Abstract
Gln3 and Gat1/Nil1 are GATA-family transcription factors responsible for transcription of nitrogen catabolic genes in S. cerevisiae. Intracellular Gln3 localization and Gln3-dependent transcription respond in parallel to the nutritional environment and inhibitors of Tor1/2 (rapamycin) and glutamine synthetase (methionine sulfoximine, MSX). However, detectable Gln3 phosphorylation, though influenced by nutrients and inhibitors, does not correlate with Gln3 localization or NCR-sensitive transcription in a consistent way. Little such data are available for Gat1. To eliminate this gap and establish relationships between Gln3 and Gat1 regulation, we performed experiments parallel to those we previously reported for Gln3. Gat1 and Gln3 localization are similar during steady state growth, being cytoplasmic and nuclear with good and poor nitrogen sources, respectively. Localization correlates with Gat1- and Gln3-mediated transcription. In contrast, three characteristics of Gat1 and Gln3 differ significantly: (i) the kinetics of their localization in response to nutritional transitions and rapamycin-treatment, (ii) their opposite responses to MSX-treatment, i.e., cytoplasmic Gln3 becomes nuclear following MSX addition, while nuclear Gat1 becomes cytoplasmic, and (iii) their phosphorylation levels in the above situations. In instances where Gln3 phosphorylation can be straightforwardly demonstrated to change, Gat1 phosphorylation (in the same samples) appears invariant. The only exception was following carbon starvation where Gat1, like Gln3, is hyperphosphorylated in a Snf1-dependent manner. However, neither carbon-starvation, nor MSX-treatment elicits Snf1-independent Gat1 hyperphosphorylation as observed for Gln3.
Introduction
An ability to respond appropriately to changing environments is critical to the survival of free living organisms. Studies in Saccharomyces cerevisiae have contributed much to our understanding of how environmental signals are sensed in eucaryotic cells and identified many of the components in signal transduction pathways responding to them. The major challenge at present is to elucidate the precise structures of such signal transduction pathways and how their component branches interact in a complex web of control that regulates gene expression and protein activity. Regulated transcription of the nitrogen catabolic genes is being increasingly used as a sensitive reporter for the Tor1/2 signal transduction pathway, which is broadly influenced by a cell’s nutritional status and regulates many processes required for cell division in response to it (Beck and Hall, 1999; Crespo et al., 2002; Bertram et al., 2002). The GATA-family transcription activators, Gln3 and Gat1/Nil1, are required for transcription of these genes in response to nitrogen source availability, a regulatory process designated nitrogen catabolite repression (NCR) (Hoffman-Bang, 1999; ter Schure et al., 2000; Cooper, 1996; Cooper, 2002). When supplied with nitrogen sources supporting rapid growth (glutamine or in some strains, ammonia), genes required for the transport and degradation of poorer ones (proline) are expressed at minimal levels. In the absence of or when only limited supplies of good nitrogen sources are available, transcription of the NCR-sensitive genes increases markedly thereby permitting a cell to exploit a wide variety of poor nitrogen sources that might be available (Hoffman-Bang, 1999; ter Schure et al., 2000; Cooper, 1996; Cooper, 2002).
Intracellular localization of Gln3 and Gat1 is a central mechanism through which NCR-sensitive transcription is regulated. Both proteins are nuclear and bound to GATAAG-containing promoter elements when transcription is high (Cox et al., 2000). In contrast, when NCR-sensitive transcription is low, the GATA elements are free of transcriptional activator proteins with Gln3 and Gat1 being situated in the cytoplasm (Cox et al., 2000). The observation that treating cells with rapamycin (a specific inhibitor of Tor proteins from yeast to humans) elicits nuclear localization of Gln3 and increased transcription of NCR-sensitive genes in cells cultured in rich nitrogen sources implicated the Tor signal transduction pathway in NCR-sensitive transcriptional regulation (Beck and Hall, 1999; Cardenas et al., 1999; Hardwick et al. 1999; Bertram et al., 2000). This implication was extended by the finding that mutations in components of the Tor regulatory pathway altered rapamycin-induced nuclear accumulation of Gln3 (Shamji et al., 2000; Jacinto et al., 2001; Carvalho et al., 2001; see Ref. Thomas et al., 2004 for a collection of reviews on Tor and rapamycin). Since rapamycin-treatment also resulted in dephosphorylation of Gln3, relative to untreated cells, Tormediated control of Gln3 phosphorylation levels was suggested as the mechanism of regulating Gln3 intracellular localization (Beck and Hall, 1999; Bertram et al., 2000). Furthermore, glutamine or one of its metabolites has been suggested to be the nitrogen-related signal to which Tor1/2 respond, because like rapamycin treatment, incubating cells with the glutamine synthetase inhibitor, L-methionine sulfoximine (MSX), elicited Gln3 dephosphorylation and nuclear localization (Crespo et al., 2002).
Much more remains to be learned about the relationship between Gln3 phosphorylation and intracellular localization, however, because emerging results are difficult to rectify with the above model describing Tor regulation of Gln3 (Cooper, 2004; Di Como and Arndt, 1996; Jiang and Broach, 1999; Wang et al., 2003; Cox et al., 20003). The tight association between the nitrogen source provided and NCR-sensitive transcription led us to assess the correlation between these parameters, Gln3 phosphorylation and intracellular localization. Although Gln3 localization correlates well with NCR-sensitive transcription, neither characteristic correlates with Gln3 dephosphorylation levels (Cox et al., 20003). A similar lack of correlation was observed between Gln3 localization following carbon and nitrogen starvation, and the dephosphorylated form of Gln3. In fact, the only time Gln3 dephosphorylation and localization correlate is shortly (30 mins.) after the onset of rapamycin-treatment of glutamine-grown cells; at longer times this correlation disappears as well (Cox et al., 20003). In addition, a recent study of Gln3 phosphorylation and localization, following MSX-treatment, carbon and nitrogen starvation, reported that MSX- and rapamycin-treatment both elicit nuclear Gln3 accumulation, but generate opposite responses with respect to Gln3 phosphorylation. Rapamycin-treatment decreased Gln3 phosphorylation levels, while MSX-treatment increased it (Tate et al., 2005). Further, there were at least two sources of Gln3 phosphorylation following carbon-starvation, one is Snf1-dependent and the other Snf1-independent. Snf1 is a well-studied protein kinase that regulates carbon-responsive gene expression (Tate et al., 2005). Increased Gln3 phosphorylation in response to MSX-treatment is Snf1-independent and exhibits a profile identical to that seen in a carbon-starved snf1Δ (Tate et al., 2005).
Although Gln3 and Gat1 are both, in varying ratios, required for transcription of most NCR-sensitive genes (Coffman et al., 1996; Kuruvilla et al., 2001), much less is known about Gat1. Gat1, like Gln3 is a GATA-family transcriptional activator, but its production is regulated quite differently from Gln3. Unlike Gln3, GAT1 transcription is Gln3- and Gat1-dependent, Dal80-regulated, and NCR-sensitive (Coffman et al., 1996; Rowen et al., 1997; Cunningham et al., 2000). GAT1 expression is not as NCR-sensitive as many nitrogen catabolic genes, which is consistent with the fact that GAT1 transcription is activated by promoter elements beyond those containing GATA sequences (Coffman et al., 1996; R. Andhara and T.G. Cooper, unpublished observations). In contrast to its own production, Gat1 mediates gene expression that is more highly NCR-sensitive (Coffman et al., 1996). In agreement with this observation, Gat1, like Gln3, is nuclear during times of high NCR-sensitive transcription (Coffman et al., 1996), in rapamycin-treated (Crespo et al., 2002), ure2Δ and npr1Δ cells (Crespo et al., 2004), the latter with ammonia as nitrogen source. However, unlike Gln3, Gat1 localization is reported to be unaffected when cells are treated with MSX.
In contrast to the above reports of Gat1 localization, there are no published data depicting Gat1 phosphorylation levels. The conclusions that Gat1 phosphorylation and localization are similar to those of Gln3 appear in the initial report of rapamycin-induced Gln3 dephosphorylation, but are based on data not shown (Beck and Hall, 1999). Given differenes in transcriptional regulation of GAT1 and GLN3, i.e., (i) the fact that the ratios of Gln3:Gat1 required for transcription are gene-specific, and (ii) proposals (based on transcriptome analyses) that some Gat1 functions are different from those of Gln3, we were prompted to characterize and correlate Gat1 phosphorylation and localization in a manner parallel to that we performed with Gln3 (Cox et al., 20003).
The data obtained suggest Gat1-Myc13 intracellular localization responds similarly to that of Gln3-Myc13 during steady state growth in good vs. poor nitrogen sources, but quite differently during nutritional transitions. Gat1-Myc13 responses to such transitions were more rapid than those of Gln3-Myc13 exhibiting a rapid recovery from the perturbation similar to that seen in many stress responses. Moreover, treating cells with MSX caused opposite effects on Gat1-Myc13 and Gln3-Myc13 localization. MSX-treatment elicited Gln3-Myc13 accumulation in the nuclei of nearly all cells, whereas Gat1-Myc13 became cytoplasmic in nearly all cells. Finally, Gat1-Myc13 could not be demonstrated to be phosphorylated (even in the same samples) under any of the conditions, for example, following MSX-treatment, where such phosphorylation could be straightforwardly demonstrated for Gln3-Myc13. The notable exception occurred during carbon-starvation, where Gat1 phosphorylation increased, just as that of Gln3, in a Snf1-dependent manner. However, the Snf1-independent component of carbon-starvation-induced Gln3-Myc13 phosphorylation could not be demonstrated in Gat1-Myc13.
Materials and Methods
Strains and growth conditions
Saccharomyces cerevisiae strains used in this work (Table I) were grown to midlog phase (A600 = 0.4 to 0.6) in YNB medium containing 0.1% glutamine, ammonium sulfate or proline as sole nitrogen source. Appropriate auxotrophic supplements were added where needed: uracil 20 mg/L, histidine 20 mg/L, arginine 20 mg/L, lysine 40 mg/L, leucine 120 mg/L, tryptophan 20 mg/L). Where indicated, rapamycin (dissolved in a solution of 10% Tween20:90% ethanol) or MSX (dissolved in water) was added to a final concentration of 200 ng/ml or 2 mM, respectively. Unless indicated otherwise, cultures were treated with rapamycin or MSX for 30 min at 30°C. Transfer of cultures to nitrogen or carbon starvation media were as described earlier (Cox et al., 20003, Tate et al., 2005).
Table I.
S. cerevisiae strains used in this work
| Strain | Genotype |
|---|---|
| TCY1 | MATα lys2 ura3 |
| yKHC2 | MATα lys2 ura3 gln3Δ::hisG |
| YKHC7 | MATα lys2 ura3 gln3Δ::hisG gat1Δ::hisG |
| RJ715 | MATα lys2 ura3 gat1Δ::hisG |
| TB123 | MATa leu2–3,112, ura3–52,rme1,trp1,his4,GAL+, HMLa, GLN3-Myc13 [KANmx |
| TB106-2a | MATa leu2–3,112, ura3–52,rme1,trp1,his4,GAL+, HMLa, GAT1- HA[KANmx] |
| RR183 | MATa leu2–3,112, ura3–52,rme1,trp1,his4,GAL+, HMLa, GAT1- HA[KANmx], snf1Δ::TRP1 |
Construction and validation of pGAT1-Myc
pRS316, digested with HindIII and NotI, was used as the CEN-vector backbone for construction of pGAT1-Myc. A 775 bp HindIII-PacI fragment from pFA6a (Longtine et al., 1998), containing a Myc13 repeat and the ADH1 transcriptional terminator, was cloned into pRS316 to yield pKA-Myc/pKA61. A DNA fragment containing wild type GAT1, including 675 bp upstream of the start codon, as well as NotI and HindIII sites at its 5′ and 3′ termini was prepared using PCR amplification with TCY1 genomic DNA as template. Primers used in these constructions were 5′-TTCCAAGCTTATATTACCCTGTTATCCCTA-3′, 5′-GTCGACGGATCCCCGGGTTAATTAAC-3′, 5′-ATATGAATGCGGCCGCGTGCATTAACCACAAGTACTGCGT-3′, and 5′-CGCGGATCCGTAAATTCAGATTCAACCAATCCAGGCTCAG-3′. The PCR-generated DNA fragment was cloned into pKA61, digested with NotI and HindIII, to yield pKA62, also designated pGAT1-Myc). The construction was verified by DNA sequence analysis.
Northern blot analysis
RNA was isolated by the method of Schmitt et al. (1990). Northern blots were performed as described earlier (Cox et al., 1999).
Western blot analysis
Cells were harvested by centrifugation, extracts prepared and resolved by electrophoresis, transferred to nitrocellulose membranes and Gat1-Myc13 or Gln3-Myc13 visualized as described earlier (Cox et al., 2002). Similarly, phosphatase treatment of cell extracts were performed as described (Cox et al., 20003).
Indirect immunofluorescence imaging
Methods used for indirect immunofluorescence imaging of Gat1-Myc13 were the same as those reported earlier for Gln3-Myc13 (Cox et al., 20003, Cox et al., 2002). The percentage of cells containing nuclear vs. cytoplasmic Gat1-Myc13 or Gln3-Myc13 was determined by double-blind counting of five or more random fields, accumulating 100 or more cells as described earlier (Cox et al., 20003, Cox et al., 2002). Values observed for duplicate samples of transformants grown on different days varied by 10% or less.
Results
We initiated investigation of Gat1 intracellular localization and phosphorylation, by preparing a GAT1-MYC13 construct in a CEN-based plasmid (pGAT1-Myc), permitting us to move the tagged gene conveniently into various strains. To be useful, it was necessary that expression of GAT1-MYC13 and genes whose expression depends on Gat1 retain their normal sensitivity to NCR. This was assessed by transforming pGAT1-Myc (or the parental vector) into gat1Δ, gln3Δ gat1Δ and gln3Δ strains and comparing the expression profiles of GAT1-Myc13 and representative reporter genes to those observed in an untransformed wild type.
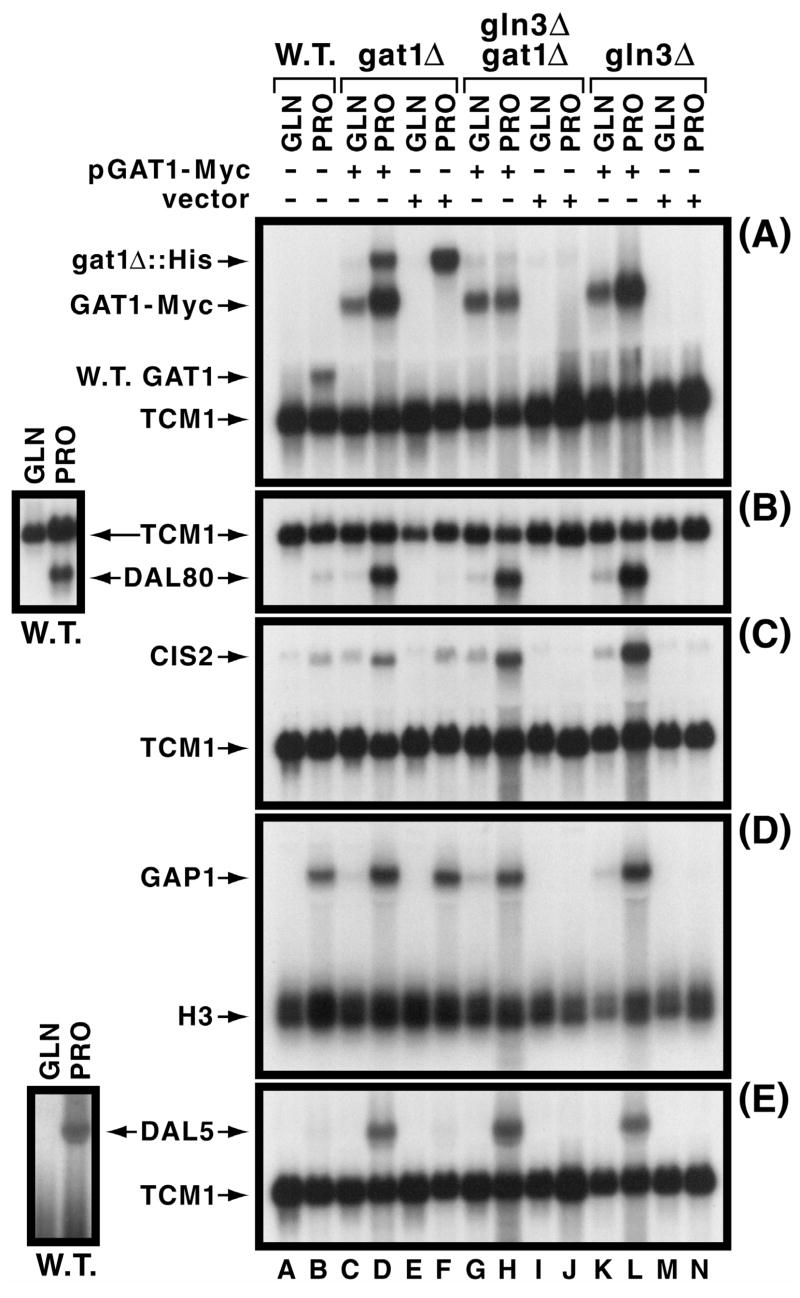
NCR-sensitivity of gat1Δ::His-URA3-His (mRNA from the deletion/insertion of wild type GAT1) and GAT1-MYC13 expression in the transformed gat1Δ strain (used for most subsequent experiments) was similar to that of GAT1 expression in the untransformed wild type (Fig. 1A, lanes A-F). NCR-sensitivity was largely lost in the gat1Δgln3Δ double mutant (Fig. 1A, lanes G and H), a characteristic of GAT1 basal level expression reported earlier (Coffman et al., 1996). In the gln3Δ, containing both chromosomal and plamid copies of GAT1, NCR-sensitivity of GAT1 expression was once again observed (Fig. 1A, lanes K and L). This is consistent with the observation that when expressed at sufficiently high levels, GAT1 is able to partially suppress the phenotype of a gln3Δ (Cunningham et al., 2000; Cunningham et al., 2000).
Fig. 1.
Characterization of NCR-sensitive gene expression supported by Gat1-Myc13. Panel A. GAT1 and GAT1-MYC13 expression supported by pGAT1-Myc (pKA62) or vector (pKA61) as indicated. Untransformed wild type (TCY1) and gat1Δ [(RJ715) (lanes C, D, E and F)], gln3Δ gat1Δ [(yKHC7) (lanes G, H, I and J)] or gln3Δ [(yKHC2) (lanes K, L, M and N)] transformed with pKA62 or pKA61 were grown to mid-log phase in YNB medium with either 0.1% glutamine (GLN) or proline (PRO). A GAT1-specific probe was used to detect expression of GAT1-MYC, with the TCM1 probe being used to assess uniform loading and transfer efficiencies. Panels B–E. Assessment of pGAT1-Myc’s ability to support NCR-sensitive expression of DAL80, CIS2, GAP1 or DAL5. Strains and conditions were as in Panel A. TCM1 and H3 were used as in Panel A. Small images left of the main figure depict a second experiment characterizing wild type NCR-sensitive DAL80 and DAL5 expression. This was done because proper exposure of the weak signals for these two genes would have resulted in over-exposure of the other species in the figure to the point of being impossible to critically assess NCR-sensitivity. A similarly longer exposure would likely also be required to demonstrate GAP1 expression that occurs in proline-grown gln3Δ cultures (lane N).
The only significant difference between untransformed and transformed strains was an overall greater level of expression. Increased GAT1-Myc13 expression in proline-grown gln3Δ and gat1Δ strains is mediated by the Gat1 and Gln3 transcription factors. The GAT1 promoter, containing multiple functional GATAA sequences (R. Andhara and T.G. Cooper, unpublished observations) which bind Gln3 and Dal80 (Coffman et al., 1996; Coffman et al., 1997), is autogenously regulated in addition to regulation imposed by the other GATA-family transcription factors (Coffman et al., 1996; Coffman et al., 1997; Soussi-Boudekou et al., 1997). Transcription factors supporting GAT1-Myc13 expression in glutamine-grown gln3Δ and gln3Δgat1Δ mutants isn’t known. The UAS elements responsible may be situated in the plasmid upstream of the GAT1-MYC13 insert or alternatively may have derived from other UAS elements in the cloned native GAT1 promoter itself. In this regard, it is pertinent that the GAT1 promoter contains multiple STRE-homologous sequences whose deletion as part of a 45 bp DNA fragment decreases GAT1 expression in vivo under some circumstances (R. Andhare and T.G. Cooper, unpublished observations).
Similarly, DAL80, CIS2, GAP1 and DAL5 expression used as reporters of native physiological regulation being supported by the tagged Gat1 protein were normally NCR-sensitive in pGAT1-Myc transformants of the gat1Δ and dependent upon presence of the plasmid (Fig. 1B–E, lanes A–F). The main difference observed between untransformed wild type and pGAT1-Myc transformed strains was again an overall increased level of gene expression which, notwithstanding, remained highly NCR sensitive.
Increased NCR-reporter gene expression likely derived from moderately increased GAT1-MYC13 expression supported by the plasmid-borne GAT1-MYC13 gene. As previously characterized in detail (Cunningham et al., 2000), overproduction of Gat1 will increase expression of GATA-factor dependent genes and partially suppress the gln3Δ phenotype, but NCR-sensitivity is not adversely affected unless GAT1 is expressed at very high levels under control of the GAL1,10 promoter. Even then, NCR-sensitivity is only diminished rather than eliminated as long as URE2 remains functional (Cunningham et al., 2000).
Together, these data indicate GAT1-MYC effectively complements a gat1Δ and supports NCR-sensitive expression of reporter genes for which Gat1 serves as a transcriptional activator. They also point to the conclusion that merely increasing the level of GAT1 expression is alone insufficient to increase its ability to support gene expression. Otherwise, much higher DAL80, CIS2, GAP1 and DAL5 expression should have occurred in glutamine medium. This however, did not occur (Fig 1 B–E, lanes C, G, and K).
Intracellular localization and phosphorylation of Gat1-Myc13 in various nitrogen sources
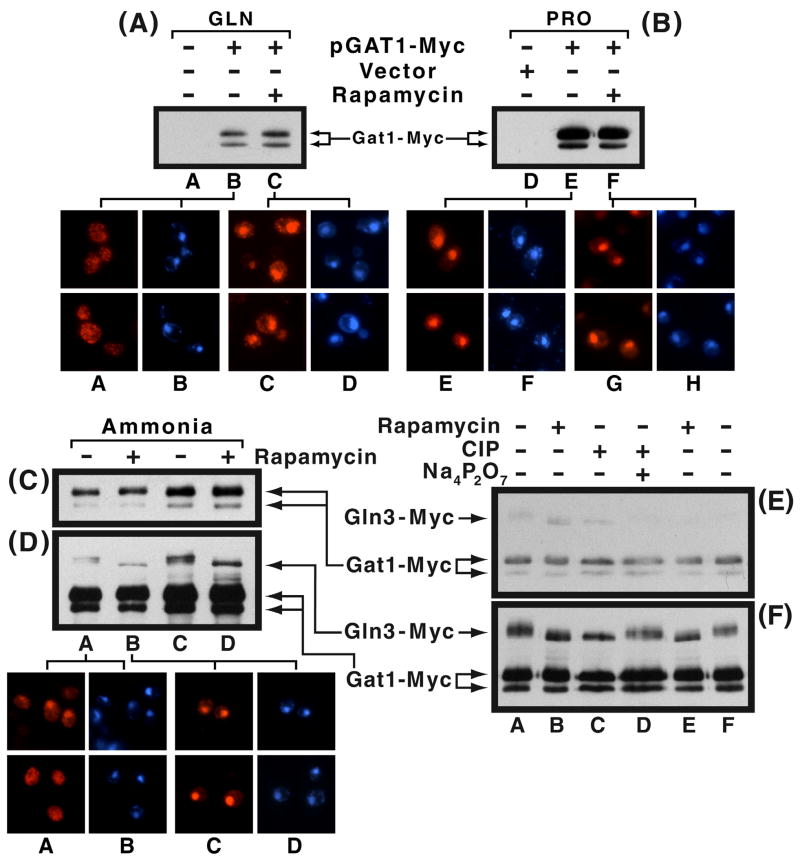
Intracellular localization of Gat1-Myc13 is cytoplasmic in minimal glutamine medium, moderately cytoplasmic with ammonia, and largely nuclear with proline as nitrogen sources (Fig. 2A–D, microscopic images). In proline- and ammonia-grown rapamycin-treated cells, Gat1-Myc13 was nuclear. Glutamine-grown cells gave the least response, but even in this instance the fraction of cells with nuclear Gat1-Myc13 increased to more than half (Fig. 2A–C, microscopic images). These responses are similar to those observed with Gln3-Myc13 (Cox et al., 20003). They are also in agreement with an earlier report of Gat1 distribution in ammonia-containing medium (Crespo et al., 2002), and clearly those expected of a transcription factor whose localization and ability to function are NCR-sensitive. To assess the correlation between Gat1-Myc13 phosphorylation and intracellular localization, we performed Western blot analyses under the same conditions used for immunofluorescence imaging. As shown in Fig. 2A–C, the Gat1-Myc13 mobility profile was the same irrespective of the nitrogen source provided, the presence or absence of rapamycin, or the intracellular localization of Gat1-Myc13 protein. In most preparations, there were two Gat1-Myc13 species with the minor one exhibiting greater mobility and the most preparation-to-preparation variability in the amount of it observed. In some preparations there were two such minor species.
Fig. 2.
Panels A and B. Comparison of Gat1-Myc13 phosphorylation and intracellular localization. RJ715 (gat1Δ) was transformed with pGAT1-Myc (pKA62) or vector (pKA61). Transformants were grown to mid-log phase in YNB-glutamine (lanes A–C) or proline (lanes D–F) medium. The cultures were divided into two portions: one remained untreated, while the other received rapamycin (30 min., 200 ng/ml). Western blot analysis and indirect immunofluorescence were used to determine phosphorylation and intracellular localization of Gat1-Myc13. Gat1-Myc13 and DAPI-positive material appear in red and blue, respectively. Panels C and D. Comparison of Gln3-Myc13 and Gat1-Myc13 phosphorylation in YNB-ammonia-grown cells incubated in the presence or absence of rapamycin (30 min.). TB123 was transformed with pGAT1-Myc13. Therefore, transformants contained both Gln3-Myc13 and Gat1-Myc13. Extracts were prepared from the transformed cultures. 2μg and 3 μg of extract protein were loaded into Panels C and D, lanes A, B and C, D, respectively. The film in panel D is a longer exposure of the one in panel C. This was necessitated because the Gln3-Myc13 signal is weaker than that of Gat1-Myc13. The weak bands between the Gln3-Myc13 and Gat1-Myc13 species in Panel D are Gln3-Myc13 degradation products (independently verified, data not shown). RJ715 transformed with pGAT1-Myc was used to prepare the microscopic images of ammonia-grown cells using the same conditions as the Western blots above. Gat1-Myc13 and DAPI-positive material appear as red and blue, respectively. Panels E and F. Effect of calf intestine alkaline phosphatase (CIP, 50 U) on the migration of Gln3-Myc13 and Gat1-Myc13 in extracts derived from untreated and rapamycin-treated ammonia-grown cells (TB123 transformed with pGAT1-Myc). Short and long film exposures appear in Panels E and F, respectively. The phosphatase inhibitor, sodium pyrophosphate (Na4P2O7, 40 mM) was added to the extract where indicated.
The unexpected failure of Gat1-Myc13 mobility to increase following rapamycin addition, prompted us to question whether Gat1-Myc13 was artifactually dephosphorylated in vitro during extract preparation. To test this possibility, TB123, containing Gln3-Myc13, was transformed with pGAT1-Myc and grown in minimal-ammonia medium. Therefore, transformants contained two Myc13-tagged proteins, permitting us to compare phosphorylation of the two molecules directly in the same samples. This was possible because the molecular weights of Gln3-Myc13 and Gat1-Myc13, calculated from their amino acid sequences are 107 and 84 kDa, respectively. Mobility of Gln3-Myc13 increased in response to rapamycin addition, whereas that of Gat1-Myc13 did not (Fig. 2E and 2F, lanes A vs. B and F vs. E). [Fig. 2F is a longer exposure of Fig. 2E required to visualize the weaker Gln3-Myc13 signal. This was done because subtle changes in mobility are easily masked by the “blooming” that occurs with high-level signals.]
A similar comparison of Gat1-Myc13 and Gln3-Myc13 mobilities was made using extracts treated with calf intestine alkaline phosphatase (CIP). Gln3-Myc13 mobility increased in CIP-treated cells as occurred following rapamycin-treatment (Fig. 2F, lanes A–C). This increase was prevented by adding the phosphatase inhibitor, sodium pyrophosphate, to the extract (Fig. 2F, lane D). In contrast, Gat1-Myc13 mobility was not affected by phosphatase treatment or the phosphatase inhibitor (Fig. 2E and F, lanes A, C and D). The fact that phosphatase treatment did not alter migration rates of the two Gat1-Myc13 species or their ratio to one another, supports the suggestion that the minor species derives from something other than phosphorylation, most likely in vitro proteolysis. Together, these data argued that Gat1-Myc13 could not be demonstrated to exhibit changes in its phosphorylation profile related to either the nitrogen source provided or to the presence or absence of rapamycin under conditions in which such changes were clearly visible for Gln3-Myc13. The Gln3-Myc13 data also demonstrate that Ure2 and NCR-sensitive control operates normally in the transformed strain.
Gat1-Myc13 phosphorylation and intracellular localization during nitrogen and carbon starvation
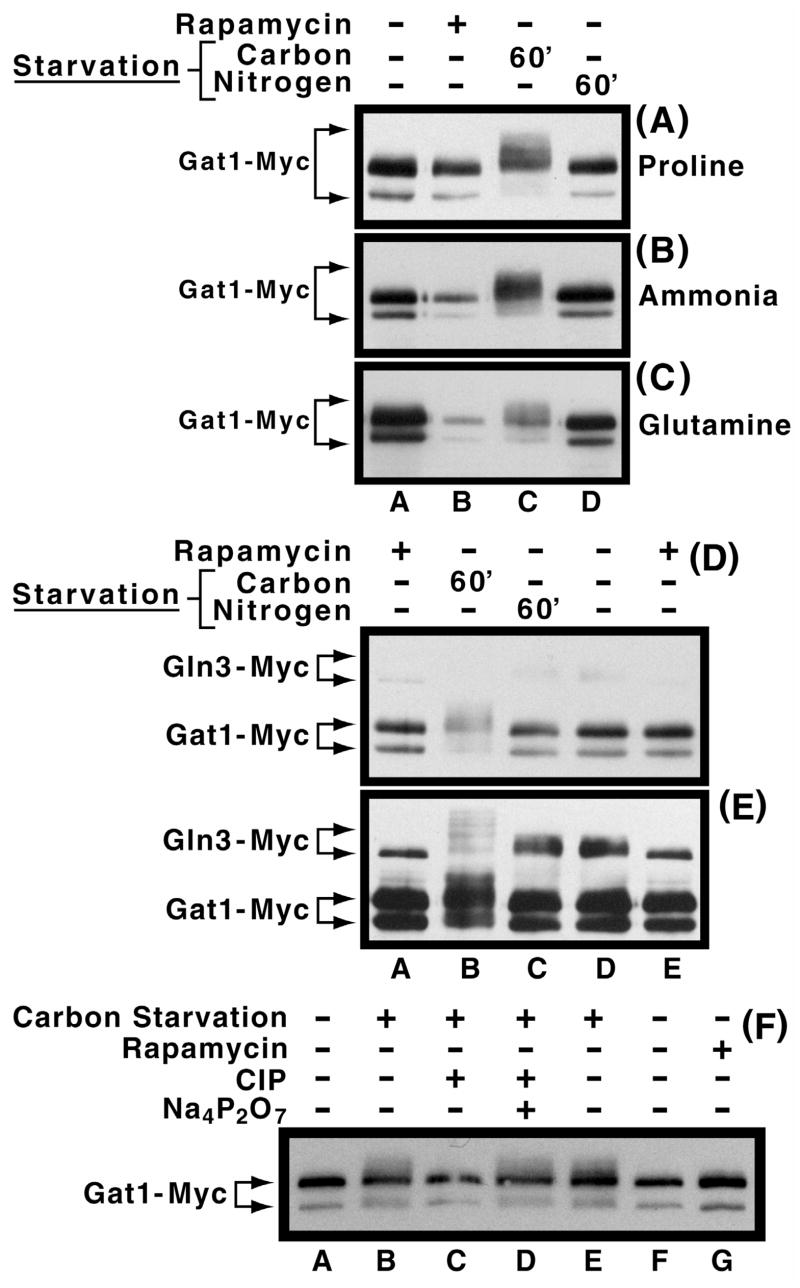
We next determined the effect of carbon and nitrogen starvation on Gat1-Myc13 mobility, because Gln3-Myc13 mobility decreases markedly during the former condition and slightly during the latter (Cox et al., 20003). Gat1-Myc13 mobility clearly decreased following carbon starvation irrespective of the nitrogen source provided (Fig. 3A–C, lanes A vs. C). Three additional observations merit comment: (i) In some preparations, the amount of the minor rapidly migrating species decreased during carbon starvation and rapamycin-treatment (Fig. 3B and C, lanes B and C), but not during nitrogen starvation or in the control extract (Fig. 3B and C, lanes A and D). The source of differing mobilities for the two Gat1-Myc13 species observed throughout this work is not known. (ii) There was a hint of a band whose mobility was slightly less than that of the major Gat1-Myc13 species (for example in Fig. 3A, lanes A, B and D). However, this band was not reproducibly present nor could it be correlated with any experimental perturbation. (iii) Although the major species in Fig. 3B and C, lanes A and B appear to possess different migration rates, this apparent difference was shown, using multiple exposures, to derive from blooming of the signals in Fig. 3A and B, lane A.
Fig. 3.
Panels A–C. Effect of nitrogen and carbon starvation on Gat1-Myc13 phosphorylation levels in cells provided with various nitrogen sources. Cultures of RJ715, transformed with pGAT1-Myc, were grown in YNB-proline (Panel A), ammonia (Panel B), or glutamine (Panel C) medium to mid-log phase. Cells were then transferred to the same medium devoid of either glucose (lane C) or the nitrogen source (lane D). After 60 min. of starvation, cells were harvested and extracts prepared as described in Materials and Methods. Untreated and rapamycin-treated cultures were included as controls (lanes A and B). Panels D and E. Comparison of the effects of nitrogen and carbon starvation on Gat1-Myc13 and Gln3-Myc13 phosphorylation. Experimental conditions were as in Panel B except that TB123 transformed with pGAT1-Myc was used in place of the RJ715 transformant. As described earlier, Panels D and E depict short and long exposures of the same gel. Panel F. Sensitivity of carbon starvation induced Gat1-Myc13 phosphorylation to phosphatase treatment. pGAT1-Myc transformants of RJ715 were grown in YNB-ammonia medium in the presence or absence of rapamycin as described earlier. Calf intestional alkaline phosphatase (CIP) in the presence or absence of its inhibitor, sodium pyrophosphate (Na4P2O7), were added, as described in Fig. 2, to extracts where indicated.
When directly compared in the same sample, Gat1-Myc13 and Gln3-Myc13 mobilities responded similarly to carbon starvation in wild type cells, i.e., they decreased (Fig. 3D and E, lane B). Decreased Gat1-Myc13 mobility during carbon starvation derived from increased phosphorylation as the smear of slower mobility Gat1-Myc13 species collapsed into a single band when extracts were treated with CIP (Fig. 3F, lanes E vs. C), a response inhibited by addition of sodium pyrophosphate (Fig 3F, lane D).
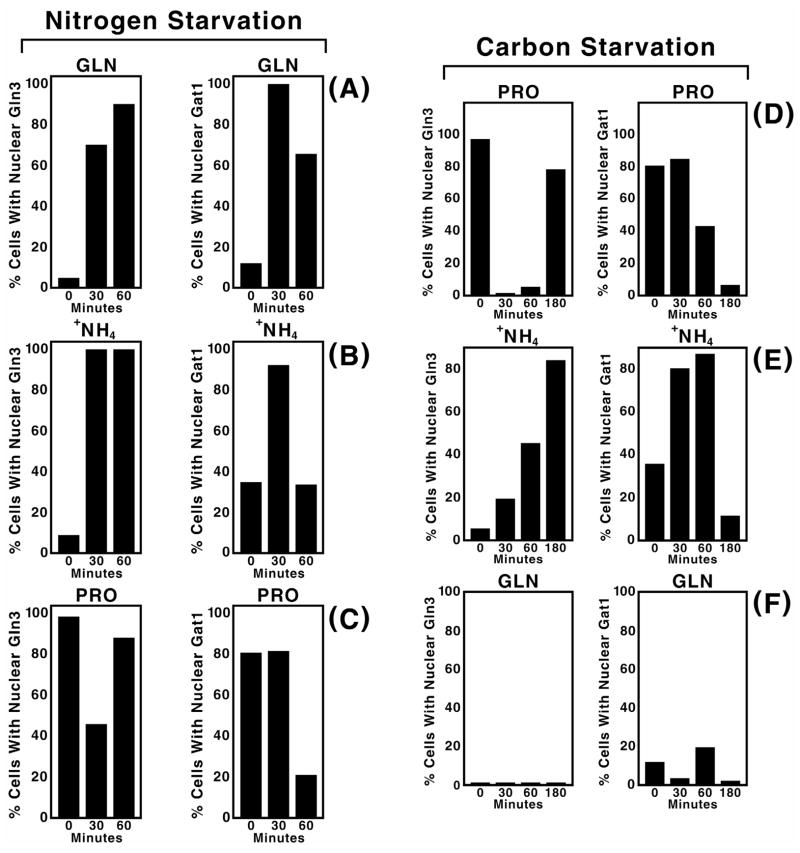
Gln3 and Gat1 are both GATA-family transcription activators mediating NCR-sensitive gene expression (Hoffman-Bang, 1999; ter Schure et al., 2000; Cooper, 1996; Cooper, 2002). Although over-produced Gat1 can, to an extent, functionally suppress the effects of deleting GLN3 increasing evidence points to distinct as well as redundant functions for the two proteins (Shamji et al., 2000). Since distinct physiological functions might differentially influence the kinetics of Gln3 and Gat1 localization in response to environmental changes, we monitored Gat1-Myc13 localization following nitrogen and carbon starvation in cells pre-grown with glutamine, ammonia or proline as sole nitrogen source and compared it with earlier analogous Gln3-Myc13 experiments (Cox et al., 20003). Within 30 minutes of the onset of nitrogen starvation, Gat1-Myc13 became nuclear in more than 80% of cells pregrown in ammonia or glutamine medium, behavior similar to that of Gln3-Myc13 (Fig. 4A and B). However, accumulation was transitory with Gat1-Myc13 exiting from the nucleus over the next 30 min. irrespective of the nitrogen source used for pre-growth (Fig. 4A–C, right panels). In contrast, Gln3-Myc13 remained nuclear in 80% or more of the cells 60 min after the onset of nitrogen starvation (Fig. 4A–C, left panels; these data are taken from Cox et al., 20003).
Fig. 4.
Comparison of Gat1-Myc13 and Gln3-Myc13 intracellular localization during nitrogen and carbon starvation of cells grown in YNB medium with glutamine, ammonia or proline as sole nitrogen source. Strains and culture conditions were similar to those used in Fig. 3, Panels A–C. Starvation times (min.) are indicated at the bottom of each histogram. Gat1-Myc13 was visualized by indirect immunofluorescence as described in Materials and Methods. Gln3-Myc13 data (all left panels) are reproduced from Ref. 20, describing a parallel analysis under the same conditions except that strain TB123 was used in place of RJ715 transformed with pGAT1-Myc. They are included here to facilitate the comparison between Gat1-Myc13 and Gln3-Myc13.
The response of Gat1-Myc13 localization to carbon starvation was less uniform than with nitrogen starvation. Gat1-Myc13 was largely nuclear at the outset of carbon-starvation in proline-grown cells and remained so for 30 min. It then decreased to the point of being almost entirely cytoplasmic by 180 min. (Fig. 4D, right panel). This was distinct from Gln3-Myc13, which was nuclear at the onset of carbon starvation, was localized to the cytoplasm between 30 and 60 min., and again accumulated in the nuclei of most cells at 180 min. (Fig. 4D, left panel). In contrast with proline, the localization profiles for Gat1-Myc13 and Gln3-Myc13 were similar in that both became more nuclear after ammonia-grown cells were carbon starved, albeit with Gat1-Myc13 responding more quickly (Fig. 4E). These responses are reflective of those seen with nitrogen starvation of ammonia grown cells (Fig. 4B). This is consistent with earlier results demonstrating carbon starvation quickly generates nitrogen starvation due to the loss of α-ketoglutarate needed to assimilate the ammonia (Cox et al., 2002). Between 60 and 180 min., the fraction of cells with nuclear Gln3-Myc13 continued to increase, whereas Gat1-Myc13 became predominantly cytoplasmic. Both transcription factors remained cytoplasmic in carbon-starved glutamine-grown cells (Fig. 4F). Overall the time courses of Gat1-Myc13 responses exhibited characteristics more like those expected of a transitory stress response than steady state responses to NCR observed with Gln3-Myc13.
Only limited correlations can be drawn with respect to the response of Gat1-Myc13 alone to carbon starvation. Gat1-Myc13 phosphorylation increased in all carbon starved cells (60 min. after the onset of starvation) regardless of the nitrogen source provided (Fig. 3A–C). This phosphorylation did not, however, closely parallel Gat1-Myc13 intracellular localization. The most uniform Gat1-Myc13 intracellular localization response occurred at 180 minutes post starvation in which case Gat1-Myc13 was localized to the cytoplasm of most cells. Earlier in starvation, for example at 60 min. when Gat1-Myc13 phosphorylation was measured, Gat1-Myc13 localization was nitrogen source specific, ranging from nuclear in nearly 90% of the cells provided with ammonia as nitrogen source to 40% and 20% for proline and glutamine respectively (Fig. 4D–F). At 30 min. after starvation Gat1-Myc13 was nuclear in most proline or ammonia grown cells, but not those grown in glutamine.
Gat1 phosphorylation and intracellular localization following rapamycin and MSX-treatment
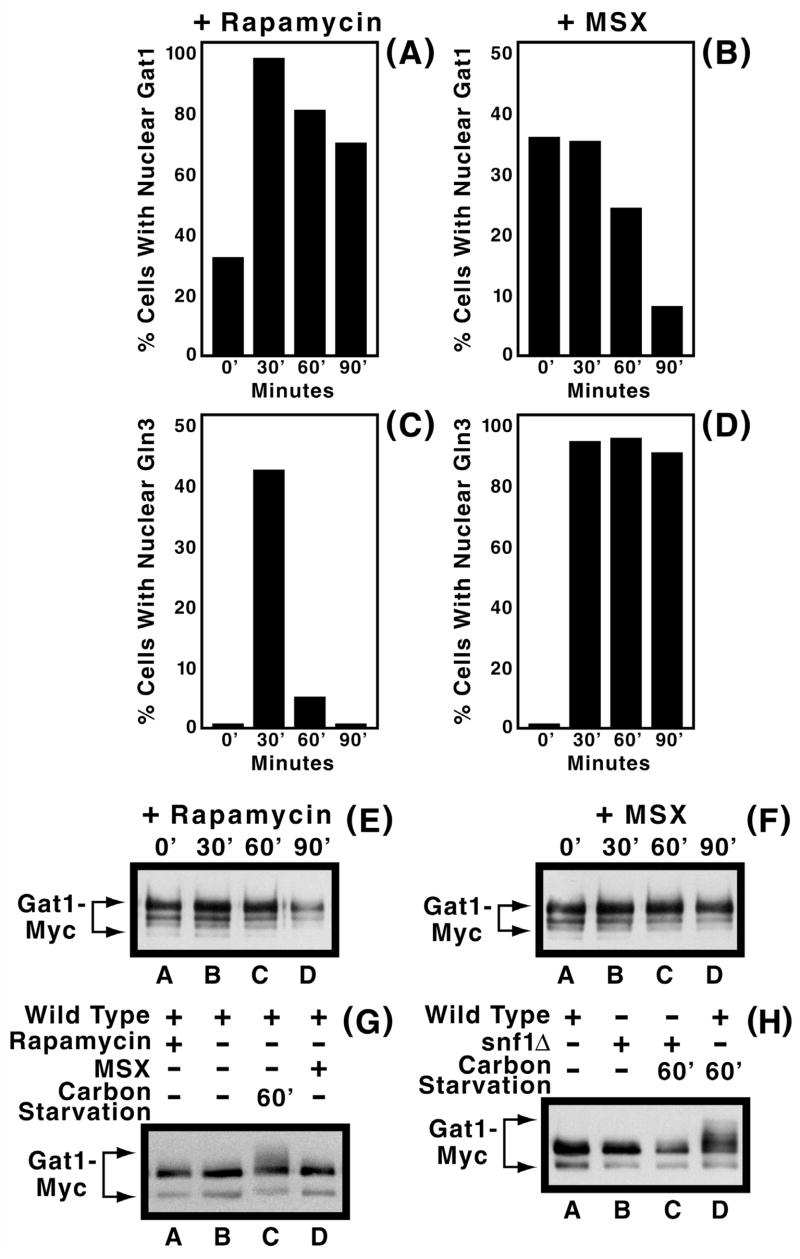
Differing localization profiles of Gln3-Myc13 following rapamycin- and MSX-treatment prompted us to investigate these responses for Gat1-Myc13 also. Rapamycin-treatment increased nuclear accumulation of Gat1-Myc13, as reported for Gln3-Myc13. However, in contrast to Gln3-Myc13 and the starvation response described above, the decrease in fraction of cells containing nuclear Gat1-Myc13 was much less precipitous than with Gln3-Myc13 (Compare Figs. 5A and 5C and Fig. 8 of Cox et al., 20003). On the other hand, the responses of Gat1-Myc13 and Gln3-Myc13 localization to MSX treatment were surprising. Given Gln3 and Gat1 both support highly NCR-sensitive gene expression, and that MSX-inhibits glutamine production (the molecule suggested to be sensed for nitrogen sufficiency (Crespo et al., 2002)), one would a priori, expect the two transcriptional activators to respond similarly, but they did not. MSX-treatment resulted in nuclear Gln3-Myc13 localization in nearly all cells (Fig 5D). In contrast, nuclear Gat1-Myc13 observed in 30–40% of ammonia-grown cells became largely cytoplasmic over the 90 min. time course of the experiment (Fig. 5B). Equivalency of the 0 and 30 min. values in Fig. 5B easily account for the earlier conclusion that MSX-treatment did not affect the intracellular distribution of Gat1 in SD medium (Crespo et al., 2002).
Fig. 5.
Panels A–F. Effects of rapamycin (200 ng/ml, panels A, C and E) and MSX (2 mM, panels B, D and F) treatment on intracellular Gat1-Myc13 and Gln3-Myc13 distribution (panels A–D) and Gat1-Myc13 phosphorylation (panels E and F) in RJ715 transformed with pGAT1-Myc grown in YNB-ammonia medium. Culture conditions and methods of cell preparation and counting were as described in earlier figures. The times of treatment are indicated below the histograms and above the Western blots, respectively. Panels G and H. Carbon starvation induced Gat1-Myc13 phosphorylation is Snf1-dependent. However, MSX- and carbon starvation induced Snf1-independent phosphorylation observed with Gln3-Myc13 (Tate et al., 2005) was not demonstrable for Gat1-Myc13. Further experimental conditions and Western blot analysis are the same as in earlier figures. pGAT1-Myc transformed RJ715 grown in ammonia-YNB medium were used for preparation of cell extracts. Cells were treated with MSX for 30 min. Strains used in panel H were TB106-2a and RR183.
In contrast to the situation with Gln3-Myc13, where MSX- and rapamycin-treatment elicited opposite phosphorylation responses, Gat1-Myc13 phosphorylation levels for these two inhibitors were identical for at least 90 min. (Figs. 5E and F). In neither case did phosporylation levels differ from those in inhibitor-free cells. This observation did not arise from an inability to detect phosphorylated Gat1-Myc13, because it was clearly visible with extracts from carbon starved cells (Fig. 5G, lane C and 5H, lane D). Note that the blots in Fig. 5E and F exhibit greater Gat1-Myc13 degradation than seen in earlier figures, as indicated by appearance of additional minor truncated species. Failure to observe any difference in the phosphorylation profiles following MSX addition prompted us to ask whether carbon starvation induced changes in Gat1-Myc13 phosphorylation could be detected in a snf1Δ because Gln3-Myc13 phosphorylation following carbon starvation consists of both Snf1-dependent and –independent components, the former being absent following MSX-treatment (Tate et al., 2005). Carbon-starvation generated Gat1-Myc13 phosphorylation was eliminated in the snf1Δ (Fig. 5H, lane C), but in contrast with Gln3-Myc13, there was no evidence of a Snf1-indepndent phosphorylation component with Gat1-Myc13. Also, the Gat1-Myc13 minor species that sometimes appeared, migrating slightly more slowly than the major species, can be seen in Fig. 5H, lanes A and B. As mentioned earlier, when this species was present, it appeared in all or nearly all of the lanes and did not correlate with a particular experimental perturbation.
Discussion
Experiments described above provide concrete data comparing phosphorylation and intracellular localization of GATA-family transcription activator Gat1 with that of Gln3. The evidence shows a positive correlation of Gln3-Myc13 and Gat1-Myc13 localization in the presence (30 min.) vs. absence of rapamycin and during steady state growth conditions supporting high vs. low-level NCR-sensitive gene expression. These data agree with, as well as extend, those reported earlier (Crespo et al., 2002). However, the similarity of Gat1 and Gln3 localization in steady state conditions does not extend to the environmental transitions, carbon and nitrogen starvation. In such transitions, intracellular Gat1 localization responds more transiently than Gln3. Only during carbon starvation of glutamine-grown cells, where localization is largely unaffected, do Gat1 and Gln3 responses correlate. These results suggest overlapping as well as protein-specific functions during steady state growth vs. nutrient transitions, respectively; conclusions that are consistent with those from genomic data of Kuruvilla et al. (2001).
Gat1-Myc13 localization in MSX-treated cells observed in our experiments support the results of Crespo et al. (2002), in that we saw no effect of the inhibitor 30 min. after its addition to the culture compared with a 20 min. incubation in the published report. There is, however, a very marked effect opposite to that with Gln3 when MSX-treatment is extended to 90 min. The fraction of cells in which Gat1 is nuclear decreases four fold, while that of Gln3 increases to nearly 100% and remains there throughout the experiment. Whether this decrease is a direct or indirect effect of MSX is not clear.
MSX and rapamycin inhibitor data from the present and earlier reports may be interpreted in two ways: (i) as concluded earlier, components of the Tor signal transduction pathway may regulate Gat1 differently from Gln3 (Crespo et al., 2002), and/or alternatively, (ii) MSX and rapamycin do not inhibit tandem steps in a single regulatory pathway, but steps in different pathways or different branches of a single pathway with downstream events affecting Gat1 and Gln3 differently. The essential difference between the interpretations is whether the results reported by others and ourselves derive from direct cause-effect relationships or alternatively are only correlative, deriving from secondary effects. We favor the latter alternative over the former because according to the model derived from the initial experiments using MSX as a perturbant of the Tor pathway (Crespo et al., 2002), MSX inhibits glutamine synthetase thereby preventing production of glutamine or a metabolite of it, which is in turn sensed by Tor. In other words, the MSX inhibited step of the posited single regulatory model is situated above that inhibited by rapamycin, i.e. Tor1/2. Therefore, if the intracellular localizations of both Gln3 and Gat1 similarly respond to rapamycin-inhibition of Tor1/2, then both would be expected to respond similarly to a regulatory molecule upstream of Tor1/2. i.e., the role envisioned for glutamine. It is the lack of a common response of Gln3-Myc13 and Gat1-Myc13 localization to rapamycin and MSX that prompts our support of the two pathway or two branch alternative in preference to the tandem one pathway model.
Less similarity was seen in Gln3 and Gat1 phosphorylation profiles. Although, like Gln3-Myc13 (Cox et al., 20003), Gat1-Myc13 phosphorylation failed to correlate in a consistent way with the nitrogen source provided, under all conditions but carbon starvation-induced Snf1-dependent phosphorylation, we were unable to elicit demonstrable changes in Gat1-Myc13 phosphorylation levels. Most important, we could not demonstrate decreased Gat1-Myc13 phosphorylation in response to rapamycin inhibition of Tor1,2 even though Gln3-Myc13 phosphorylation levels decreased in the same samples. With the one exception noted above, Gat1-Myc13 mobility remained unchanged and unaffected by the presence or absence of CIP. While it would be imprudent to argue the negative, i.e., that Gat1-Myc13 is not phosphorylated in response to changes in nitrogen supply or Tor pathway activity, we can safely conclude that changes in Gat1-Myc13 phosphorylation levels could not be detected in samples where changes in Gln3-Myc13 phosphorylation were easily demonstrated. These results, which are the first Western blots to appear in the literature, differ from the earlier conclusion that Gln3 and Gat1 localization and phosphorylation levels responded similarly to rapamycin addition and nature of the nitrogen source (Beck and Hall, 1999, as data not shown).
Given our results, it would not be unreasonable to argue that we were merely unable to detect Gat1-Myc13 phosphorylation because of a technical defect in our assay. Although possible, this seems somewhat unlikely because during glucose starvation, Snf1-dependent Gat1-Myc13 phosphorylation, similar to that observed with Gln3-Myc13, was easily and repeatedly demonstrated. Carbon starvation induced Gat1-Myc13 phosphorylation did differ from that with Gln3-Myc13 (Tate et al., 2005) in that the Snf1-independent component of Gln3-Myc13 phosphoryla-tion did not occur with Gat1-Myc13. The smear of hyper-phosphorylated Gln3-Myc13 and Gat1-Myc13 species generated by carbon starvation argues in favor of generalized phosphorylation of both proteins at many sites.
Failure to observe changes in Gat1-Myc13 phosphorylation levels might arise if: (i) In contrast to Gat1-Myc13 sites phosphorylated in response to carbon starvation, those associated with nitrogen supply, rapamycin-, and/or MSX-treatment, may be more sensitive to in vitro dephosphorylation and hence lost during extract preparation. (ii) Phosphorylation-dephosphorylation of Gln3-Myc13 brought about sufficiently large changes in the protein’s conformation to be detected on an SDS gel (Smith et al., 1989), whereas such changes may have been too small in Gat1-Myc13 to be detected. (iii) Only a small number of physiologically significant Gat1-Myc13phosphorylation events are associated with changes in nitrogen source or inhibitor presence and are below our level of detection. (iv) The Gat1-Myc13 product of the plasmid borne GAT1-MYC13 gene was aberrantly regulated due to the tag or fact that it is expressed at a higher level than wild type from the plasmid. This possibility is considered unlikely because both Gat1-Myc13 localization and the transcription it supports exhibit a normal NCR response. If NCR-sensitive localization and transcription were dictated by: (i) Gat1-Myc13 in vivo phosphorylation/dephosphorylation profiles or (ii) defects in normal operation the NCR regulatory system elicited by overproduction of GAT1-MYC13, then they would have affected both physiological Gat1-Myc13 responses to the nitrogen source as well as those of Gln3-Myc13. This, however, did not occur arguing against this possibility.
Feller and Dubois have independently assayed the migration of Gat1-Myc13 from strain BY4709 transformed with a low copy number plasmid (pFL38) containing the gene GAT1-Myc13 grown in buffered minimal medium with glutamine as nitrogen source, glutamine plus rapamycin or proline media. They observed identical Gat1-Myc13 phosphorylation profiles leading them to conclude that Gat1 was either (i) not phosphorylated, (ii) was phosphorylated independently of the nitrogen source, or (iii) was differentially phosphorylated in a manner that could not be detected under their experimental conditions. These data did not support extension of the model of phosphosphorylation/dephosphorylation-dependent Gln3 localization proposed by Beck and Hall in the regulation of Gat1 (E. Dubois, personal communication).
Acknowledgments
We thank Dr. Michael Hall for strain TB123, Dr. M.S. Longtine for plasmid pFA-6a, Dr. Evelyne Dubois for her sharing her unpublished Gat1-Myc13 data, Tim Higgins for preparing the artwork, and the UT Yeast Group for suggestions to improve the manuscript. This work was supported by NIH grant GM-35642.
References
- Beck T, Hall MN. The TOR signaling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- Bertram PG, Choi JH, Carvalho J, Ai W, Zeng C, Chan TF, Zheng XFS. Tripartite regulation of Gln3 by TOR, Ure2, and phosphatases. J Biol Chem. 2000;275:35727–35733. doi: 10.1074/jbc.M004235200. [DOI] [PubMed] [Google Scholar]
- Bertram PG, Choi JH, Carvalho J, Chan TF, Ai W, Zheng XF. Convergence of TOR-nitrogen and Snf1-glucose signaling pathways onto Gln3. Mol Cell Biol. 2002;22:1246–1252. doi: 10.1128/MCB.22.4.1246-1252.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas ME, Cutler NS, Lorenz MC, Di Como CJ, Heitman J. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 1999;13:3271–3279. doi: 10.1101/gad.13.24.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho J, Bertram PG, Wente SR, Zheng XF. Phosphorylation regulates the interaction between Gln3p and the nuclear import factor Srp1p. J Biol Chem. 2001;276:25359–25365. doi: 10.1074/jbc.M103050200. [DOI] [PubMed] [Google Scholar]
- Coffman JA, Rai R, Cunningham T, Svetlov V, Cooper TG. Gat1p, a GATA family protein whose production is sensitive to nitrogen catabolite repression, participates in transcriptional activation of nitrogen-catabolic genes in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:847–858. doi: 10.1128/mcb.16.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman JA, Rai R, Loprete DM, Cunningham T, Svetlov V, Cooper TG. Cross regulation of four GATA factors that control nitrogen catabolic gene expression in Saccharomyces cerevisiae. J Bacteriol. 1997;179:3416–3429. doi: 10.1128/jb.179.11.3416-3429.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TG. In: Allantion degradative system - an integrated transcriptional response to multiple signals. Mycota III, Marzluf G, Bambrl R, editors. Springer Verlag; Berlin, Heidelberg: 1996. pp. 139–169. [Google Scholar]
- Cooper TG. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: connecting the dots. FEMS Microboil Rev. 2002;26:223–238. doi: 10.1111/j.1574-6976.2002.tb00612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TG. Integrated regulation of the nitrogen-carbon interface. Top in Curr Genet. 2004;7:225–257. [Google Scholar]
- Cox KH, Kulkarni A, Tate JJ, Cooper TG. Gln3 phosphorylation and intracellular localization in nutrient limitation and starvation differ from those generated by rapamycin inhibition of Tor1/2 in Saccharomyces cerevisiae. J Biol Chem. 2003;279:10270–10278. doi: 10.1074/jbc.M312023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox KH, Pinchak AB, Cooper TG. Genome-wide transcriptional analysis in S. cerevisiae by mini-array membrane hybridization. Yeast. 1999;15:703–713. doi: 10.1002/(SICI)1097-0061(19990615)15:8<703::AID-YEA413>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Cox KH, Rai R, Distler M, Daugherty JR, Coffman JA, Cooper TG. GATA sequences function as TATA elements during nitrogen catabolite repression and when Gln3 is excluded from the nucleus by overproduction of Ure2. J Biol Chem. 2000;275:17611–17618. doi: 10.1074/jbc.M001648200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox KH, Tate JJ, Cooper TG. Cytoplasmic compartmentation of Gln3 during nitrogen catabolite repression and the mechanism of its nuclear localization during carbon starvation in Saccharomyces cerevisiae. J Biol Chem. 2002;277:37559–37566. doi: 10.1074/jbc.M204879200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo JL, Helliwell SB, Wiederkehr C, Demougin P, Fowler B, Primig M, Hall MN. NPR1 kinase and RSP5-BUL1/2 ubiquitin ligase control GLN3-dependent transcription in Saccharomyces cerevisiae. J Biol Chem. 2004;279:37512–37517. doi: 10.1074/jbc.M407372200. [DOI] [PubMed] [Google Scholar]
- Crespo JL, Powers T, Fowler B, Hall MN. The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine. Proc Natl Acad Sci USA. 2002;99:6784–6789. doi: 10.1073/pnas.102687599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham TS, Andhare R, Cooper TG. Nitrogen catabolite repression of DAL80 expression depends on the relative levels of Gat1p and Ure2p production in Saccharomyces cerevisiae. J Biol Chem. 2000;275:14408–14414. doi: 10.1074/jbc.275.19.14408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham TS, Rai R, Cooper TG. The level of DAL80 expression down-regulates GATA factor-mediated transcription in Saccharomyces cerevisiae. J Bacteriol. 2000;182:6584–6591. doi: 10.1128/jb.182.23.6584-6591.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Como CJ, Arndt KT. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 1996;10:1904–1916. doi: 10.1101/gad.10.15.1904. [DOI] [PubMed] [Google Scholar]
- Hardwick JS, Kuruvilla FG, Tong JK, Shamji AF, Schreiber SL. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc Natl Acad Sci USA. 1999;96:14866–14870. doi: 10.1073/pnas.96.26.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman-Bang J. Nitrogen catabolite repression in Saccharomyces cerevisiae. Mol Bio-technol. 1999;12:35–73. doi: 10.1385/MB:12:1:35. [DOI] [PubMed] [Google Scholar]
- Jacinto E, Guo B, Arndt KT, Schmelzle T, Hall MN. TIP41 interacts with TAP42 and negatively regulates the TOR signaling pathway. Mol Cell. 2001;8:1017–1026. doi: 10.1016/s1097-2765(01)00386-0. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Broach JR. Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J. 1999;18:2782–2792. doi: 10.1093/emboj/18.10.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuruvilla FG, Shamji AF, Schreiber SL. Carbon- and nitrogen-quality signaling to translation are mediated by distinct GATA-type transcription factors. Proc Natl Acad Sci USA. 2001;98:7283–7288. doi: 10.1073/pnas.121186898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Rowen DW, Esiobu N, Magasanik B. Role of GATA factor Nil2p in nitrogen regulation of gene expression in Saccharomyces cerevisiae. J Bacteriol. 1997;179:3761–3766. doi: 10.1128/jb.179.11.3761-3766.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt ME, Brown TA, Trumpower BL. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucl Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamji AF, Kuruvilla FG, Schreiber SL. Partitioning the transcriptional program induced by rapamycin among the effectors of the Tor proteins. Curr Biol. 2000;10:1574–1581. doi: 10.1016/s0960-9822(00)00866-6. [DOI] [PubMed] [Google Scholar]
- Smith CL, Debouck C, Rosenberg M, Culp JS. Phosphorylation of serine residue 89 of human adenovirus E1A proteins is responsible for their characteristic electrophoretic mobility shifts, and its mutation affects biological function. J Virol. 1989;63:1569–1577. doi: 10.1128/jvi.63.4.1569-1577.1989. Erratum in: J Virol (1989) 63: 3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soussi-Boudekou S, Vissers S, Urrestarazu A, Jauniaux JC, Andre B. Gzf3p, a fourth GATA factor involved in nitrogen-regulated transcription in Saccharomyces cerevisiae. Mol Microbiol. 1997;23:1157–1168. doi: 10.1046/j.1365-2958.1997.3021665.x. [DOI] [PubMed] [Google Scholar]
- Tate JJ, Rai R, Cooper TG. Methionine Sulfoximine Treatment and Carbon Starvation Elicit Snf1-independent Phosphorylation of the Transcription Activator Gln3 in Saccharomyces cerevisiae. J Biol Chem. 2005;280:27195–27204. doi: 10.1074/jbc.M504052200. Epub 2005 May 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Schure EG, van Riel NA, Verrips CT. The role of ammonia metabolism in nitrogen catabolite repression in Saccharomyces cerevisiae. FEMS Microbiol Rev. 2000;24:67–83. doi: 10.1111/j.1574-6976.2000.tb00533.x. [DOI] [PubMed] [Google Scholar]
- Thomas G, Sabatini DM, Hall MN, editors. Curr Top Microbiol and Immunol. Vol. 279. Springer; 2004. TOR Target of Rapamycin. [Google Scholar]
- Wang H, Wang X, Jiang Y. Interaction with Tap42 is required for the essential function of Sit4 and type 2A phosphatases. Mol Biol Cell. 2003;14:4342–4351. doi: 10.1091/mbc.E03-02-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]