Powell S M, Jen J, Hamilton S R, et al. Science. 1993;260:812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 4.Thibodeau S N, Bren G, Schaid D. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 5.Peltomaki P, de la Chapelle A. Adv Cancer Res. 1997;71:93–119. doi: 10.1016/s0065-230x(08)60097-4. [DOI] [PubMed] [Google Scholar]
- 6.Papadopoulos N, Lindblom A. Hum Mutat. 1997;10:89–99. doi: 10.1002/(SICI)1098-1004(1997)10:2<89::AID-HUMU1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 7.Thibodeau S N, French A J, Roche P C, Cunningham J M, Tester D J, Lindor N M, Moslein G, Baker S M, Liskay R M, Burgart L J, et al. Cancer Res. 1996;56:4836–4840. [PubMed] [Google Scholar]
- 8.Borresen A L, Lothe R A, Meling G I, Lystad S, Morrison P, Lipford J, Kane M F, Rognum T O, Kolodner R D. Hum Mol Genet. 1995;4:2065–2072. doi: 10.1093/hmg/4.11.2065. [DOI] [PubMed] [Google Scholar]
- 9.Bubb V J, Curtis L J, Cunningham C, Dunlop M G, Carothers A D, Morris R G, White S, Bird C C, Wyllie A H. Oncogene. 1996;12:2641–2649. [PubMed] [Google Scholar]
- 10.Liu B, Nicolaides N C, Markowitz S, Willson J K, Parsons R E, Jen J, Papadopoulos N, Peltomaki P, de la Chapelle A, Hamilton S R, et al. Nat Genet. 1995;9:48–55. doi: 10.1038/ng0195-48. [DOI] [PubMed] [Google Scholar]
- 11.Wu Y, Nystrom-Lahti M, Osinga J, Looman M W, Peltomaki P, Aaltonen L A, de la Chapelle A, Hofstra R M, Buys C H. Genes Chromosomes Cancer. 1997;18:269–278. [PubMed] [Google Scholar]
- 12.Herman J G, Merlo A, Mao L, Lapidus R G, Issa J P J, Davidson N E, Sidransky D, Baylin S B. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- 13.Merlo A, Herman J G, Mao L, Lee D J, Gabrielson E, Burger P C, Baylin S B, Sidransky D. Nat Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- 14.Herman J G, Latif F, Weng Y, Lerman M I, Zbar B, Liu S, Samid D, Duan D S, Gnarra J R, Linehan W M, Baylin S B. Proc Natl Acad Sci USA. 1994;91:9700–9704. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lengauer C, Kinzler K W, Vogelstein B. Proc Natl Acad Sci USA. 1997;94:2545–2550. doi: 10.1073/pnas.94.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahuja N, Mohan A L, Li Q, Stolker J M, Herman J G, Hamilton S R, Baylin S B, Issa J P. Cancer Res. 1997;57:3370–3374. [PubMed] [Google Scholar]
- 17.Boyer J C, Umar A, Risinger J I, Lipford J R, Kane M, Yin S, Barrett J C, Kolodner R D, Kunkel T A. Cancer Res. 1995;55:6063–6070. [PubMed] [Google Scholar]
- 18.Kane M F, Loda M, Gaida G M, Lipman J, Mishra R, Goldman H, Jessup J M, Kolodner R. Cancer Res. 1997;57:808–811. [PubMed] [Google Scholar]
- 19.Liu B, Parsons R, Papadopoulos N, Nicolaides N C, Lynch H T, Watson P, Jass J R, Dunlop M, Wyllie A, Peltomaki P, et al. Nat Med. 1996;2:169–174. doi: 10.1038/nm0296-169. [DOI] [PubMed] [Google Scholar]
- 20.Hemminki A, Peltomaki P, Mecklin J P, Jarvinen H, Salovaara R, Nystrom-Lahti M, de la Chapelle A, Aaltonen L A. Nat Genet. 1994;8:405–410. doi: 10.1038/ng1294-405. [DOI] [PubMed] [Google Scholar]
- 21.Willson J K, Bittner G N, Oberley T D, Meisner L F, Weese J L. Cancer Res. 1987;47:2704–2713. [PubMed] [Google Scholar]
- 22.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan R S, Zborowska E, Kinzler K W, Vogelstein B. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 23.Umar A, Boyer J C, Thomas D C, Nguyen D C, Risinger J I, Boyd J, Ionov Y, Perucho M, Kunkel T A. J Biol Chem. 1994;269:14367–14370. [PubMed] [Google Scholar]
- 24.Herman J G, Graff J R, Myohanen S, Nelkin B D, Baylin S B. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bankfalvi A, Navabi H, Bier B, Bocker W, Jasani B, Schmid K W. J Pathol. 1994;174:223–228. doi: 10.1002/path.1711740312. [DOI] [PubMed] [Google Scholar]
- 26.Thomas D C, Umar A, Kunkel T A. Genomethods. 1995;7:187–197. [Google Scholar]
- 27.Bird A. Cell. 1992;70:5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- 28.Hampson R, Humbert O, Macpherson P, Aquilina G, Karran P. J Biol Chem. 1997;272:28596–28606. doi: 10.1074/jbc.272.45.28596. [DOI] [PubMed] [Google Scholar]
- 29.Prowse A H, Webster A R, Richards F M, Richard S, Olschwang S, Resche F, Affara N A, Maher E R. Am J Hum Genet. 1997;60:765–771. [PMC free article] [PubMed] [Google Scholar]
- 30.Ohtani-Fujita N, Dryja T P, Rapaport J M, Fujita T, Matsumura S, Ozasa K, Watanabe Y, Hayashi K, Maeda K, Kinoshita S, et al. Cancer Genet Cytogenet. 1997;98:43–49. doi: 10.1016/s0165-4608(96)00395-0. [DOI] [PubMed] [Google Scholar]
- 31.Rampino N, Yamamoto H, Ionov Y, Li Y, Sawai H, Reed J C, Perucho M. Science. 1997;275:967–969. doi: 10.1126/science.275.5302.967. [DOI] [PubMed] [Google Scholar]