Abstract
Oxygen homeostasis is an essential regulation system for cell energy production and survival. The oxygen-sensitive subunit α of the hypoxia inducible factor-1 (HIF-1) complex is a key protein of this system. In this work, we analyzed mouse and rat HIF-1α protein and mRNA expression in parallel to energetic metabolism variations within skeletal muscle. Two physiological situations were studied using HIF-1α–specific Western blotting and semiquantitative RT-PCR. First, we compared HIF-1α expression between the predominantly oxidative soleus muscle and three predominantly glycolytic muscles. Second, HIF-1α expression was assessed in an energy metabolism switch model that was based on muscle disuse. These two in vivo situations were compared with the in vitro HIF-1α induction by CoCl2 treatment on C2C12 mouse muscle cells. HIF-1α mRNA and protein levels were found to be constitutively higher in the more glycolytic muscles compared with the more oxidative muscles. Our results gave rise to the hypothesis that the oxygen homeostasis regulation system depends on the fiber type.
INTRODUCTION
Eukaryotic cells sense oxygen and adapt to hypoxia by regulating a certain number of genes. This was shown many years ago for various mammalian cell types, including C2C12 myoblasts (Webster, 1987). The HIF-1 (hypoxia inducible factor-1) transcription factor is a key element of this pleiotropic response. Wang and Semenza (1993) observed that HIF-1 DNA binding activity is specifically present under low oxygen conditions for different cell lines, including muscle cells. Activated HIF-1 comprises two subunits, i.e., HIF-1α and HIF-1β. HIF-1α plays a central role as it is subjected to drastic O2-dependent proteasomal control. At normoxia, HIF-1α and HIF-1β are constitutively expressed but HIF-1α is mainly ubiquitinated and degraded by the proteasome (Jaakkola et al., 2001). In severe hypoxia, the HIF-1α protein level increases by relaxing its degradation, and HIF-1α can form an active complex with HIF-1β. Activated HIF-1 induces the expression of genes involved, for example, in angiogenesis, erythropoiesis, glucose uptake, and energy metabolism (Semenza, 2000a; Wenger, 2000). HIF-1α can also be regulated at the mRNA level, but this is less documented than the regulation at protein level (Semenza, 2000b). Induction of HIF-1α mRNA following acute hypoxia was reported in vivo in rodent ferret brain, lung, or kidney (Wiener et al., 1996; Yu et al., 1998) and in cultured cell lines (Wang et al., 1995). Similarly, chronic ischemia was found to be associated with HIF-1α mRNA up-regulation in rat brain (Bergeron et al., 1999). Considering the striated muscle tissue, treatment with phorbol ester was shown to increase rat cardiomyocyte HIF-1α mRNA in tissue culture (Ladoux and Frelin, 1997); increase in the steady-state levels of cardiac HIF-1α mRNA has also been shown to be part of the early response to myocardial ischemia or infarction in humans (Lee et al., 2000) and to occur under high glucose concentrations in rats (Marfella et al., 2002). Several studies have reported an increase of skeletal muscle HIF-1α mRNA in chronic hypoxia conditions such as adaptation to altitude (Hoppeler and Vogt, 2001a,b). Finally, a recent study involving a global analysis by cDNA arrays of transcription profiles in chronic critical and acute-on-chronic human skeletal muscle ischemia conditions demonstrated up-regulation of genes involved in the HIF-1 system, including HIF-1 itself (Tuomisto et al., 2004).
HIF-1 studies in skeletal muscle are difficult due to the presence of a number of energy metabolism systems that include different O2 supplies and homeostasis, with different proportions of oxidative, glycolytic, and intermediate fibers. Indeed, it has been shown that skeletal muscle presents a continuum of mechanical and metabolic properties from the slow contractile speed type I fibers, which have a low fatigability, to the fast contractile speed type IIb fibers, which have a high fatigability. Type I fiber energy is mainly sustained by an oxidative metabolism, while type IIb energy is mainly generated by a glycolytic metabolism (Booth and Baldwin, 1996). Moreover, this metabolic/mechanical status is not fixed, but adapts to environmental changes such as chronic activity modifications. For instance, slow-twitch muscles modify their phenotype from a predominantly oxidative slow- to a glycolytic fast-type metabolism under different physiological or experimental conditions of muscle atrophy induced by restriction of muscle activity (Diffee et al., 1991; Ohira et al., 1992; Cros et al., 2001).
In this study, we investigated HIF-1α mRNA and protein expression in relation with muscular energy metabolism types. Our observations strongly suggest that HIF-1α expression is fiber type dependent.
MATERIALS AND METHODS
Animals and Tissues
All procedures were approved by the local Centre National de la Recherche Scientifique ethics committee. Entire soleus and tibialis anterior, lateral gastrocnemius, and middle region of quadriceps muscles were quickly excised from anesthetized (intraperitoneal injection of pentobarbital, 1 μL/g of body mass) 10 wk-old Ico: OF1 (Caw) mice (Charles Rivers Laboratories), and then cleaned and immediately frozen in liquid nitrogen for RNA and protein immunoblotting analysis. Rat gastrocnemius and soleus muscles atrophied by 2 wk of hindlimb suspension and control muscles were derived from a previous study (Cros et al., 2001).
Cell Culture
C2C12 mouse skeletal muscle cells were obtained from the American Type Culture Collection (ATCC no. CRL-1772), and grown in Dulbecco's modified Eagle's medium with 4.5 g/liter glucose and 10% FCS. Myotube C2C12 differentiation was induced by withdrawing FCS and adding 10 μg/ml insulin, 5 μg/ml transferrin, and 2% horse serum. Cells were stressed by adding 200 μM CoCl2 to the culture medium for indicated lengths of time.
RNA Analysis
Total RNA was isolated using Tri-reagent (Euromedex) according to manufacturer's instructions, from nine-rat or -mouse pool frozen muscles or from three fresh C2C12-culture dishes, to eliminate individual variations. Semi-quantitative RT-PCR (reverse transcriptase using Invitrogen M-MMLV reverse transcriptase and polymerase chain reaction using CLONTECH Laboratories, Inc. Advantage Klen Taq polymerase) cDNA amplification and quantification were performed as described previously (Pisani et al., 2004). In brief, ethidium bromide intensity of agarose electrophoretic bands was quantified with PCBas software in the linear amplification range determined from 15 to 35 PCR cycle amplifications. HIF-1α cDNA fragments were amplified as described by Marfella et al. (2002), and 18S rRNA fragments were amplified as described by Pisani et al. (2004).
Protein Analysis
For immunoblotting analysis, nuclear proteins were extracted from fresh excised skeletal muscle (Ulukan et al., 2001). In brief, muscles were disrupted with a Polytron homogenizer in 5 ml of ice-cold buffer A (100 mM NaCl, 20 mM KCl, 0.5 mM Na2PO4, 20 mM Tris-HCl, pH 7.5), and then centrifuged at 350 g for 10 min. Pellets were resuspended in 2 ml of ice-cold buffer B (100 mM NaCl, 50 mM KCl, 0.1 mM EDTA, 20 mM Tris-HCl, pH 7.5, 10% glycerol, 0.2% NP-40, 0.1% Triton X-100), incubated 10 min on ice and centrifuged at 350 g for 10 min. Nuclei from pellets were lysed with 200 μl of ice-cold buffer B supplemented with 1% SDS and a protease inhibitor cocktail (Sigma-Aldrich). Total soluble proteins were prepared from fresh cell cultures by immediate extraction in ice-cold lysis buffer (50 mM Hepes, pH 7.4, 150 mM NaCl, 100 mM NaF, 10 mM EDTA, 10 mM Na4P2O7, 2 mM Na3VO4, 1% Triton X-100, supplemented with protease inhibitors cocktail) (Yeow et al., 2002). All protein concentrations were determined with the Bio-Rad Laboratories protein assay reagents. Equal amounts of proteins were separated by SDS-PAGE on 6% acrylamide gels. Proteins were transferred to Hybond-C extra membranes (Amersham Biosciences) and stained with ponceau red to check whether the transfers were uniform. Blots were then incubated for 4 h at room temperature in TBS with 0.1% Tween-20 and 5% nonfat powdered milk. Membranes were probed overnight at 4°C with anti-HIF-1α polyclonal antibody (provided by C. Brahimi-Horn and J. Pouysségur, CNRS UMR 6543, Nice, France), at 1:2,000 dilution in TBS, 0.1% Tween-20, 1% nonfat powdered milk, and then for 1 h at room temperature with an HRP-conjugated swine anti-rabbit IgG (Dako) used at 1:2,000 dilution in TBS, 0.1% Tween-20, and 5% nonfat powdered milk. After each incubation, membranes were washed three times in TBS, 0.1% Tween-20, for 10 min per wash. Proteins were then revealed using the Amersham Biosciences ECL system.
Statistical Analysis
t tests were performed using SigmaStat software to compare differences between two groups of values. Statistical differences were noted * (P < 0.05).
RESULTS
Difference in HIF-1α Expression between Oxidative- and Glycolytic-energy Metabolism Muscle Fibers
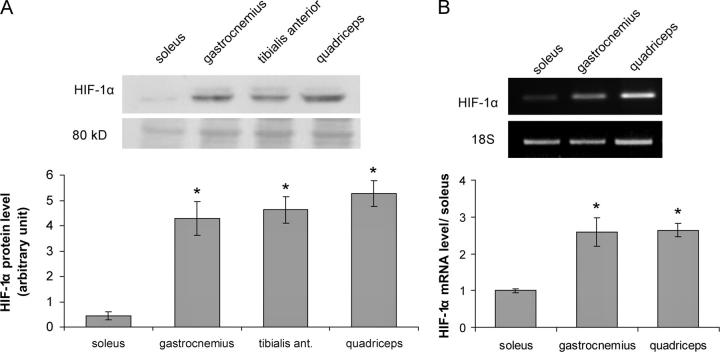
We compared HIF-1α expression between a predominantly oxidative muscle (soleus) and three predominantly glycolytic muscles (gastrocnemius, tibialis anterior, and quadriceps). The analysis was performed on nuclear protein extracts from mouse muscles, which were immediately snap frozen in liquid nitrogen after excision to avoid any modifications induced by tissue hypoxia. With the same goal, the first step of protein or RNA preparations were carefully performed directly using frozen tissues. First, we investigated the HIF-1α protein level by Western blotting (Fig. 1 A). A specific 120-kD signal was detected with a monoclonal anti–HIF-1α antibody on nuclear protein extracts. HIF-1α quantities were reproducibly found to be 8- to 10-fold higher in glycolytic muscles than in the oxidative soleus muscle. Despite the care taken in tissue storage and protein preparation for all muscles, we could not rule out that the drastic difference noted between glycolytic muscles and the oxidative muscle was caused by very short-term hypoxic conditions for glycolytic muscle sample preparations. Therefore, we compared HIF-1α mRNA quantities in the oxidative soleus muscle with gastrocnemius and quadriceps glycolytic muscles by semi-quantitative RT-PCR, since no mRNA variations were expected under such short-term hypoxia. HIF-1α mRNA expression was found to be threefold higher in glycolytic muscles than in the oxidative muscle (Fig. 1 B). The higher HIF-1α protein quantity observed in glycolytic muscles was thus found to be associated with a higher HIF-1α mRNA content.
Figure 1.
Comparison of HIF-1α expression in slow (soleus) versus fast (gastrocnemius, tibialis anterior, quadriceps) mouse skeletal muscles. (A) Protein analysis by Western blotting on skeletal muscle nuclear protein extracts. HIF-1α was detected as a specific 120-kD band, and an 80-kD nonspecific band stained with amido black is shown as an even-lane loading control. Lanes were evenly loaded with 25 μg of proteins. (B) RNA analysis performed by semiquantitative RT-PCR on skeletal muscle RNAs. Partial HIF-1α mRNAs were amplified and quantified and 18S rRNAs were used for normalization. The band intensity was measured, in linear PCR amplification range, and the results are expressed in arbitrary units after normalization. Histograms represent mean ± SEM of three independent experiments from two independent RNA or protein preparations. *, significantly different from soleus muscle (A, P < 0.001; B, P < 0.03).
CoCl2 Stress Induces an HIF-1α Protein Increase but no mRNA Modifications in Differentiated C2C12 Muscle Cells
We wondered whether HIF-1α regulation by hypoxia would also involve mRNA regulation in skeletal muscle, unlike other tissues. CoCl2 is known to mimic acute hypoxia upon in vitro HIF-1 activity induction (Wang et al., 1995). It inhibits HIF-1α ubiquitination and consequently its degradation by the proteasome, thus allowing formation of the active HIF-1α/HIF-1β complex. Myotube-differentiated C2C12 cells were stressed for 3 h with 200 μM CoCl2 treatments. As expected (Wang et al., 1995), Western blot analysis revealed an increase in HIF-1α protein (Fig. 2 A), but no variations in mRNA contents were detected by semiquantitative RT-PCR (Fig. 2, B and C). This was in agreement with the hypothesis that the increase in HIF-1α protein was only due to a decrease in HIF-1α degradation, as very well documented in other tissues (Wenger, 2000).
Figure 2.
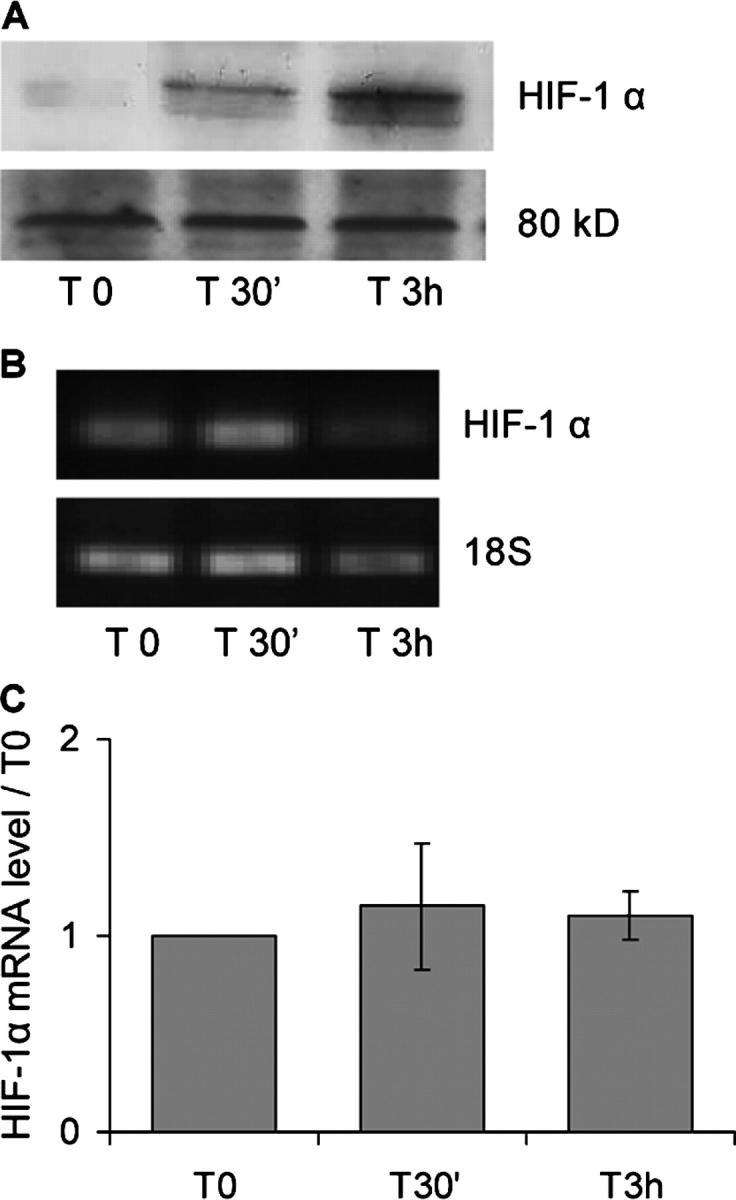
HIF-1α expression in differentiated C2C12 myotubes treated with CoCl2. HIF-1α expression was assessed before treatment (T0) and after 30 min (T30') and 3 h (T 3h) in the presence of 200 μM CoCl2. (A) Total cell proteins were extracted and analyzed by Western blotting using anti–HIF-1α antibody. An HIF-1α–specific 120-kD band was detected, and an 80-kD nonspecific band stained with amido-black is shown as an even-lane loading control. Lanes were evenly loaded with 20 μg of proteins. (B) RNAs were prepared and analyzed by semiquantitative RT-PCR. Primers specific to HIF-1α mRNA (for analysis) and to 18S rRNA (for normalization) were used. Band intensities were measured in the linear amplification range, and the results are expressed as ratios between the different time points and the T0-band intensities after normalization by 18S rRNA variations. The results represent mean ± SEM of three independent experiments.
Variations in HIF-1a mRNA in a Model of Muscle Energy Metabolism Switch
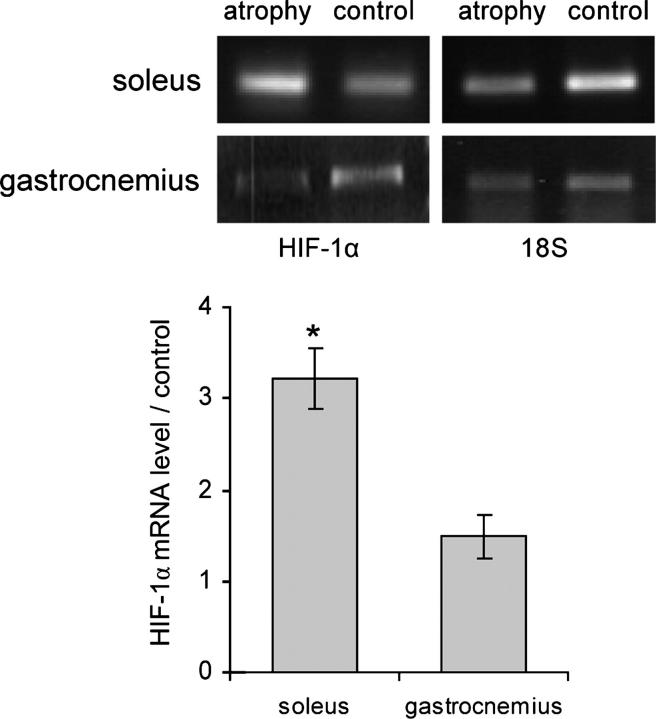
To further test the hypothesis that the HIF-1α protein level differences reported above between oxidative and glycolytic muscles was due to constitutive differences in HIF-1α mRNA quantities between the two kinds of muscles and not to artifactual hypoxic conditions, we analyzed another physiological situation. Instead of comparing different muscles, we used a procedure that induced an energy metabolism switch within the same muscle. Thus, HIF-1α mRNA expression was analyzed by semiquantitative RT-PCR in rat soleus and gastrocnemius muscles submitted to 2 wk of activity restriction by hindlimb suspension (Diffee et al., 1991). The anti-gravity oxidative soleus muscles were >40% atrophied, and ∼20% of the oxidative fibers were replaced by glycolytic fibers, as assessed via the myosin isoform contents that were studied in one of our previous works (Cros et al., 1999). In this situation of oxidative to glycolytic energy metabolism transition within soleus muscle, HIF-1α mRNA was found to be threefold up-regulated (Fig. 3). Conversely, the glycolytic gastrocnemius muscles, which underwent only 25% atrophy, showed no variation in energy metabolism (Jiang et al., 1992; Ohira et al., 1994), and the HIF-1α mRNA content was not significantly modified (Fig. 3).
Figure 3.
Comparison of HIF-1α mRNA expression in soleus and gastrocnemius muscles atrophied by unloading versus control muscles. Total RNAs isolated from rat muscle pools were analyzed by semiquantitative RT-PCR. HIF-1α mRNAs were amplified for analysis and 18S rRNAs were amplified for normalization. Band intensity was measured in the linear amplification range, and the results are expressed in an arbitrary unit after normalization by 18S rRNA variations. Histograms represent mean ± SEM of three independent RT-PCR experiments from two independent RNA preparations. *, significantly different from control (soleus muscle, P < 0.007).
DISCUSSION
Energy for muscle contraction is provided by oxidative and glycolytic metabolisms. Skeletal muscles are made of fibers that differ according to the relative involvement of each of these energy metabolisms. These metabolism differences are associated with different mechanical properties that allow muscles to meet various functional demands. As HIF-1 is a critical sensor of cellular oxygen, we investigated expression of the regulatory HIF-1α subunit in accordance with the oxidative/glycolytic energy balance in skeletal muscle. Variations in this balance naturally occur in different muscles or can be experimentally forced by modifying the mechanical constraints. In this hypoxia-free study, we have shown (a) higher HIF-1α expression in predominantly glycolytic muscles than in the predominantly oxidative soleus muscle, and (b) an HIF-1α expression increase in the oxidative to glycolytic energy transition, which occurs in soleus muscle activity reduction imposed by hindlimb suspension as shown in different previous works including ours (Diffee et al., 1991; Ohira et al., 1992; Cros et al., 2001). These HIF-1α variations were noted at the protein level but also at the mRNA level. The existence of HIF-1α mRNA variations ruled out the possibility of simple hypoxia artifacts in RNA preparations from glycolytic muscles. Indeed, as confirmed in the present work by the experimental CoCl2 treatment of C2C12 myoblasts, muscle cells react like other cells by regulating HIF-1α protein and not HIF-1α mRNA.
It was not surprising to detect HIF-1α expression in normoxic skeletal muscle. Indeed, contrary to most tissues, it has been shown that HIF-1α is present under normoxic conditions in a few mouse tissues, including brain and skeletal muscle, where its level increases in response to systemic hypoxia (Stroka et al., 2001) or ischaemia (Milkiewicz et al., 2004). In the present work, we confirmed these observations, suggesting that HIF-1 could have a potential function in muscle oxygen homeostasis under normoxia. Variations in HIF-1α content according to skeletal muscle fiber types in normoxia conditions should be compared with the myoglobin content. According to our results, oxidative fibers have a low HIF-1α content and are known to express a high level of myoglobin (Mb) (Lin et al., 2002), which is responsible for their red color. Mb is a cytoplasmic hemoprotein able to link O2 in the interstitial compartment to carry it inside the fiber (Brunori, 2001). Mb forms an intracellular oxygen reserve that to a certain extent enables muscular fibers to buffer variations in blood O2 supply (i.e., hypoxia) or to meet fiber metabolism needs. In contrast, glycolytic fibers were shown here to have a high HIF-1α content but they express a low level of Mb (Vogt et al., 2001). In this case, the Mb level is not efficient enough to compensate for alterations in O2 homeostasis (i.e., hypoxia), and the higher HIF-1α content in glycolytic fibers would be necessary to provide satisfactory hypoxia protection for these fibers. Moreover, a balance between HIF-1α and Mb was recently documented in a study of Mb−/− knocked-out mice (Grange et al., 2001). Indeed, these mice were viable and showed significant phenotypic modifications to compensate for the Mb absence, including HIF-1α up-regulation and metabolism transitions from oxidative to glycolytic muscle fibers (Grange et al., 2001). Conversely, a recent study of HIF-1α−/− knocked-out mice showed no variation in fiber type composition in soleus and gastrocnemius muscles (Mason et al., 2004). However, a clear shift from glycolytic to oxidative muscle metabolism was detected during exercise of these modified mice in comparison to wild-type mice. Taken together, these points could be explained by the presence of an O2 regulation system based mainly on Mb in oxidative fibers and on an HIF-1 complex in glycolytic fibers.
Finally, it also should be considered than HIF-1 has a wider role than oxygen sensing. Its expression higher in normoxic glycolytic muscles than in oxidative muscles may also be related to other HIF-1 functions. HIF-1 is involved in many cellular pathways, such as glucose uptake and carbohydrate metabolism. For instance, HIF-1 activated genes include hexokinase 1 and 2, enolase 1, lactate dehydrogenase A, pyruvate kinase, etc. (Semenza, 2001). The proteins encoded by these genes play key roles in energy metabolism. They act at important steps of energy supply process, especially for the anaerobic metabolism of muscle glycolytic fibers. In these fibers, an important pathway required for ATP production is the degradation of endogenous glycogen through the glycolytic pathway to pyruvate and/or lactic acid. HIF-1 also activates glucose transporter 1, one of the glucose transporter isoforms expressed in skeletal muscle (Booth and Baldwin, 1996; Semenza, 2001). This is of special interest since the rate-limiting step for glucose use by muscle is the glucose transport. Altogether, these considerations make very conceivable that higher content of the transcription factor HIF-1 in glycolytic muscles actively contributes to the adequate expression of several genes involved in the energy glycolytic supply.
In conclusion, this work confirms differences in the HIF-1–mediated cellular pathway regulations between normoxic oxidative and glycolytic skeletal muscle fibers. The higher HIF-1α expression observed under the lower O2 supply conditions in glycolytic fibers underlines the importance of HIF-1 for maintaining muscle homeostasis and HIF-1 involvement in muscle plasticity to tailor the metabolism, and therefore the mechanical properties, to meet the functional demand.
Acknowledgments
We thank Dr. Christiane Brahimi-Horn and Dr. Jacques Pouysségur for providing anti–HIF-1α polyclonal antibody, Dr. Annie Ladoux for constructive discussions on the HIF-1 system, Anne-Sophie Coldefy for the statistical analysis and David Manley for the English revision of the manuscript.
D.F. Pisani is a recipient of grants from the Association Française contre les Myopathies and the Fondation pour la Recherche Médicale.
Olaf S. Andersen served as editor.
Abbreviations used in this paper: HIF-1, hypoxia inducible factor-1; Mb, myoglobin.
References
- Bergeron, M., A.Y. Yu, K.E. Solway, G.L. Semenza, and F.R. Sharp. 1999. Induction of hypoxia-inducible factor-1 (HIF-1) and its target genes following focal ischaemia in rat brain. Eur. J. Neurosci. 11:4159–4170. [DOI] [PubMed] [Google Scholar]
- Booth, F.W., and K.M. Baldwin. 1996. Muscle plasticity: energy demand and supply processes. In Handbook of Physiology: Integration of Motor, Circulatory, Respiratory, and Metabolic Control during Exercise. American Physiological Society, Bethesda, MD. 1075–1123.
- Brunori, M. 2001. Nitric oxide, cytochrome-c oxidase and myoglobin. Trends Biochem. Sci. 26:21–23. [DOI] [PubMed] [Google Scholar]
- Cros, N., J. Muller, S. Bouju, G. Pietu, C. Jacquet, J.J. Leger, J.F. Marini, and C.A. Dechesne. 1999. Upregulation of M-creatine kinase and glyceraldehyde3-phosphate dehydrogenase: two markers of muscle disuse. Am. J. Physiol. 276:R308–R316. [DOI] [PubMed] [Google Scholar]
- Cros, N., A.V. Tkatchenko, D.F. Pisani, L. Leclerc, J.J. Leger, J.F. Marini, and C.A. Dechesne. 2001. Analysis of altered gene expression in rat soleus muscle atrophied by disuse. J. Cell. Biochem. 83:508–519. [DOI] [PubMed] [Google Scholar]
- Diffee, G.M., V.J. Caiozzo, R.E. Herrick, and K.M. Baldwin. 1991. Contractile and biochemical properties of rat soleus and plantaris after hindlimb suspension. Am. J. Physiol. 260:C528–C534. [DOI] [PubMed] [Google Scholar]
- Grange, R.W., A. Meeson, E. Chin, K.S. Lau, J.T. Stull, J.M. Shelton, R.S. Williams, and D.J. Garry. 2001. Functional and molecular adaptations in skeletal muscle of myoglobin-mutant mice. Am. J. Physiol. Cell Physiol. 281:C1487–C1494. [DOI] [PubMed] [Google Scholar]
- Hoppeler, H., and M. Vogt. 2001. a. Hypoxia training for sea-level performance. Training high-living low. Adv. Exp. Med. Biol. 502:61–73. [DOI] [PubMed] [Google Scholar]
- Hoppeler, H., and M. Vogt. 2001. b. Muscle tissue adaptations to hypoxia. J. Exp. Biol. 204:3133–3139. [DOI] [PubMed] [Google Scholar]
- Jaakkola, P., D.R. Mole, Y.M. Tian, M.I. Wilson, J. Gielbert, S.J. Gaskell, A. Kriegsheim, H.F. Hebestreit, M. Mukherji, C.J. Schofield, et al. 2001. Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 292:468–472. [DOI] [PubMed] [Google Scholar]
- Jiang, B., Y. Ohira, R.R. Roy, Q. Nguyen, E.I. Ilyina-Kakueva, V. Oganov, and V.R. Edgerton. 1992. Adaptation of fibers in fast-twitch muscles of rats to spaceflight and hindlimb suspension. J. Appl. Physiol. 73:58S–65S. [DOI] [PubMed] [Google Scholar]
- Ladoux, A., and C. Frelin. 1997. Cardiac expressions of HIF-1α and HLF/EPAS, two basic loop helix/PAS domain transcription factors involved in adaptative responses to hypoxic stresses. Biochem. Biophys. Res. Commun. 240:552–556. [DOI] [PubMed] [Google Scholar]
- Lee, S.H., P.L. Wolf, R. Escudero, R. Deutsch, S.W. Jamieson, and P.A. Thistlethwaite. 2000. Early expression of angiogenesis factors in acute myocardial ischemia and infarction. N. Engl. J. Med. 342:626–633. [DOI] [PubMed] [Google Scholar]
- Lin, J., H. Wu, P.T. Tarr, C.Y. Zhang, Z. Wu, O. Boss, L.F. Michael, P. Puigserver, E. Isotani, E.N. Olson, et al. 2002. Transcriptional co-activator PGC-1α drives the formation of slow-twitch muscle fibres. Nature. 418:797–801. [DOI] [PubMed] [Google Scholar]
- Marfella, R., M. D'Amico, C. Di Filippo, E. Piegari, F. Nappo, K. Esposito, L. Berrino, F. Rossi, and D. Giugliano. 2002. Myocardial infarction in diabetic rats: role of hyperglycaemia on infarct size and early expression of hypoxia-inducible factor 1. Diabetologia. 45:1172–1181. [DOI] [PubMed] [Google Scholar]
- Mason, S.D., R.A. Howlett, M.J. Kim, I.M. Olfert, M.C. Hogan, W. McNulty, R.P. Hickey, P.D. Wagner, C.R. Kahn, F.J. Giordano, and R.S. Johnson. 2004. Loss of skeletal muscle HIF-1α results in altered exercise endurance. PLoS Biol. 2:e288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkiewicz, M., C.W. Pugh, and S. Egginton. 2004. Inhibition of endogenous HIF inactivation induces angiogenesis in ischaemic skeletal muscles of mice. J. Physiol. 560:21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohira, Y., B. Jiang, R.R. Roy, V. Oganov, E. Ilyina-Kakueva, J.F. Marini, and V.R. Edgerton. 1992. Rat soleus muscle fiber responses to 14 days of spaceflight and hindlimb suspension. J. Appl. Physiol. 73:51S–57S. [DOI] [PubMed] [Google Scholar]
- Ohira, Y., W. Yasui, F. Kariya, T. Wakatsuki, K. Nakamura, T. Asakura, and V.R. Edgerton. 1994. Metabolic adaptation of skeletal muscles to gravitational unloading. Acta Astronaut. 33:113–117. [DOI] [PubMed] [Google Scholar]
- Pisani, D.F., P.M. Pierson, A. Massoudi, L. Leclerc, A. Chopard, J.F. Marini, and C.A. Dechesne. 2004. Myodulin is a novel potential angiogenic factor in skeletal muscle. Exp. Cell Res. 292:40–50. [DOI] [PubMed] [Google Scholar]
- Semenza, G.L. 2000. a. Oxygen-regulated transcription factors and their role in pulmonary disease. Respir. Res. 1:159–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza, G.L. 2000. b. HIF-1: mediator of physiological and pathophysiological responses to hypoxia. J. Appl. Physiol. 88:1474–1480. [DOI] [PubMed] [Google Scholar]
- Semenza, G.L. 2001. Hypoxia-inducible factor 1: oxygen homeostasis and disease pathophysiology. Trends Mol. Med. 7:345–350. [DOI] [PubMed] [Google Scholar]
- Stroka, D.M., T. Burkhardt, I. Desbaillets, R.H. Wenger, D.A. Neil, C. Bauer, M. Gassmann, and D. Candinas. 2001. HIF-1 is expressed in normoxic tissue and displays an organ-specific regulation under systemic hypoxia. FASEB J. 15:2445–2453. [DOI] [PubMed] [Google Scholar]
- Tuomisto, T.T., T.T. Rissanen, I. Vajanto, A. Korkeela, J. Rutanen, and S. Yla-Herttuala. 2004. HIF-VEGF-VEGFR-2, TNF-α and IGF pathways are upregulated in critical human skeletal muscle ischemia as studied with DNA array. Atherosclerosis. 174:111–120. [DOI] [PubMed] [Google Scholar]
- Ulukan, H., M.T. Muller, and P.W. Swaan. 2001. Downregulation of topoisomerase I in differentiating human intestinal epithelial cells. Int. J. Cancer. 94:200–207. [DOI] [PubMed] [Google Scholar]
- Vogt, M., A. Puntschart, J. Geiser, C. Zuleger, R. Billeter, and H. Hoppeler. 2001. Molecular adaptations in human skeletal muscle to endurance training under simulated hypoxic conditions. J. Appl. Physiol. 91:173–182. [DOI] [PubMed] [Google Scholar]
- Wang, G.L., B.H. Jiang, E.A. Rue, and G.L. Semenza. 1995. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. USA. 92:5510–5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G.L., and G.L. Semenza. 1993. General involvement of hypoxia-inducible factor 1 in transcriptional response to hypoxia. Proc. Natl. Acad. Sci. USA. 90:4304–4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster, K.A. 1987. Regulation of glycolytic enzyme RNA transcriptional rates by oxygen availability in skeletal muscle cells. Mol. Cell. Biochem. 77:19–28. [DOI] [PubMed] [Google Scholar]
- Wenger, R.H. 2000. Mammalian oxygen sensing, signalling and gene regulation. J. Exp. Biol. 203:1253–1263. [DOI] [PubMed] [Google Scholar]
- Wiener, C.M., G. Booth, and G.L. Semenza. 1996. In vivo expression of mRNAs encoding hypoxia-inducible factor 1. Biochem. Biophys. Res. Commun. 225:485–488. [DOI] [PubMed] [Google Scholar]
- Yeow, K., C. Cabane, L. Turchi, G. Ponzio, and B. Derijard. 2002. Increased MAPK signaling during in vitro muscle wounding. Biochem. Biophys. Res. Commun. 293:112–119. [DOI] [PubMed] [Google Scholar]
- Yu, A.Y., M.G. Frid, L.A. Shimoda, C.M. Wiener, K. Stenmark, and G.L. Semenza. 1998. Temporal, spatial, and oxygen-regulated expression of hypoxia-inducible factor-1 in the lung. Am. J. Physiol. 275:L818–L826. [DOI] [PubMed] [Google Scholar]