Abstract
The absorptive Na+-K+-Cl− cotransporter (NKCC2) is a polytopic protein that forms homooligomeric complexes in the apical membrane of the thick ascending loop of Henle (TAL). It occurs in at least four splice variants (called B, A, F, and AF) that are identical to one another except for a short region in the membrane-associated domain. Although each of these variants exhibits unique functional properties and distributions along the TAL, their teleological purpose and structural organization remain poorly defined. In the current work, we provide additional insight in these regards by showing in mouse that the administration of either furosemide or an H2O-rich diet, which are predicted to alter NKCC2 expression in the TAL, exerts differential effects on mRNA levels for the variants, increasing those of A (furosemide) but decreasing those of F and AF (furosemide or H2O). Based on a yeast two-hybrid mapping analysis, we also show that the formation of homooligomeric complexes is mediated by two self-interacting domains in the COOH terminus (residues 671 to 816 and 910 to 1098), and that these complexes could probably include more than one type of variant. Taken together, the data reported here suggest that A, F, and AF each play unique roles that are adapted to specific physiological needs, and that the accomplishment of such roles is coordinated through the splicing machinery as well as complex NKCC2–NKCC2 interactions.
INTRODUCTION
The second isoform of the Na+-K+-Cl− cotransporter (NKCC2) is restricted in its localization to the apical membrane of the thick ascending loop of Henle (TAL; see Payne and Forbush, 1994; Kaplan et al., 1996; Yang et al., 1996). It plays a critical role in salt reabsorption along this nephron segment, thereby participating in the maintenance of extracellular fluid volumes and excretion of free H2O (Greger, 1985; Gimenez and Forbush, 2003; Fernandez-Llama et al., 2005). The physiological importance of NKCC2 in mammals is suggested by the occurrence of salt wasting, uncompensated polyuria, or systemic hypertension when the transporter is functionally impaired (Simon et al., 1996; Bartter et al., 1998; Takahashi et al., 2000; Alvarez-Guerra and Garay, 2002).
NKCC2 belongs to the cation-Cl cotransporter (CCC) family, which is comprised of several isoforms that mediate cell surface Cl−-dependent Na+ and/or K+ cotransport. Members within this family are very homologous to one another, possessing 12 transmembrane domains (tm) flanked by cytosolic termini (see model of NKCC2 in Fig. 1). Previous studies have revealed that a number of CCC isoforms also occur in splice variants (Payne and Forbush, 1994, Igarashi et al., 1995; Race et al., 1999; Plata et al., 2001; Gagnon et al., 2002; Hebert et al., 2004). Among terrestrial vertebrates, e.g., at least four types of NKCC2s have been described: NKCC2F, A, and B, which are identical to one another except for a 96-bp exon that encodes the second tm and part of the following connecting segment, and NKCC2AF, which possesses both the A and F exons in tandem (Payne and Forbush, 1994; Igarashi et al., 1995; Plata et al., 2001; Gagnon et al., 2002).
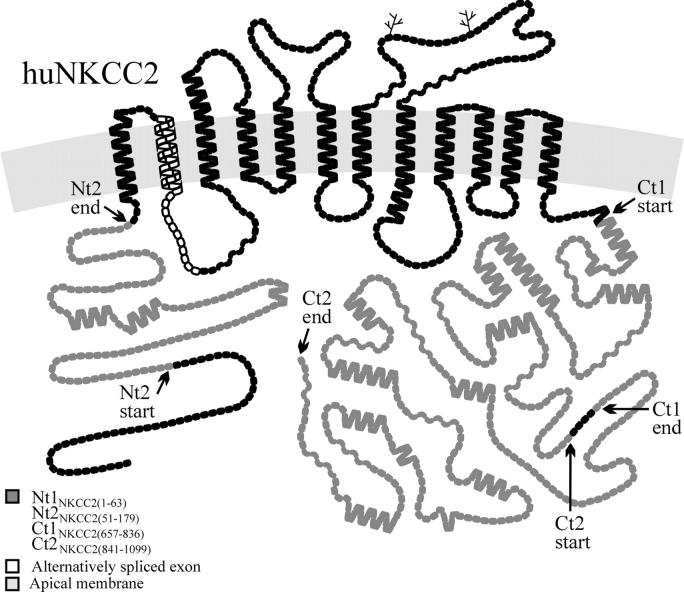
Figure 1.
Hydropathy plot model of huNKCC2. The symbols represent amino acid residues, showing the protein segments Nt2NKCC2(51–179), Ct1NKCC2(657–836), and Ct2NKCC2(841–1099) in gray and the localization of the alternatively spliced exon in white. The nomenclature used to designate these protein segments is explained in MATERIALS AND METHODS and in Table III. The model of huNKCC2 was drawn using the program PLOT (Biff Forbush).
In situ hybridization studies in lagomorphs have revealed that the NKCC2 variants are differentially distributed along the TAL but with some degree of overlap in their site of expression; A is to a large extent in the outer stripe of the outer medulla (OM) and cortex, and F in the inner stripe of the OM (Payne and Forbush, 1994; Igarashi et al., 1995). These studies have also demonstrated that A and F are not produced in equal proportions within the diluting segment. For instance, F is the most abundant transcript whereas B is the least abundant. As for AF, indirect evidence suggests that this variant is present in the OM (Payne and Forbush, 1994; Gagnon et al., 2002, 2003), and that it could constitute a large fraction of the NKCC2-derived mRNAs in some species (Gagnon et al., 2002).
In mammals, the NKCC2 splice variants differ not only in their distribution and expression levels, but also in their kinetic features (Gimenez et al., 2002; Plata et al., 2002; Gagnon et al., 2003). By way of illustration, F displays the lowest affinity for ions under controlled conditions and B the highest (Kms: F > A > B), whereas A displays the highest transport capacity and B the lowest (Vmax: for A ≥ F > B); for convenience, Km values based on two of these studies are summarized in Table I. Regarding AF, intriguingly, influx studies in the Xenopus laevis oocyte expression system have shown that this variant is nonfunctional even if it is able to reach the cell surface (Gagnon et al., 2002).
TABLE I.
Functional Characteristics of the Rabbit NKCC2 Splice Variants Expressed in X. laevis Oocytes
| Km values (in mM)
|
||||
|---|---|---|---|---|
| Na+ | Rb+ | Cl− | Distribution along the TAL | |
| NKCC2B | 10 | 2 | 10 | Macula densa cells |
| NKCC2A | 10 | 2 | 25 | Cortex and outer stripe of the OM |
| NKCC2F | 60 | 5 | 100 | Inner stripe of the OM |
The Kms shown represent estimated values based on two independent studies (Gimenez et al., 2002; Gagnon et al., 2003) and the localization data is based on in situ hybridization studies (Igarashi et al., 1995). Here, TAL = thick ascending limb of the loop of Henle, and OM = outer medulla.
Despite the information that has been acquired on the localization, expression, and function of the renal Na+-K+-Cl− symporters, the physiological basis for the existence of multiple NKCC2 variants remains uncertain at present. Likewise, the mechanisms that underlie the normal operation of the variants have still not been examined in great detail. For example, the hormone ADH and the loop diuretic furosemide have been shown to alter the function and expression of NKCC2 in lagomorphs (Ecelbarger et al., 1996, 2001; Knepper et al., 1999; Gimenez and Forbush, 2003), but whether they exert the same effects on all of the variants is unknown.
Coimmunoprecipitation and cross-linking studies have suggested that NKCC2 is organized at the surface of renal tubular cells as a homooligomer (Ichinose et al., 1999; Moore-Hoon and Turner, 2000; Starremans et al., 2003). The protein domains that lead to the formation of such structures have not been identified but recent two-hybrid mapping analyses using NKCC1 as a model revealed that the COOH terminus (Ct) could play an important role in this regard (Simard et al., 2004a). The mechanisms by which monomers assemble with each other are of particular interest given that nonfunctional NKCCs have been shown to exert dominant-negative effects on their functional counterparts (Isenring et al., 1998b; Jacoby et al., 1999; Caron et al., 2000; Casula et al., 2001).
In this research, studies were performed to elucidate further the functional and structural properties of A, F, and AF. Based on our findings, we propose that the variants each play unique roles in the TAL not only because of differences in their localizations and kinetic features but also because of differences in the mechanisms by which their expression is regulated. We propose, in addition, that the inclusion of different types of NKCC2 variants in the same oligomer could endow AF with unexpectedly important tasks in salt transport along the TAL.
MATERIALS AND METHODS
Overview
The approaches that were exploited in this research include yeast two-hybrid studies, influx assays, and expression analyses. Chemicals, reagents, and kits were from different suppliers or collaborators, whereas oligonucleotides and E. coli strains were from Sigma-Aldrich and Stratagene, respectively.
NKCC Constructs/pGilda or pB42AD
These constructs were used for the two-hybrid studies. They consisted of NKCC-derived DNA fragments that were cloned in pGilda (CLONTECH Laboratories, Inc.), a vector that possesses coding sequences for the LexA DNA-binding domain and a His transformation marker (His™), or in pB42AD (CLONTECH Laboratories, Inc.), a vector that possesses coding sequences for the activating domain of a transcription factor, the HA epitope, and a Trp™. The inserts were obtained by reverse transcriptase (RT)-PCR using human kidney RNA and primers to which restriction sites were added, or they were generated by bp deletions from existing constructs after creating convenient restriction sites with the Quick Change Mutagenesis Kit (Stratagene) when necessary (see Table II for primers). The constructs were termed according to the NKCC protein segments encoded, using prefixes to designate the specific domains from which the segments are derived (Nt, NH2 terminus; Ct, COOH terminus; 1, proximal portion; 2, distal portion) and numbers in parentheses to designate their position in human (hu) NKCC2. The characteristics of these protein segments are shown in Table III.
TABLE II.
Oligonucleotides Used in this Study
| Name of DNA | Oligonucleotides | Restriction sites |
|---|---|---|
| For constructs | ||
| a) Nt2NKCC2(51–179)/pGilda | forward: gg GAA TTC ggg gat gaa gct cag aaa ag | EcoRIb |
| reverse: cc CTC GAG cca tcc aaa ctt cac aac acc | XhoI | |
| b) Ct1NKCC2(657–836)/pGilda | forward: cc GAA TTC gct ctt tcc tac gtg agt gc | EcoRIb |
| reverse: tg CTC GAG cct gtt cta atc tct cta att cc | XhoI | |
| d) Ct1NKCC2(671–836)/pGilda | forward: c aat gct ctg GAA TTC acc aca gtg gaa gac | EcoRIc |
| reverse: gtc ttc cac tgt ggt GAA TTC cag agc att g | EcoRI | |
| e) Ct1NKCC1(759–947)/pGilda | forward: tc acC CCG GGa tcc tct aca caa gca | XmaIb |
| reverse: ca tgC CAT GGt gcc agg aga ttt cta | NcoI | |
| f) Ct1NKCC1(834–926)/pGilda | forward: ca GAA TTC atg tcc atc gat caa gcca | EcoRIb |
| reverse: c ACT CGA gtc cag acc ttc ttt tag gcga | BsRI | |
| h) Ct2NKCC2(830–1099)/B42AD | forward: gg GAA TTC gaa tta gag aga tta gaa cag gag | EcoRIb |
| reverse: gg CTC GAG agt taa aat gta ttc caa tct ttc | XhoI | |
| i) Ct2NKCC2(910–1058)/B42AD | forward: c att gtc ctg agc ctt CCC GGG caa gaa agg g | XmaIc |
| reverse: c cct ttc ttg CCC GGG aag gct cag gac aat c | XmaI | |
| For sequencing | ||
| j) pGilda | forward: cgt cag cag agc ttc acc att ga | |
| k) pB42AD | forward: cc agc ctc ttg ctg agt gga gat ga | |
| reverse: gca aag tag aca agc cga caa cc | ||
| For expression studies | ||
| l) “A” exon (RT-PCR) | forward: cg gga att ggt ctt gga g | |
| reverse: cc tcc acg aac aaa ccc g | ||
| m) “F” exon (RT-PCR) | forward: cg gga att ggt ctg ggc g | |
| reverse: cc tcc tcg cac cac tcc g | ||
| n) “AF” exon (RT-PCR) | forward: cg gga att ggt ctt gga g | |
| reverse: cc tcc tcg cac cac tcc g | ||
| o) “A” oligoprobe (in situ) | forward: tt ctt ctt tcc acc atg gta acc tct at | |
| reverse: at aga ggt tac cat ggt gga aag aag aa | ||
| p) “F” oligoprobe (in situ) | forward: tt ggc ctg agc gta gtt gtg aca aca ct | |
| reverse: ag tgt tgt cac aac tac gct cag gcc aa |
They are all written 5′ to 3′. Added restriction sites are designated by capital letters. Note that Ct1NKCC2(657–782)/pGilda is not shown in the list because it was produced by deleting a portion of Ct1NKCC2(657–836) between BplI, a naturally occurring restriction site in huNKCC2 and XhoI in the pGidla cloning site.
Primers were available from a previous study (Simard et al., 2004a).
Primers were used for RT-PCR.
Primers were used to introduce substitutions with the Quick Change Mutagenesis Kit (Stratagene).
TABLE III.
Characteristics of the Hybrid Protein Segments
| Name of construct | Length of fused proteins (n residues) |
Length of NKCC protein segment (n residues) |
Total length of hybrid protein (n residues) |
Position in the huNKCC2 Ct (1–457) or Nt (1–183) |
|---|---|---|---|---|
| In the vector pGilda | ||||
| Ct1NKCC2(657–836) | 202 | 180 | 400 | 15–194 |
| Ct1NKCC2(657–782) | 202 | 126 | 328 | 15–140 |
| Ct1NKCC2(671–836) | 202 | 166 | 368 | 29–195 |
| Ct1NKCC1(759–947) | 202 | 189 | 391 | 10–196 |
| Ct1NKCC1(834–926) | 202 | 93 | 295 | 85–177 |
| Nt2NKCC2(51–179) | 202 | 129 | 331 | 51–179 |
| In the vector pB42AD | ||||
| Ct2NKCC2(818–1099) | 108 | 282 | 390 | 176–457 |
| Ct2NKCC2(830–1099) | 108 | 270 | 378 | 188–457 |
| Ct2NKCC2(841–1099) | 108 | 259 | 267 | 199–457 |
| Ct2NKCC2(843–1099) | 108 | 257 | 365 | 201–457 |
| Ct2NKCC2(846–1099) | 108 | 264 | 372 | 204–457 |
| Ct2NKCC2(867–1099) | 108 | 233 | 341 | 225–457 |
| Ct2NKCC2(892–1099) | 108 | 208 | 316 | 250–457 |
| Ct2NKCC2(899–1099) | 108 | 201 | 309 | 257–457 |
| Ct2NKCC2(910–1058) | 108 | 149 | 257 | 268–457 |
| Ct2NKCC1(970–1212) | 108 | 243 | 351 | 206–457 |
For each construct, the numbers in parentheses indicate the position of the encoded protein segment relative to the huNKCC1 or huNKCC2 amino acid sequence. For the Ct1NKCC1(759–947), Ct1NKCC1(834–926), and Ct2NKCC1(970–1212) protein segments, this position in huNKCC2 is between residue 652 and 838, 727 and 819, and 848 and 1099, respectively.
pGilda Constructs
One such construct was used for a two-hybrid screen; the insert consisted of a 540-bp fragment that encodes the huNKCC2 Ct1 from residue 657 to 836 (see Fig. 1). Based on the nomenclature outlined above, it was termed Ct1NKCC2(657–836). Five other constructs were used for yeast two-hybrid mapping analyses. They are termed Nt2NKCC2(51–179), Ct1NKCC2(657–782), Ct1NKCC2(671–836), Ct1NKCC1(834–926), and Ct1NKCC1(759–947) (protein segments encoded by two of these constructs are also shown in Fig. 1). Here, the Ct1NKCC1(759–947) and Ct1NKCC1(834–926) protein segments are designated based on their position in huNKCC1. The corresponding protein segments in huNKCC2 would be termed Ct1NKCC2(652–840) and Ct1NKCC2(727–819), respectively. Note that all of the constructs that encode these various NKCC1 protein segments were generated for a recently published study (Simard et al., 2004a).
pB42AD Constructs
One of the interacting proteins identified during the screen is derived from a 777-bp fragment that encodes the huNKCC2 distal Ct; it was called Ct2NKCC2(841–1099). Two other pB42AD-derived constructs, which were employed for the mapping analyses, consisted of Ct2NKCC2(830–1099) and Ct2NKCC1(970–1212). Here, the latter construct was also generated for a recently published study (Simard et al., 2004a), it would encode the protein fragment C2tNKCC2(848–1099) in huNKCC2.
Shark (sa) NKCC2/Pol1 Constructs
Three such constructs (saNKCC2A, F, and AF) were used for influx and expression studies. They were already available from previously published work (Gagnon et al., 2002, 2003). The vector in which they are cloned, Pol1, contains a T7 promoter, a cloning site flanked by the X. laevis β-globin untranslated regions and a polyA tract. It should be noted here that the kinetic features of any given splice variant are very similar among various species such as mouse, rabbit, and shark (Gimenez et al., 2002; Plata et al., 2002; Gagnon et al., 2003).
Yeast Two-hybrid Screen
EGY48 yeast (Y) were first transformed with the construct p8op-lacZ (CLONTECH Laboratories, Inc.) and seeded on −Ura plates, generating the strain Yop; p8op-LacZ is a plasmid that contains LexA operators destined to interact with the pB42AD-derived activating domain, and that encodes two reporter genes (Leu and LacZ) controlled by LexA and Ura. After this first selection step, Yop cells were transformed a second time with the bait Ct1NKCC2(657–836) and seeded on −Ura−His (−UH) plates, generating YopCt1NKCC2(657–836). The latter strain was subsequently tested for expression of the hybrid protein (see below and Fig. 2 A, lane 7), and tested for autonomous reporter gene activation on −Ura−His−Leu + X-galactosidase (−UHL+X) plates.
Figure 2.
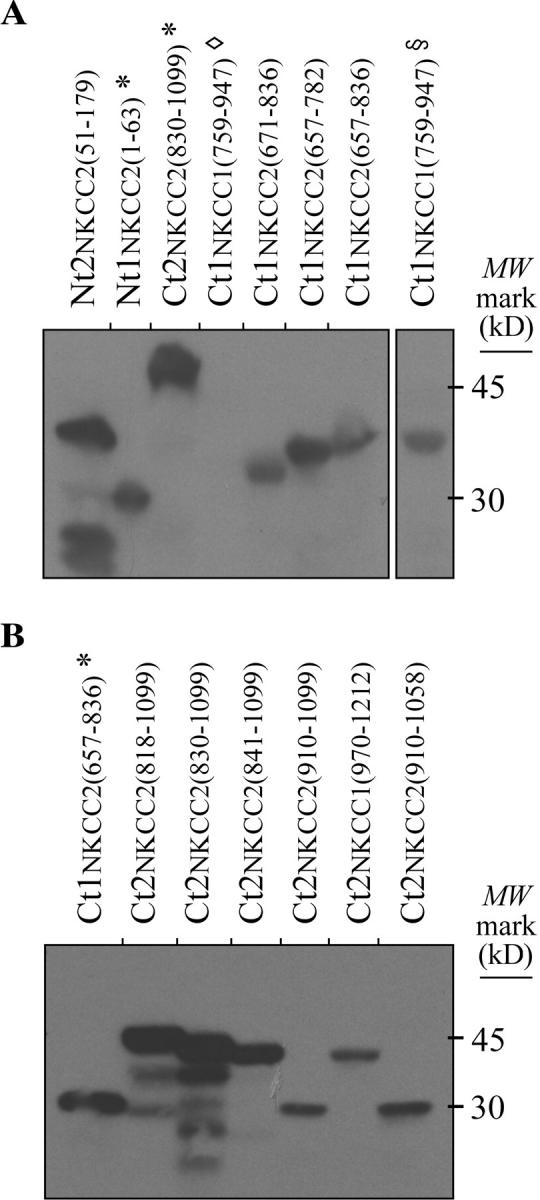
Western analyses. Proteins were extracted from yeast with a lysis solution containing glass beads, 8 M urea, 40 mM Tris-HCl, 0.1 mM EDTA, 125 µM β-mercaptoethanol, 5% SDS, and protease inhibitors (final pH 6.8). In the top panel, the protein extracts were from NKCC/pGilda-transformed cells and the analyses were performed with ∼2 mg of bait proteins using an anti-LexA Ab. In the bottom panel, the protein extracts were from NKCC/pB42AD-transformed cells and the analyses were performed with ∼0.4 mg of prey proteins using an anti-HA Ab. Here, * indicates that the protein segments expressed by these transformants are currently being studied, ◊, that the protein fragment of interest had apparently degraded, and §, that the protein fragment was from another preparation of Ct1NKCC1(759–947)-transformed yeast but that it was run in a distant lane within the same gel.
A search for interactors was performed by transforming YopCt1NKCC2(657–836) with a human kidney library cloned in pB42AD and subjecting the transformants to two rounds of selection, first on −Ura−His−Trp (−UHT) plates and subsequently on −Ura−His−Trp−Leu (−UHTL) plates. Resistant colonies were transferred on −UHT+X plates, and those expressing β-gal activity were amplified in a regular yeast medium. Prey plasmids were extracted according to well-established procedures and inserts were analyzed by automated sequencing (Simard et al., 2004a).
Two procedures were performed to confirm the specificity of the identified interactions. First, Yop cells were transformed with a single prey/pB42AD and tested for expression of a hybrid protein by Western blotting after selection of colonies on −Ura−Trp (−UT) plates (see below and examples in lanes 2, 4, and 5 of Fig. 2 B). Second, Yop cells were cotransformed with pairs of plasmids (single prey + empty pGilda versus single prey + LAM/pGilda) and tested for reporter gene activation on −UHT+X plates. Note that LAM, which stands for human lamin C, has been reported not to form complexes or interact with most other proteins (Bartel et al., 1993).
Yeast Two-hybrid Mapping Analysis
These studies were performed to determine (a) whether the Ct1–Ct2 interacting sites can be identified more precisely, (b) whether they are similar to those of NKCC1, and (c) whether Ct1–Ct1, Ct2–Ct2, or Nt–Ct associations are also possible. To these ends, Yop cells were transformed with different types of NKCC-bearing constructs and tested for their ability to express a hybrid protein after selection on −UH or −UT plates (see examples in Fig. 2 A, lanes 1, 4, 5, 6, 7, and Fig. 2 B, lanes 3, 4, 6, and 7) and for reporter gene activation on −UHT+X plates. After this step, the cells were retransformed with another NKCC-bearing construct and tested for their ability to grow on −UHTL plates or to express β-gal activity on −UHT+X plates.
Expression of saNKCC2s in Oocytes
To obtain synthesis of saNKCC2s in this system, defolliculated stage V–VI oocytes were injected with saNKCC2/Pol1-derived cRNA (∼2–20 ng/oocyte diluted in H2O) and maintained for 3 d at 18°C in Barth's (B) medium (see Table IV) + 125 μM furosemide. Groups of oocytes were also injected with H2O alone as controls (ctls).
TABLE IV.
Composition of Flux Solutions
| Solution (in mM) | Na+ | Rb+ or K+ | Cl− | Ca2+ | Mg2+ | PO4 2− | SO4 2− | HEP | GLU | SUC | OSM |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A. Regular medium (R) | 87 | 5 | 86 | 2 | 2 | 1 | 1 | 10 | 0 | 0 | 200 |
| B. Modified R (RΔ) for 22Na+ influxes | 43 | 5 | 86 | 2 | 2 | 1 | 1 | 10 | 0 | 0 | 200 |
| B. Hyperosmolar (R-SUC) | 87 | 5 | 86 | 2 | 2 | 1 | 1 | 10 | 0 | 84 | 284 |
| C. Wash | 19 | 73 | 8 | 2 | 2 | 1 | 1 | 10 | 78 | 0 | 200 |
| D. Barth's medium | 90.4 | 1 | 90 | 0.7 | 0.8 | 0 | 0.8 | 10 | 0 | 0 | 200 |
In medium RΔ, NaCl was replaced by an equimolar concentration of NMG-Cl. Barth's medium contains, in addition, 0.7 mM NO3 2− and 2.4 mM HCO3 −. All solutions are at pH 7.4. Here, R = regular, GLUC = gluconate, HEP = HEPES, SUC = sucrose, and OSM = osmolality.
Immunofluorescence Studies of saNKCC2-expressing Oocytes
They were performed as previously described (Caron et al., 2000; Gagnon et al., 2002). The anti-P-NKCC immuno-globulin (Ig), which is specific to the phosphorylated form of various NKCCs (Flemmer et al., 2002) and which was a gift from B. Forbush (Yale University, New Haven, CT), was used as primary antibody, whereas the goat Alexa Fluor 488–conjugated goat anti-rabbit anti-IgG (Invitrogen) was used as secondary antibody.
Flux Studies in saNKCC2-expressing Oocytes
Furosemide was first removed through several rinses in a regular (R) medium (see Table IV). Eggs were then subjected to two consecutive incubation steps: one of 60 min in medium R + 84 mOsM sucrose (R-SUC) to activate the cotransporter, and another of 45 min in one of two solutions to measure NKCC-mediated accumulation of the isotope. The latter solutions were (a) medium R + 1 μCi/ml 86Rb+ ± 250 μM bumetanide + 10 μM ouabain (for 86Rb+ influxes) or (b) a modified medium R (RΔ) (in which [Na+] was decreased to 43 mM) + 1 μCi/ml 22Na+ + 10 μM ouabain + 125 μM hydrochlorothiazide + 125 μM amiloride (for 22Na+ influxes). Fluxes were terminated with several washes in a high K+ medium + 250 μM bumetanide + the inhibitors used during the prior incubation, and oocytes were transferred afterwards to 96-well plates (one oocyte/well) prefilled with 2% SDS and scintillation fluid. 86Rb+ was detected with the TopCountNXT counter (Packard).
Experimental Animals
Male C3H mice (age: 200–400 d, weight: 25–30 g) were kept in the animal care facility of our Center. After a brief stabilization period, they were submitted for 7 d to one of three regimens: (1) a standard diet consisting of Teklad global 2018 mouse (ms) chow (5 g/day) and H2O (10 ml/day); (2) mouse chow (5 g/day) mixed with furosemide (5 mg/day), and H2O (10 ml/day); (3) mouse chow (5 g/day) and H2O (∼100 ml/day) supplemented with sucrose (600 mM). At the end of this 7-d period, all mice were killed by neck dislocation.
RNA Studies
Expression levels of A, F, and AF were first determined by RT-PCR. Templates consisted of first cDNA strands that were produced from oligodT-primed human kidney RNA (2 μg/reactions) while primers consisted of sequences that are derived from the 5′ end of exon A or F (sense primers), from the 3′ end of these exons (antisense primers), or from β-actin. The exon-specific primers were used in three combinations to amplify exon A (sense A + antisense A), F (sense F + antisense F). or AF (sense A + antisense F), whereas the β-actin–specific primers were used as ctls. PCR conditions were titrated to generate DNA strands over a linear range of amplification versus time using simple arithmetic plots. All samples were migrated on ethidium bromide–stained agarose gels.
Expression levels of A, F, and AF were also determined by in situ hybridization. For these studies, freshly harvested mouse kidneys were frozen in liquid N and mounted on cryostat supports. From there, they were cut sagittally into 12-μm cryosections, applied directly on glass slides, and fixed in 4% paraformaldehyde for 10 min. The prehybridization and hybridization steps were performed in a Denhardt's-based solution for 2 and 16 h, respectively. RNA messages were probed with end-labeled oligonucleotides derived from exon 4 (Table II). The hybridization step was followed by several washes in SSC (2×, 1×, 0.5×) at a maximum of 37°C.
Western Analyses
They were performed with yeast protein extracts prepared in a special lysis solution (detailed in Fig. 2 legend) as previously described (Simard et al., 2004a). In brief, extracts were separated on SDS-polyacrylamide tricine gels and transferred by capillarity to Immobilon-P nylon blots (Millipore). Later on, the blots were incubated sequentially with a primary and secondary Ab, and proteins of interest were visualized by chemiluminescence (using the ECL kit, Amersham Biosciences).
Sequence Analyses, Quantification Methods, Statistics
DNA characterizations were performed by automated sequencing using plasmid-derived primers (Table II) and restriction analyses. For blast searches, sequence alignments, and structure predictions, we used a combination of programs including PLOT (B. Forbush) and DNAStar (Lasergene). Whole-cell extracts were quantified with the DC protein assay (Bio-Rad Laboratories) and DNA or RNA-specific signals by densitometry. When appropriate, statistical dispersion within datasets was determined by calculating SEMs, and differences between groups of variables were analyzed by Student's two-tail t tests, rejecting the null hypothesis for P > 0.05.
RESULTS
Two-hybrid Screen
Transformation of YopCt1NKCC2(657–836) with a human kidney library led to the formation of over 4 million −UHT-resistant colonies, 41 of which were also −UHTL resistant. After the quadruple selection, however, only 32 of the colonies expressed above-background β-gal activity on X-gal plates, and after the confirmation studies, 31 were suspected to enclose Ct1NKCC2 partners.
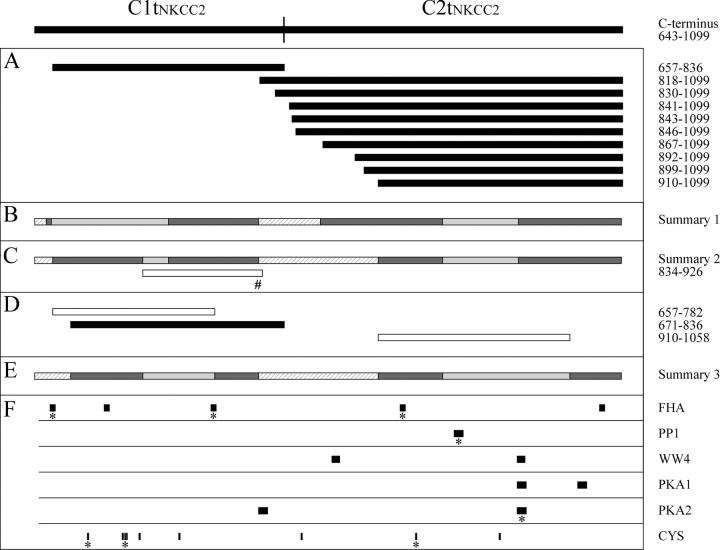
Detailed DNA analyses revealed that 13 of the colonies identified expressed Ct2NKCC2 protein segments, 8 of which also differed in amino acid sequence. These distinct prey are illustrated in Fig. 3 A by horizontal bars aligned with the region in huNKCC2 to which they correspond. It should be noted that all of the Ct2NKCC2 prey were characterized exhaustively in this work after noting that they tended to vary in length and could therefore be exploited for the mapping studies described below.
Figure 3.
huNKCC2 protein segments represented schematically according to their position in the COOH terminus and their ability to support an interaction. Each horizontal bar corresponds to a huNKCC2 residue or protein segment aligned with the region of Ct to which it corresponds. (A, B, C#, and D) Localization of Ct1 segments coexpressed in yeast with Ct2NKCC2(830–1099) or Ct2NKCC2(841–1099) and of Ct2 segments coexpressed with Ct1NKCC2(657–836). Here, black bars are used to indicate that the protein segment supports an interaction, white bars, that it does not, and the pound sign, that it was analyzed in a previous study. (C) Properties of various protein segments based on previous mapping analyses (Simard et al., 2004a) and on the double hybrid screen. Such properties are illustrated as follows: hatched bars indicate that the protein segment is not required for the Ct1–Ct2 association, light gray bars, that it may or may not be required, and dark gray bars, that it is minimally required but not sufficient; the dark gray bars are referred to in the manuscript as essential interacting regions (EIRs). (E) Properties of protein segments based on the behavior of three additional cotransformants. (F) Positions of residues or consensus binding sites in the huNKCC2 Ct. Here, the asterisks indicate that the residues or consensus sites are conserved between NKCC1 and 2, and the abbreviations FHA, PP1, WW4, PKA1, and PKA2 signify forkhead-associated binding domain, protein phosphatase type 1, [ST]P-containing binding domain, protein kinase A type, and protein kinase A type 2.
Two-hybrid Mapping Analyses: Expression of the Hybrid Proteins
In these studies, the NKCC-specific regions tested for their ability to support bait–prey interactions in yeast included the distinct Ct2NKCC2 protein segments shown in Fig. 3 (A and D), CtNKCC1 protein segments used in previous studies, and additional NtNKCC2 or CtNKCC2 protein segments. Western analyses, which were performed before the final selection procedures to confirm expression of the hybrid protein, are shown in Fig. 2. They reveal bands of the adequate molecular weight mark for each of the transformants tested, using the anti-LexA Ab to detect bait and the anti-HA Ab to detect prey. Although some bands are fainter than others, we have observed in previous studies that the activation of Leu and LacZ through a bait–prey interaction can occur even if the partners are expressed at low levels. Note that Western analyses in Fig. 2 show only a few examples of the cotransformants that were tested (see lanes 2, 4, and 5 in Fig. 2 B).
Two-hybrid Mapping Analyses: Interaction between Ct1NKCC2(657–836) and the Distinct Preys
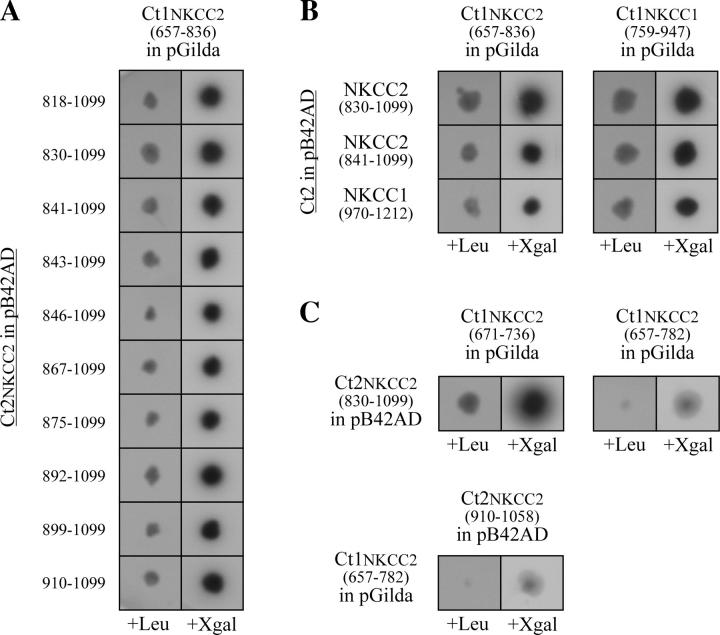
These analyses were performed to confirm the bait–Ct2NKCC2 associations identified during the initial two-hybrid screen and results are summarized in Fig. 4 A. As can be seen here, YopCt1NKCC2(657–836) cells retransformed with either of the distinct prey uncovered, including the 190-residue NKCC protein segment Ct2NKCC2(910–1099), are able to grow on −UHTL plates (left) and generate strong β-gal activity on −UHT+X plates (right). For convenience, we have also illustrated these results graphically by reusing Fig. 3 A and showing the protein segments that have supported an interaction as black bars.
Figure 4.
Two-hybrid mapping analyses. Cotransformed yeast were tested for their ability to grow on −UHTL plates (columns +Leu) or generate strong β-gal activity on −UHT + X plates (columns +X-gal). (A) Each cell expresses a different Ct2NKCC2 prey (among those identified during the initial two-hybrid screen) but the same Ct1NKCC2(657–836) bait. (B) Each cell was cotransformed with a huNKCC1- and a huNKCC2-encoding construct using the Ct1 domain of one isoform and the Ct2 domain of the other isoform. (C) Cells coexpress the protein segments shown in Fig. 3 D along with Ct2NKCC2(830–1099) or Ct1NKCC2(657–836), respectively. All of the cotransformants were incubated at 30°C for 3–6 d.
Two-hybrid Mapping Analyses: Interaction between NKCC1 and 2
In a recent study, we have found that the huNKCC1 Ct also behaves as a self-interacting domain and have identified specific contact sites that underlie this property (Simard et al., 2004a). Such sites, which had been termed essential interacting regions (EIRs), should share high levels on homology with those of NKCC2 given that these isoforms are closely related. They are represented in Fig. 3 B as dark gray bars aligned with the regions to which they correspond in huNKCC2. Interestingly, it is seen from such illustrations that Ct1NKCC2(657–836) and Ct2NKCC2(910–1099) (see Fig. 3 A) do not include all of the NKCC1 EIRs, suggesting that these sites were partially misidentified or that they differ between the isoforms.
To determine which of these scenarios is the most likely, we analyzed the behaviors of yeast coexpressing the Ct1 domain of one isoform and the Ct2 domain of the other isoform, assuming that the oligomerization domains of NKCC1 and 2 would probably interact with each other if they are highly conserved. As shown in Fig. 4 B, the behaviors of the cotransformants suggest that the former scenario is in fact the correct one. For instance, all of the combinations tested support cell growth on −UHTL plates (left) and generate strong β-gal activity on −UHT+X plates (right).
The bar in Fig. 3 C represents a summary of the mapping analyses that were conducted thus far for NKCC1 and 2, assuming that the EIRs of both isoforms are highly homologous. It shows where EIRs in huNKCC2 are now localized relative to those identified in huNKCC1 (Fig. 3 B). Based on this summary, EIR3 appears to be shorter than initially predicted but, reassuringly, the residues no longer included in this previously defined contact site are in reality poorly matched between NKCC1 and 2. EIR1, conversely, now appears to be at a different position, as suggested by the behaviors of Ct1NKCC2(657–836), which was used for the two-hybrid screen in this research, and of Ct1NKCC1(834–926), which had been used previously to identify EIR2 in huNKCC1. Indeed, Ct1NKCC2(657–836) interacts with Ct2 but begins just after the previously defined EIR1, whereas Ct1NKCC1(834–926) was not able to interact with Ct2NKCC1 but corresponds to the longest EIR2-containing protein segment tested to date.
Two-hybrid Mapping Analyses: Interaction between Other NKCC2s
The behaviors of various transformants that coexpress a truncated Ct1 or Ct2 with a full-length Ct2 and Ct1 are shown in Fig. 4 C (the protein segments are also illustrated graphically in Fig. 3 D). Here, interestingly, it is seen that the only truncated protein segment capable of supporting cell growth or inducing β-gal activity is Ct2NKCC2(671–836). When these new results are interpreted in relation with the combined analyses shown in Fig. 3 C, the previously defined EIR2 and EIR4 are now seen to be more precisely mapped and the localization of EIR1 remain slightly different from that predicted initially (see Fig. 3 E).
A number of putative binding sites or residues in huNKCC2 could account for the EIRs' properties or play complementary roles in the formation of Ct1–Ct2 complexes; their localization relative to the newly defined EIRs is shown in Fig. 3 F. Interestingly, several of these sites/residues (highlighted by an asterisk) are conserved between NKCC1 and 2. As will be discussed later, however, the current analysis has placed a conserved, presumably important FHA-related site outside of one of the newly relocalized EIR.
Two-hybrid Mapping Analyses: a Search for Other Types of Self-interactions
The segments used for these studies, Nt2NKCC2(51–179), Ct1NKCC2(657–836), and Ct2NKCC2(830–1099), are shown in Fig. 1. The only conclusion that could be drawn from the behavior of various cotransformants is that Ct1NKCC2 does not interact with Ct1NKCC2 (unpublished data). The coexpression of a bait Ct2NKCC2 with a prey Ct2NKCC2 yielded equivocal results, whereas Nt2NKCC2(51–179) was found to induce LacZ in the absence of a prey protein and could therefore not be tested (unpublished data).
Flux Studies
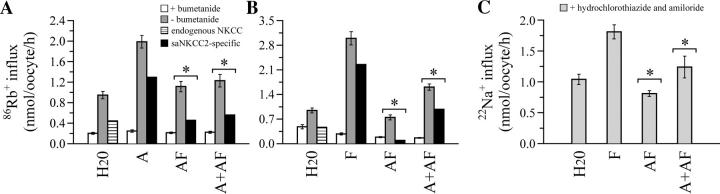
Because A, F, and AF are identical in the region of huNKCC2 that harbors EIRs, and because AF is probably expressed in the same cell types as A or F, we hypothesized that some of the variants could assemble with each other in some regions of the TAL to form heterooligomeric structures in vivo. We have thus determined the potential repercussions of such assemblies by analyzing the functional behavior of A or F coexpressed in oocytes with AF.
The results of these studies are shown in Fig. 5. Quite interestingly, 86Rb fluxes in oocytes that express both A and AF together are ∼50% lower than in oocytes that express A alone, approaching those measured in H2O-injected ctls (Fig. 5 A). Similar results are observed when A is replaced by F in these coexpression experiments (Fig. 5 B) or when 86Rb is replaced by 22Na (Fig. 5 C). In conjunction with our previous findings, these studies suggest that AF exerts a dominant-negative effect on A or F by forming nonfunctional heterodimers with these variants.
Figure 5.
Flux studies. X. laevis oocytes were injected with saNKCC2A, saNKCC2F, saNKCCC2AF, saNKCC2A + saNKCC2AF, or saNKCC2F + saNKCC2AF (∼2 ng for A or F, and ∼20 ng for AF). Groups of eggs were also injected with H2O alone to determine background cotransporter activity. (A and B) 86Rb influx measured in 13–22 oocytes expressed as means ± SEM. Bumetanide and ouabain were added to the influx and wash solution at a concentration of 250 μM and 10 μM. (C) 22Na influx measured in 9–10 oocytes expressed as means ± SEM. Hydrochlorothiazide, amiloride, and ouabain were added to the influx and wash solution at concentrations of 250, 125, and 125 μM. Here, * indicates that within any given panel, the values are significantly different from those of A or F (P < 0.01).
Immunofluorescence
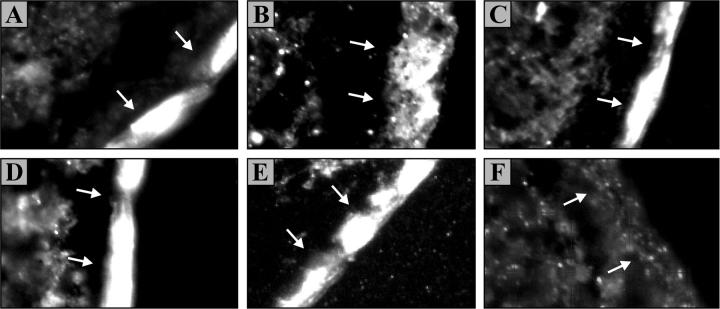
These studies, which are summarized in Fig. 6, show that all of the variants tested are able to reach the oocytes' cell surface whether they are expressed individually (Fig. 6, A–C) or in combination (D and E). They also show that the signal obtained is of similar intensity among the variants. For AF, hence, lack of transport activity is apparently not due to lack of cell surface delivery, supporting further the idea of a physical interaction between AF and other variants.
Figure 6.
Immunofluorescence studies of X. laevis oocytes expressing saNKCC2 variants. The cotransporters were immunolocalized with the anti-P-NKCC antibody (Gagnon et al., 2002; Flemmer et al., 2002) and signals were microphotographed under immunofluorescence microscopy using similar exposure times and representative membrane section among >3 oocytes. The variants shown in this figure are (A) saNKCC2A, (B) saNKCC2F, (C) saNKCC2AF, (D) saNKCC2AF + A, and (E) saNKCC2AF + F. In F, oocytes were injected with H2O as ctl. In each micrograph, the plasma membrane is shown by arrows and lie on the right side of the cytosol.
RNA Studies
The effects of different maneuvers on the expression of individual variants in mouse are shown in Fig. 7. They were assessed through RT-PCR (Fig. 7, A and B) and in situ hybridization (C). It should be mentioned, here again, that the cDNA products analyzed were not from the initial exponential phase of amplification (as such, they were quantified in ethidium bromide-stained agarose gels) and that this semiquantitative assay may therefore not be sufficiently precise to monitor small (less than 1.5-fold) differences in mRNA abundance. It should be mentioned, in addition, that even if exon A- or exon F-specific antisense oligoprobes were used to identify msNKCC2 by in situ hybridization, they will not allow distinguishing among all of the variants perfectly. Indeed, the A-specific probe will hybridize with both A and AF, and the F-specific probe with both F and AF. Thus, changes in A or F expression will be underestimated or overestimated if concomitant changes in AF expression occur. Importantly, however, the ratios of changes between A and F will not be altered under such circumstances.
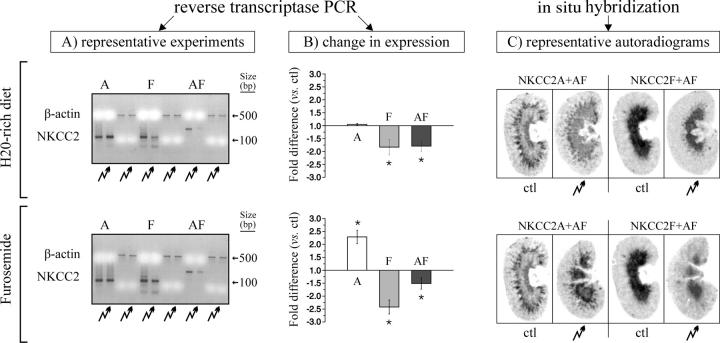
Figure 7.
RNA expression studies. Semiquantitative RT-PCR experiments using mouse kidney cDNAs as templates and huNKCC2A-, huNKCC2F-, or huβ-actin–specific oligonucleotides as primers. Mice were subjected to a ctl diet, an H2O-rich diet (top panels), or a furosemide-enriched diet (bottom panels) for 7 d (two animals per condition). PCR products were generated three to five times for each primer set over a linear range of DNA amplification vs. time and were electrophoresed on ethidium bromide–stained gels. (A) Representative gels using broken arrows to designate the test lane. Here, the absence of bands at the 200-bp molecular weight mark in the first four lanes indicates that the A or F primers, which share high levels of homology, are nevertheless exon specific. (B) Mean fold-changes in band densities (BD) between ctl and test diets where * indicates that the difference was statistically significant (P < 0.01). Here, values from which means are derived were calculated as follows: BDhuNKCC2 (test diet) ÷ BDhuβ-actin (test diet) ÷ BDhuNKCC2 (ctl diet) ÷ BDhuβ-actin (ctl diet). (C) In situ hybridization studies using two sets of NKCC2 oligoprobes, one that hybridizes with both NKCC2A and NKCC2AF, and another that hybridizes with both NKCC2F and NKCC2AF. These oligoprobes were labeled with [α-35S] dATP and purified on G25-Quick spin RNA columns (Roche). The specificity of these probes is suggested by the localization of the signals in the expected regions of the renal cryosections and was further confirmed by carrying out parallel studies with sense A and F oligoprobes (Table II), both of which revealed no signals (not depicted). All of the cryosections were exposed on Kodak MR-1 films during the same period of time. As in A, broken arrows are used to indicate the test lanes.
The effect of an H2O-rich diet (mean ingestion ∼100 ml/d versus ∼10 ml/d in the ctl) is presented in the top row of Fig. 7. Here, interestingly, PCR analyses illustrated by representative gels in Fig. 7 A, and by densitometry measurements in B, show that the expression levels of A are unchanged following the administration of the H2O-rich diet, whereas those of F and AF decrease substantially (by a factor of 1.8 based on Fig. 7 B, P < 0.01, n = 2–6). Results obtained by in situ hybridization (Fig. 7 C) are consistent with these findings, showing an apparent decrease in msNKCC2-specific signals with both oligoprobes.
The effect of furosemide is presented in the bottom row. Here again, PCR analyses show that the variants are affected differentially by this maneuver. As illustrated in Fig. 7 (A and B), e.g., the expression levels of A are seen to increase substantially (by a factor of 2.3, P < 0.01, n = 3) following the administration of furosemide, whereas those of F and AF are seen to decrease (by a factor of 2.4 and 1.5, respectively, P < 0.01, n = 5). As for the in situ hybridization studies (Fig. 7 C), results are once more consistent with those obtained by PCR, showing an expected rise in msNKCC2-specific signals with the A oligoprobe but a decrease in these signals with the F oligoprobe.
DISCUSSION
In this work, yeast two-hybrid studies led to the identification of separate domains in huNKCC2 that can interact with each other; one is in the proximal portion of the COOH terminus (Ct1) and the other in the distal portion (Ct2). They revealed, moreover, that the corresponding domains in huNKCC1, which were recently shown to exhibit similar properties (Simard et al., 2004a), can also interact with those of huNKCC2. In conjunction with previous studies demonstrating that the structural unit of both NKCC1 and 2 is oligomeric (Ichinose et al., 1999; Moore-Hoon and Turner, 2000; Starremans et al., 2003), and that the Ct1NKCC1–Ct2NKCC1 interaction can be reproduced in pull down assays (Simard et al., 2004a), the current data strongly suggest that the identified self-interacting domains mediate NKCC2 assembly at the cell surface.
An alternative role that could be played by Ct1 and Ct2 is to promote the formation of molecular interactions within the Ct of one monomer instead of between the Cts of two monomers. Although such a possibility cannot be ruled out with certainty, we still favor the hypothesis that the primary purpose of Ct1 and Ct2 is to support intermonomeric associations. Indeed, the organization of a protein into a tertiary structure, that is, the mechanism by which intramolecular folding occurs, is to a large extent mediated by the action of chaperones, foldases, or isomerases (Fink, 1999), whereas the organization of a protein into a homooligomeric structure generally relies on the presence of self-interacting domains as those described here (Fink, 1999; Engelman et al., 2003).
A significant outcome of the mapping analyses that were conducted in this study is the identification of <200 candidate residues that may underlie the Ct1–Ct2 association. As depicted in Fig. 4 E, these candidate residues are regrouped into four clusters (called EIRs) that are dispersed along the COOH terminus; EIR1 corresponds to Ct2NKCC2(671–726), EIR2 to Ct2NKCC2(783–816), EIR3 to Ct2NKCC2(910–959), and EIR4 to Ct2NKCC2(1059–1098). This level of resolution was obtained by showing that the Ct1–Ct2 domains, which had already been mapped partially in huNKCC1, are functionally conserved between the isoforms, by sequencing all of the huNKCC2 prey identified in the library, and by analyzing the behavior of yeast expressing truncated versions of Ct1 or Ct2 protein segments.
The importance of mapping the oligomerization domains of NKCC2 stems from the possibility of exploiting certain interacting residues as molecular targets for future structure function studies, and from the insight that could be gained into the molecular principles of oligomerization for this transport system. By way of illustration, it may become possible through various interventions, e.g., site-directed mutagenesis or peptide interference (Bjornson et al., 2003; Polo et al., 2004), to alter carrier assembly as a means of determining whether the monomeric structure is functional or correctly targeted at the cell membrane and if so, whether its kinetic features differ from those of the oligomer. Along the same line, the mapping analyses performed here could lead to the identification of novel motifs or mechanisms that are involved in protein–protein interactions or carrier assembly.
A potential shortcoming of the method employed to map the oligomerization domains of huNKCC2 comes from the widely dispersed localization of the candidate interacting residues. In fact, the specific manner in which these residues are disposed suggests that some level of tertiary organization may be required for Ct1–Ct2 associations to occur in cells and implies that some of the unmapped regions (e.g., those represented by light gray boxes in Fig. 4) will include long stretches of residues flanked by EIRs on both sides. Hence, certain deletions could result in false negatives by generating conformational artifacts and erroneous interpretations of where EIRs reside, or will have to include one of the EIRs. The relocalization of EIR1 based on the current analysis may represent a good illustration of these potential limitations.
Left alone, the finding that the COOH terminus of NKCC2 can interact with that of NKCC1 is of disputable physiological importance given that these isoforms are not expressed in the same cell types. However, it does point toward the possibility that certain carriers within the same CCC subgroup or from different CCC subgroups could associate with one another in vivo. Indeed, several of these transport systems share similar amino acid sequences, tissue distributions, and subcellular localizations (Haas and Forbush, 1998 and 2000; Russell, 2000; Hebert et al., 2004). Heterooligomeric assemblies between various CCC isoforms would carry the advantage of broadening functional diversity within the family.
The existence of conserved oligomerization domains among CCC family members suggests that certain NKCC2 variants could also assemble as heterooligomeric complexes. Two lines of evidence are in fact consistent with this possibility. (1) Ct1 domains appear to interact specifically with Ct2 domains (and vice et versa) throughout a region of huNKCC2 that is identical among the variants (Fig. 3 B). (2) When expressed in X. laevis oocytes, AF can exert dominant-negative effects on A or F (Fig. 6). This variant, which probably constitutes a substantial fraction of NKCC2-derived transcripts in the mammalian TAL based on the current PCR data, and which has been isolated from the renal medulla where A and F are coexpressed, could thus play a similar role in vivo (Payne and Forbush, 1994). Collectively, these observations also suggest that the inclusion of both the A and F exons in the same protein is not simply the result of splicing artifacts as previously suspected.
If AF does play an important role in vivo, transcriptional or posttranscriptional activity for this variant should be affected by a number of stimuli or conditions that act upon NKCC2-expressing cells. According to Western analyses performed in previous studies, these stimuli or conditions could include a long-term increase in circulating ADH levels or the administration of furosemide; indeed, both maneuvers have been shown to up-regulate NKCC2 synthesis in lagomorphs (Ecelbarger et al., 1996, 2001; Knepper et al., 1999). In the present work, we showed that the expression of AF in mouse was also affected by such maneuvers, that is, the number of AF transcripts decreased after 7-d regimens of H2O-rich or furosemide-enriched diets. The data reported here thus point toward a potential role for AF in adaptation responses, which may necessitate diverse levels of NKCC2 activity along the TAL.
The potential involvement of AF in overall salt reabsorption by the TAL is perhaps easier to decipher if the effect of different maneuvers on transcriptional and posttranscriptional activities for the other variants are also known. To this effect, our studies in mouse showed that the observed modifications in expression levels were not always matched between AF and the other variants. Such behaviors imply that the number of nonfunctional AF-containing oligomers could vary substantially under certain conditions and, accordingly, that changes in expression levels and transport activities for A or for F could also be poorly matched. In this setting, the regulated production of AF variants could represent a means by which separate NKCC2-expressing nephron segments alter their transport capacities differentially in response to the same transcriptional stimulus.
Growing evidence suggests that the optional inclusion or excision of specific exons in single gene-derived transcripts is a highly regulated process that involves specific components of the splicing machinery (Beyersmann, 2000; Xie and Black, 2001). Hence, unmatched changes in the expression levels of A, F, or AF may imply that this machinery plays an important role during NKCC2-dependent adaptation responses. For instance, if a spliceosome becomes less active at the 3′ splice site of exon F in A-expressing cells or at the 5′ end of exon A in F-expressing cells, the number of AF transcripts produced would be expected to decrease and that of A or F to increase in parallel. Changes in expression levels through regulation of alternative splicing could then constitute an important posttranscriptional mechanism by which NKCC2-expressing cells coordinate their action appropriately in response to distinct sets of environmental cues.
Although we have not explored the mechanisms by which the H2O-rich diet led to changes in transcriptional of posttranscriptional activities for some of the variants, the large quantity of low salt fluid ingested by the mice and the pattern of responses observed suggest that these mechanisms are related, at least in part, to ADH suppression. In lagomorphs, for example, this hormone has been shown to increase NKCC2 expression, whereas the H2O diet administered here produced the opposite effect in that it decreased the expression of F and AF by >1.5-fold while leaving that of A relatively unchanged (Ecelbarger et al., 1996, 2001; Knepper et al., 1999). Such behaviors may also imply that A-mediated ion transport is more affected than F-mediated transport by changes in circulating ADH levels.
In this work, the mechanisms underlying the effect of loop diuresis were also not explored. It is nonetheless tempting to postulate that furosemide led to the observed changes by decreasing [Cl−] in TAL cells. Indeed, several studies have shown that the activity of both NKCC1 and NKCC2 increases when intracellular Cl− (Cl− i) is reduced nonpharmacologically (Lytle and Forbush, 1992; Haas and Forbush, 1998, 2000; Isenring et al., 1998a; Russell, 2000; Isenring and Forbush, 2001; Darman and Forbush, 2002; Gagnon et al., 2002, 2003; Simard et al., 2004b). Although regulatory events that affect the transporter at posttranslational steps could play an important role in this response (Lytle and Forbush, 1992; Darman and Forbush, 2002), such mechanisms do not exclude the possibility of Cli-dependent regulatory events acting at earlier steps, especially in the setting of long term adaptation responses. In this regard, we showed that furosemide also exerted differential effects on variant expression, increasing that of A substantially, but reducing that of F and AF. These behaviors support the idea of a Cl− i-dependent pretranslational regulatory step, and imply once more that A-mediated ion transport is more affected than F-mediated transport by changes in Cl− i.
In conclusion, we have identified novel features regarding the modus operandi of the NKCC2 splice variants. These features may endow NKCC2-expressing cells with much greater functional diversity than expected for both the normal operations and the adaptation responses. Further studies are required to understand how the NKCC2-dependent splicing machinery is regulated and determine the extent of its contribution to changes in A, F, or AF expression along TAL.
Acknowledgments
The authors are thankful to Valérie Montminy for technical assistance.
This work was supported by grants from the Canadian Institute of Health and Research (MOP-68949 and MOP-15405) and by the Kidney Foundation of Canada.
Lawrence G. Palmer served as editor.
Abbreviations used in this paper: CCC, cation-Cl cotransporter; Ct, COOH terminus; ctl, control; EIR, essential interacting region; hu, human; ms, mouse; NKCC2, Na+-K+-Cl− cotransporter; OM, outer medulla; RT-PCR, reverse transcriptase PCR; sa, shark; TAL, thick ascending loop of Henle.
References
- Alvarez-Guerra, M., and R.P. Garay. 2002. Renal Na+-K+-Cl− cotransporter NKCC2 in Dahl salt-sensitive rats. J. Hypertens. 20:721–727. [DOI] [PubMed] [Google Scholar]
- Bartel, P., C.T. Chien, R. Sternglanz, and S. Fields. 1993. Elimination of false positives that arise in using the two-hybrid system. Biotechniques. 14:920–924. [PubMed] [Google Scholar]
- Bartter, F.C., P. Pronove, J.R. Gill, and R.C. MacCardle. 1998. Hyperplasia of the juxtaglomerular complex with hyperaldosteronism and hypokalemic alkalosis, a new syndrome, 1962. J. Am. Soc. Nephrol. 9:516–528. [DOI] [PubMed] [Google Scholar]
- Beyersmann, D. 2000. Regulation of mammalian gene expression. EXS. 89:11–28. [DOI] [PubMed] [Google Scholar]
- Bjornson, K.P., L.J. Blackwell, H. Sage, C. Baitinger, D. Allen, and P. Modrich. 2003. Assembly and molecular activities of the MutS tetrAm. J. Biol. Chem. 278:34667–34673. [DOI] [PubMed] [Google Scholar]
- Caron, L., F. Rousseau, E. Gagnon, and P. Isenring. 2000. Cloning and functional characterization of a cation-Cl− cotransporter interacting protein. J. Biol. Chem. 275:32027–32036. [DOI] [PubMed] [Google Scholar]
- Casula, S., B.E. Shmukler, S. Wilhelm, A.K. Stuart-Tilley, W. Su, M.N. Chernova, C. Brugnara, and S.L. Alper. 2001. A dominant negative mutant of the KCC1 K-Cl cotransporter: both N- and C-terminal cytoplasmic domains are required for K-Cl cotransport activity. J. Biol. Chem. 276:41870–41878. [DOI] [PubMed] [Google Scholar]
- Darman, R.B., and B. Forbush. 2002. A regulatory locus of phosphorylation in the N terminus of the Na+-K+-Cl− cotransporter, NKCC1. J. Biol. Chem. 277:37542–37550. [DOI] [PubMed] [Google Scholar]
- Ecelbarger, C.A., J. Terris, J.R. Hoyer, S. Nielsen, J.B. Wade, and M.A. Knepper. 1996. Localization and regulation of the rat renal Na+-K+-2Cl− cotransporter, BSC-1. Am. J. Physiol. 271:F619–F628. [DOI] [PubMed] [Google Scholar]
- Ecelbarger, C.A., G.H. Kim, J.B. Wade, and M.A. Knepper. 2001. Regulation of the abundance of renal sodium transporters and channels by vasopressin. Exp. Neurol. 171:227–234. [DOI] [PubMed] [Google Scholar]
- Engelman, D.M., Y. Chen, C.N. Chin, A.R. Curran, A.M. Dixon, A.D. Dupuy, A.S. Lee, U. Lehnert, E.E. Matthews, Y.K. Reshetnyak, et al. 2003. Membrane protein folding: beyond the two stage model. FEBS Lett. 555:122–125. [DOI] [PubMed] [Google Scholar]
- Fernandez-Llama, P., S. Ageloff, G. Fernandez-Varo, J. Ros, X. Wang, N. Garra, C. Esteva-Font, J. Ballarin, P. Barcelo, V. Arroyo, et al. 2005. Sodium retention in cirrhotic rats is associated with increased renal abundance of sodium transporter proteins. Kidney Int. 67:622–630. [DOI] [PubMed] [Google Scholar]
- Fink, A.L. 1999. Chaperone-mediated protein folding. Physiol. Rev. 79:425–449. [DOI] [PubMed] [Google Scholar]
- Flemmer, A.W., I. Gimenez, B.F. Dowd, R.B. Darman, and B. Forbush. 2002. Activation of the Na+-K+-Cl− cotransporter NKCC1 detected with a phosphospecific antibody. J. Biol. Chem. 277:37551–37558. [DOI] [PubMed] [Google Scholar]
- Gagnon, E., B. Forbush, A.W. Flemmer, I. Giménez, L. Caron, and P. Isenring. 2002. Functional and molecular characterization of the shark renal Na+-K+-Cl− cotransporter: novel aspects. Am. J. Physiol. Renal Physiol. 283:F1046–F1055. [DOI] [PubMed] [Google Scholar]
- Gagnon, E., B. Forbush, L. Caron, and P. Isenring. 2003. Functional characterization of renal Na+-K+-Cl− cotransporters between distant species. Am. J. Physiol. Cell Physiol. 284:C365–C370. [DOI] [PubMed] [Google Scholar]
- Gimenez, I., and B. Forbush. 2003. Short-term stimulation of the renal Na+-K+-Cl− cotransporter (NKCC2) by vasopressin involves phosphorylation and membrane translocation of the protein. J. Biol. Chem. 278:26946–26951. [DOI] [PubMed] [Google Scholar]
- Gimenez, I., P. Isenring, and B. Forbush. 2002. Spatially distributed alternative splice variants of the renal Na+-K+-Cl− cotransporter exhibit dramatically different affinities for the cotransported ions. J. Biol. Chem. 277:8767–8770. [DOI] [PubMed] [Google Scholar]
- Greger, R. 1985. Ion transport mechanisms in the thick ascending limb of Henle's loop. Physiol. Rev. 65:760–797. [DOI] [PubMed] [Google Scholar]
- Haas, M., and B. Forbush. 1998. The Na+-K+-Cl− cotransporters. J. Bioenerg. Biomembr. 30:161–172. [DOI] [PubMed] [Google Scholar]
- Haas, M., and B. Forbush. 2000. The Na+-K+-Cl− cotransporter of secretory epithelia. Annu. Rev. Physiol. 62:515–534. [DOI] [PubMed] [Google Scholar]
- Hebert, S.C., D.B. Mount, and G. Gamba. 2004. Molecular physiology of cation-coupled Cl− cotransport: the SLC12 family. Pflugers Arch. 447:580–593. [DOI] [PubMed] [Google Scholar]
- Ichinose, M., A.E. Hall, S. Cheng, J.Z. Xu, and S.C. Hebert. 1999. Novel structure of the flounder thiazide-sensitive Na-Cl cotransporter (flTSC) in the flounder urinary bladder. J. Am. Soc. Nephrol. 10:34A. [Google Scholar]
- Igarashi, P., G.B. Vanden Heuvel, J.A. Payne, and B. Forbush. 1995. Cloning, embryonic expression, and alternative splicing of a murine kidney-specific Na+-K+-Cl− cotransporter. Am. J. Physiol. 269:F405–F418. [DOI] [PubMed] [Google Scholar]
- Isenring, P., and B. Forbush. 2001. Ion transport and ligand binding by the Na-K-Cl cotransporter, structure-function studies. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 130:487–497. [DOI] [PubMed] [Google Scholar]
- Isenring, P., S.C. Jacoby, J. Chang, and B. Forbush. 1998. a. Mutagenic mapping of the Na+-K+-Cl− cotransporter for domains involved in ion transport and bumetanide binding. J. Gen. Physiol. 112:549–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isenring, P., S.C. Jacoby, and B. Forbush. 1998. b. Characterization of the renal absorptive Na+-K+-Cl− (NKCC2), comparative studies including hNKCC1, sNKCC1 and the HEK-293 NKCC1. J. Biol. Chem. 273:11295–11301. [DOI] [PubMed] [Google Scholar]
- Jacoby, S.C., E. Gagnon, L. Caron, J. Chang, and P. Isenring. 1999. Inhibition of Na+-K+-Cl− cotransport by mercury. Am. J. Physiol. 277:C684–C692. [DOI] [PubMed] [Google Scholar]
- Kaplan, M.R., M.D. Plotkin, W.S. Lee, Z.C. Xu, J. Lytton, and S.C. Hebert. 1996. Apical localization of the Na+-K+-Cl− cotransporter, rBSC1, on rat thick ascending limbs. Kidney Int. 49:40–47. [DOI] [PubMed] [Google Scholar]
- Knepper, M.A., G.H. Kim, P. Fernandez-Llama, and C.A. Ecelbarger. 1999. Regulation of thick ascending limb transport by vasopressin. J. Am. Soc. Nephrol. 10:628–634. [DOI] [PubMed] [Google Scholar]
- Lytle, C., and B. Forbush. 1992. The Na+-K+-Cl− cotransport protein of shark rectal gland. J. Biol. Chem. 267:25438–25443. [PubMed] [Google Scholar]
- Moore-Hoon, M.L., and R.J. Turner. 2000. The structural unit of the secretory Na+-K+-2Cl− cotransporter (NKCC1) is a homodimer. Biochemistry. 39:3718–3724. [DOI] [PubMed] [Google Scholar]
- Payne, J.A., and B. Forbush. 1994. Alternatively spliced isoforms of the putative renal Na+-K+-Cl− cotransporter are differentially distributed within the rabbit kidney. Proc. Natl. Acad. Sci. USA. 91:4544–4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plata, C., P. Meade, A. Hall, R.C. Welch, N. Vazquez, S.C. Hebert, and G. Gamba. 2001. Alternatively spliced isoform of apical Na+-K+-Cl− cotransporter gene encodes a furosemide-sensitive Na+-Cl− cotransporter. Am. J. Physiol. Renal Physiol. 280:F574–F582. [DOI] [PubMed] [Google Scholar]
- Plata, C., P. Meade, N. Vazquez, S.C. Hebert, and G. Gamba. 2002. Functional properties of the apical Na+-K+-2Cl− cotransporter isoforms. J. Biol. Chem. 277:11004–11012. [DOI] [PubMed] [Google Scholar]
- Polo, J.M., T. Dell'Oso, S.M. Ranuncolo, L. Cerchietti, D. Beck, G.F. Da Silva, G.G. Prive, J.D. Licht, and A. Melnick. 2004. Specific peptide interference reveals BCL6 transcriptional and oncogenic mechanisms in B-cell lymphoma cells. Nat. Med. 10:1329–1335. [DOI] [PubMed] [Google Scholar]
- Race, J.E., F.N. Makhlouf, P.J. Logue, F.H. Wilson, P.B. Dunham, and E.J. Holtzman. 1999. Molecular cloning and functional characterization of KCC3, a new K+-Cl− cotransporter. Am. J. Physiol. 277:C1210–C1219. [DOI] [PubMed] [Google Scholar]
- Russell, J.M. 2000. Sodium-potassium-chloride cotransport. Physiol. Rev. 80:211–276. [DOI] [PubMed] [Google Scholar]
- Simard, C.F., G.M. Brunet, N.D. Daigle, V. Montminy, L. Caron, and P. Isenring. 2004. a. Self-interacting domains in the C-terminus of a cation-Cl− cotransporter described for the first time. J. Biol. Chem. 279:40769–40777. [DOI] [PubMed] [Google Scholar]
- Simard, C.F., N.D. Daigle, M.J. Bergeron, G.M. Brunet, L. Caron, M. Noel, V. Montminy, and P. Isenring. 2004. b. Characterization of a novel interaction between the secretory Na+-K+-Cl− cotransporter and the chaperone hsp90. J. Biol. Chem. 279:48449–48456. [DOI] [PubMed] [Google Scholar]
- Simon, D.B., F.E. Karet, J.M. Hamdan, A. DiPietro, S.A. Sanjad, and R.P. Lifton. 1996. Bartter's syndrome, hypokalaemic alkalosis with hypercalciuria, is caused by mutations in the Na+-K+-2Cl− cotransporter NKCC2. Nat. Genet. 13:183–188. [DOI] [PubMed] [Google Scholar]
- Starremans, P.G., F.F. Kersten, L.P. Van Den Heuvel, N.V. Knoers, and R.J. Bindels. 2003. Dimeric architecture of the human bumetanide-sensitive Na+-K+-2Cl− cotransporter. J. Am. Soc. Nephrol. 14:3039–3046. [DOI] [PubMed] [Google Scholar]
- Takahashi, N., D.R. Chernavvsky, R.A. Gomez, P. Igarashi, H.J. Gitelman, and O. Smithies. 2000. Uncompensated polyuria in a mouse model of Bartter's syndrome. Proc. Natl. Acad. Sci. USA. 97:5434–5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, J., and D.L. Black. 2001. A CaMK IV responsive RNA element mediates depolarization-induced alternative splicing of ion channels. Nature. 410:936–939. [DOI] [PubMed] [Google Scholar]
- Yang, T., Y.G. Huang, I. Singh, J. Schnermann, and J.P. Briggs. 1996. Localization of bumetanide- and thiazide-sensitive Na+-(K+)-Cl− cotransporters along the rat nephron. Am. J. Physiol. 271:F931–F939. [DOI] [PubMed] [Google Scholar]