Abstract
The rodent olfactory epithelium expresses more than 1000 odorant receptors (ORs) with distinct patterns, yet it is unclear how such patterns are established during development. In the current study, we investigated development of the expression patterns of different ORs in the septal organ, a small patch of olfactory epithelium predominantly expressing nine identified ORs. The presumptive septal organ first appears at about embryonic day 16 (E16) and it completely separates from the main olfactory epithelium (MOE) at about postnatal day 7 (P7). Using in situ hybridization, we quantified the densities of the septal organ neurons labeled by specific RNA probes of the nine abundant OR genes from E16 to postnatal 3 months. The results indicate that olfactory sensory neurons (OSNs) expressing different ORs have asynchronous temporal onsets. For instance, MOR256-17 and MOR236-1 cells are present in the septal organ at E16; however, MOR0-2 cells do not appear until P0. In addition, OSNs expressing different ORs show distinct developmental courses and reach their maximum densities at different stages ranging from E16 (e.g. MOR256-17) to 1 month (e.g. MOR256-3 and MOR235-1). Furthermore, early onset does not correlate with high abundance in adult. This study reveals a dynamic composition of the OSNs expressing different ORs in the developing olfactory epithelium.
Keywords: olfactory receptor, gene expression, temporal onset, olfactory sensory neuron, main olfactory epithelium
INTRODUCTION
Detection of volatile odor molecules by rodents essentially relies on G-protein coupled odorant receptors (ORs), encoded by a large gene family containing >1000 members (Young et al., 2002; Zhang and Firestein, 2002; Zhang et al., 2004a). Each olfactory sensory neuron (OSN) in the main olfactory epithelium (MOE) most likely expresses a single OR gene, although the underlying mechanisms are not fully understood (Serizawa et al., 2003; Lewcock and Reed, 2004; Shykind et al., 2004; Lomvardas et al., 2006; Fuss et al., 2007). All OSNs expressing the same OR converge their axons onto one or a few specific glomeruli in the main olfactory bulb (Mombaerts, 2006). The transcribed mRNA levels of different receptor genes can vary as many as a few hundred folds in the adult olfactory epithelium (Young et al., 2003; Zhang et al., 2004b). The onset and choice of OR expression are critical for functional maturation and glomerular targeting of the OSNs (Wang et al., 1998; Treloar et al., 2002; Zou et al., 2004).
During early development, OSNs in the olfactory epithelium are produced massively from the progenitor cells via proliferation and differentiation (Beites et al., 2005). Neurogenesis of the OSNs continues into adulthood and is offset by programmed cell death (apoptosis) to maintain a constant thickness of the epithelium (Carr and Farbman, 1993; Mahalik, 1996; Fung et al., 1997; Voyron et al., 1999; Cowan and Roskams, 2004). Expression onsets of a small subset of ORs in the olfactory epithelium have been examined using in situ hybridization, revealing a time window between E11.5 and E14 (Strotmann et al., 1995; Sullivan et al., 1995; Saito et al., 1998). Because of the vast number of ORs expressed in the MOE, it is still elusive whether OSNs expressing different ORs have similar onsets and growth rates during development.
In contrast to the MOE, the septal organ, a small island of olfactory epithelium located at the ventral base of the nasal septum, offers a much smaller and simpler system to study development of OR expression patterns. The presumptive septal organ starts to appear at about E16 in the rat and gradually separates from the MOE (Giannetti et al., 1995; Oikawa et al., 2001). In adult mice, the septal organ expresses nine abundant ORs in greater than 90% of the cells (Kaluza et al., 2004; Tian and Ma, 2004), which makes it practical to perform a detailed analysis on development of the OR expression patterns in this region. In the current study, we quantified the densities of OSNs expressing each of the nine ORs in the mouse septal organ from E16 to 3 months by in situ hybridization. The data demonstrate that the septal organ OSNs expressing different ORs have asynchronous temporal onsets. Different subtypes of OSNs also show differential developmental courses and reach their highest densities at different stages ranging from E16 to 1 month. Therefore, the expression level of a particular OR in the adult olfactory epithelium does not correlate with its temporal onset. Consequently, the relative contributions of OSNs expressing different ORs in the olfactory epithelium alter with developmental stages.
MATERIALS AND METHODS
Tissue Preparation
Wide-type C57BL/6 mice (1 and 3 months old) were purchased from Charles River. Breeding pairs of OMP-GFP mice, in which the coding region of olfactory marker protein (OMP) was replaced by green fluorescent protein (GFP), were kindly provided by Dr. Peter Mombaerts at the Rockefeller University (Potter et al., 2001). These mice at the age of E14 to 1 month were used to facilitate identification of the septal organ during development. For embryonic mice, we timed pregnancy by placing a male mouse into the cage of a single-housed female overnight. The pregnant mice were sacrificed after 14, 16, or 18 days and the age of the embryonic mice was labeled as E14, E16, or E18, respectively. Mice were deeply anesthetized by injection of overdose ketamine (P7 to 3 months) or by hypothermia (E14 to P3) in icy water slush for 20 min. After decapitation, the heads were immediately put into 4% paraformaldehyde (Sigma) overnight at 4°C. The tissues were then decalcified in 0.5 M EDTA (pH 8.0, ethylenediaminetetraacetic acid) for 2 days. The nose was cut into 20-μm coronal sections on a cryostat. The procedures of animal handling and tissue harvesting were approved by the institutional animal care and use committee of the University of Pennsylvania.
In Situ Hybridization
Digoxigenin (DIG) labeled RNA probes of the OR genes were generated using DIG RNA Labeling Kit (SP6/T7) (Roche no. 11175025, Indianapolis, IN), and the sequences are included in the supplemental data (sTable 1). The sections were hybridized with the RNA probes (~1 μg/mL) overnight at 65°C in the hybridization solution (50% deionized formamide, 10 mM Tris-Cl (pH 8.0), 10% dextran sulfate, 1X Denhardt’s solution, 200 μg/mL tRNA, 0.6 M NaCl, 0.25% SDS and 1 mM EDTA), followed by high-stringency washing steps sequentially in 2×, 0.2×, and 0.1× SSC at 65°C (20 min in each solution). The sections were then incubated with alkaline phosphatase (AP)-conjugated anti-DIG antibody (Anti-digoxigenin-AP, Roche no. 11093274) at room temperature (RT) for 1 h and the signals were detected by nitro blue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (NBT/BCIP, Roche no. 1681451) (2 h at RT).
For double in situ hybridization, we followed the procedures described previously (Ishii et al., 2004). The tissue sections were hybridized with mixed fluorescein (FLU) and DIG-labeled probes. After high-stringency washing steps, the sections were reacted with horse-radish peroxidase-conjugated antifluorescein (1:100, Roche no. 11426346) and AP-anti-DIG (1:1000) antibodies (1 h at RT) followed by incubation in Tyramide-biotin (1:50, 10 min at RT) (no. NEL700A, New England Nuclear, PerkinElmer, Boston, MA) to amplify the peroxidase signal. The signals were visualized by Streptavidine-Alexa 488 (no. S11223, Molecular Probes, Eugene, OR) (1:300, 30 min in the dark at RT) and Fast Red (2-Hydroxy-3-naphthoic acid-2′-phenyl-ani-lide phosphate (HNPP) Fluorescent Detection Set, Roche no. 11758888) (1:100, 30 min in the dark at RT), respectively. The pictures were taken under a confocal microscope (Nikon Eclipse E600). Control experiments confirmed that there were no stained cells with the secondary antibodies alone (data not shown).
Cell Counting and Analysis
For each side of a coronal section, the septal organ was outlined from the apical surface to the basement membrane, which separates the olfactory epithelium from the laminal propria, and the cross-sectional area was measured using Canvas 9.0 (Deneba) [Fig. 2(A) in Results]. The number of animals used at each stage was as follows: E16 (9), E18 (7), P0 (9), P3 (7), P7 (7), 1 month (15), and 3 months (5). One-month wild-type and OMP-GFP mice were grouped together because no significant difference was observed. To minimize variations from the tissue source, adjacent sections from each animal (E16 to P7) were tested for different OR probes and each OR probe was hybridized onto only one section from a single septal organ. The two sides of a single animal were treated as two independent septal organs, due to significant variations between them (Kaluza et al., 2004; Tian and Ma, 2004). NBT/BCIP stained cells appeared as dark brown dots in the tissue sections and ~85–90% of the dots were unequivocally individual OSNs with a characteristic cell contour. A questionable dot was included in the cell counting only if it met the following two criteria: its diameter was greater than 50% of an averaged cell (see below) and its intensity exceeded 30% of the darkest cell in that section (the background was set to 0 and the darkest cell was set to 1 in Canvas 9.0). Occasionally, a dot was significantly bigger than a single cell and counted as two cells by visual examination.
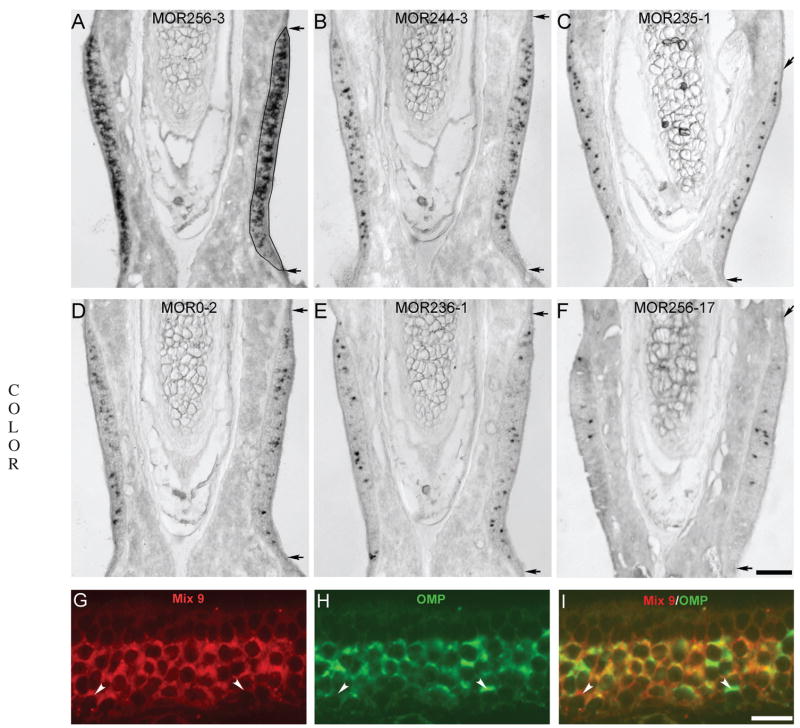
Figure 2.
Nine abundant odorant receptors are expressed in greater than 90% of the septal organ neurons in 1-month mice. A–F: Coronal sections were hybridized with the following six OR probes: MOR256-3 (A), MOR244-3 (B), MOR235-1 (C), MOR0-2 (D), MOR236-1 (E), and MOR256-17 (F). The images were taken from the region within the rectangle shown in Figure 1(L). Paired arrows mark the dorsal and ventral boundaries of the septal organ. The drawing in (A) outlines the septal organ cross-sectional area. The scale bar in (F) equals 0.1 mm and applies to (A–F). G–I: Mixed RNA probes detecting the nine OR genes (listed in Table 1, Mix 9) labeled >90% of the septal organ neurons. Coronal sections were hybridized by a mixture of nine DIG-labeled OR probes (red) (G) and a FLU- labeled OMP probe (green) (H). The combined image of G and H is shown in (I). Confocal images were taken with a z step of 1 μm. Arrowheads mark two cells labeled by either Mix 9 (left) or OMP (right) alone. Scale bar = 20 μm.
The number of NBT/BCIP cells was counted for each septal organ section (except for MOR256-3 cells at 1 and 3 months, see below) and corrected using an Abercrombie’s factor for the overcounting problem (Guillery, 2002). The ratio of the “real” number to the observed number in the Abercrombie’s formula is T/(T + h), where T = 20 μm (section thickness) and h = 8.2 μm (mean diameter of the objects along the axis perpendicular to the plane of the section). The mean diameter of the septal organ neurons in horizontal sections was measured in confocal images taken under a 60× objective from fluorescent sections as 8.2 ± 0.3 μm (n = 35). For this measurement, only the large cells (top 25% in size) were included to avoid an underestimation, because cells cut at the edge would appear smaller than those cut in the middle. The cell density for each septal organ section was then obtained as the “real” cell number/cross-sectional area in mm2.
Because NBT/BCIP labeled MOR256-3 cells in the septal organ from adult mice were too dense to be counted individually, the density of MOR256-3 cells in these sections was determined by double in situ hybridization with MOR256-3 and OMP probes (Tian and Ma, 2004). Confocal images taken with a z step of 10 μm were used to count labeled MOR256-3 cells and measure the septal organ cross-sectional area. The observed number was corrected using an Abercrombie’s factor (10/(10 + 8.2)) and then multiplied by 2 to obtain the cell density in 20 μm sections.
Statistical analysis of the cell densities of different ORs at different time points is performed using ANOVA post-hoc tests (StatView). The p value for all pairwise comparison is obtained and all averaged data are reported as mean ± standard error.
RESULTS
Development of the Septal Organ
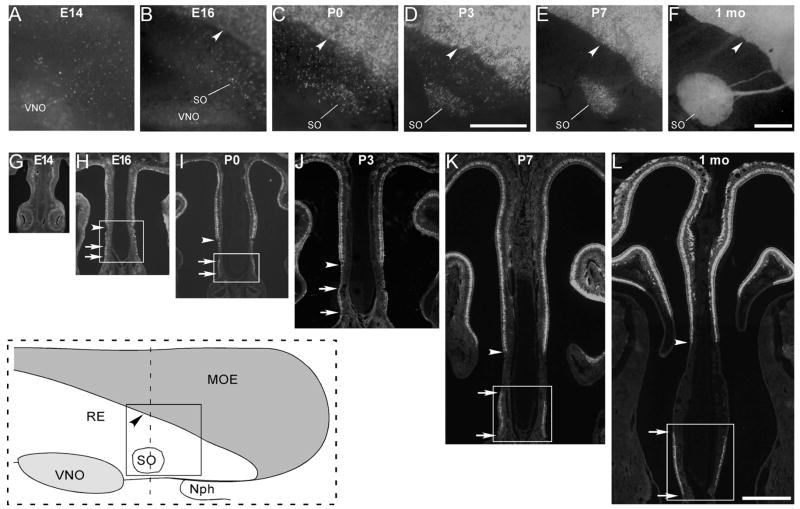
In adult mice, the septal organ is a small island of olfactory epithelium located at the ventral base of the nasal septum near the nasopharynx (inset of Fig. 1). Development of the septal organ in OMP-GFP mice was examined from E14 to postnatal 1 month in both whole-mount septal epithelia [Fig. 1(A–F)] and coronal sections [Fig. 1(G–L)]. At E14, the septal olfactory epithelium contained scattered cells with weak GFP signals, reflecting a low expression level of OMP (a marker for mature OSNs) and no evident septal organ was observed [Fig. 1(A,G)]. At E16, the MOE was separated from the respiratory epithelium with a clear boundary (marked by the arrowhead) and some cells started to cluster in the presumptive septal organ region [Fig. 1(B)]. This was more obvious in coronal sections, in which the septal organ region was distinguishable from the respiratory epithelium by the thickened olfactory-like epithelium with a higher density of OMP-GFP cells [Fig. 1(H)]. In the later stages (E18 to 1 month), OSNs continued to cumulate in the septal organ area, accompanied by gradual disappearance of the OMP-GFP cells in the transitional zone between the septal organ and the MOE (see Fig. 1). The septal organ completely separated from the MOE at about P7 [Fig. 1(E,K)], but its size at 1 month was significantly larger than that at P7 [Fig. 1(F,L)], indicating a continued expansion and development during postnatal days.
Figure 1.
The septal organ gradually separates from the main olfactory epithelium during development. A–F: Fluorescent images were taken from whole-mount septal epithelia of OMP-GFP mice at E14 (A), E16 (B), P0 (C), P3 (D), P7 (E), and 1 month (F). The inset at the lower left corner (within the dashed rectangle) is a schematic drawing of the septal epithelium from 1-month mice, and the solid square indicates the region where the image in F was taken. MOE, main olfactory epithelium; RE, respiratory epithelium; SO, septal organ; VNO, vomeronasal organ; Nph, nasophar-ynx. Arrowheads in (B–F) mark the boundary between MOE and RE in the nasal septum. Scale bars = 0.5 mm. The scale bar in (D) applies to (A–D), and that in (F) applies to (E) and (F). Due to the low fluorescent signal at E14, the image in (A) was taken with an exposure duration that was two times longer than the others. G–L: Fluorescent images were taken from coronal sections (dotted line in the inset) of OMP-GFP mice at E14 (G), E16 (H), P0 (I), P3 (J), P7 (K), and 1 month (1 mo) (L). Arrowheads in (H–L) mark the boundary between MOE and RE, and paired arrows in (H–L) mark the dorsal and ventral boundaries of the septal organ. The rectangles in (H, I, K, L) indicate the regions in which the in situ hybridization images were taken at E16 [Fig. 3(A,D,G)], P0 [Fig. 3(B,E,H)], P7 [Fig. 3(C,F,I)], and 1 month [Fig. 2(A–F)], respectively. The scale bar in L equals 0.5 mm and applies to (G–L).
Nine Predominant ORs Expressed in the Adult Septal Organ
The ORs expressed in the adult septal organ have been identified (Kaluza et al., 2004; Tian and Ma, 2004). Here, we quantified the cell densities for nine abundant ORs in 20 μm coronal sections of the septal organ from young adult mice (1 and 3 months old). Using in situ hybridization, the mRNAs of the following nine OR genes were detected in the septal organ neurons by the antisense probes: MOR256-3, MOR244-3, MOR235-1, MOR0-2, MOR236-1, MOR256-17, MOR122-1, MOR160-5, and MOR267-16 (Table 1), and six examples are shown in Figure 2(A–F). The nine probes used in this study presumably only detect their target OR genes, because seven of them share less than 40% of nucleotide homology with any other OR in the genome, and the other two, MOR235-1 and MOR236-1 sharing 75% homology to each other, detect different subsets of cells in double in situ hybridization (Tian and Ma, 2004). Together, these nine ORs were expressed in greater than 90% of the septal organ neurons, demonstrated by double in situ hybridization [Fig. 2(G-I)]. DIG-labeled Mix 9 (a mixture of the nine RNA probes) and FLU-labeled OMP probes were hybridized concurrently, and Mix 9 labeled >90% of the OMP positive cells in all six sections examined. The results indicate that a relatively complete profile of OR expression patterns is achieved in the adult septal organ.
Table 1.
The Cell Densities of Nine Abundant Odorant Receptors in the Septal Organ are Summarized for Different Developmental Stages
| OR Gene | E16 | E18 | P0 | P3 | P7 | 1 month | 3 months |
|---|---|---|---|---|---|---|---|
| MOR256-3 | 82 ± 31 (n = 6) | 320 ± 101 (n = 10) | 507 ± 55 (n = 12) | 842 ± 74 (n = 8) | 1456 ± 95 (n = 10) | 3550 ± 230 (n = 14) | 3832 ± 258 (n = 14) |
| MOR244-3 | 0 (n = 6) | 110 ± 47 (n = 8) | 134 ± 29 (n = 13) | 383 ± 30 (n = 9) | 871 ± 76 (n = 12) | 983 ± 82 (n = 20) | 1044 ± 66 (n = 20) |
| MOR235-1 | 0 (n = 6) | 23 ± 14 (n = 8) | 105 ± 20 (n = 17) | 183 ± 35 (n = 8) | 194 ± 28 (n = 12) | 447 ± 60 (n = 24) | 500 ± 54 (n = 20) |
| MOR0-2 | 0 (n = 6) | 0 (n = 6) | 137 ± 34 (n = 13) | 188 ± 35 (n = 10) | 372 ± 34 (n = 12) | 440 ± 57 (n = 22) | 454 ± 52 (n = 20) |
| MOR236-1 | 224 ± 57 (n = 8) | 218 ± 40 (n = 14) | 250 ± 38 (n = 15) | 263 ± 50 (n = 14) | 418 ± 32 (n = 12) | 424 ± 55 (n = 24) | 442 ± 49 (n = 20) |
| MOR256-17 | 222 ± 45 (n = 8) | 194 ± 26 (n = 8) | 278 ± 31 (n = 13) | 225 ± 38 (n = 8) | 307 ± 57 (n = 8) | 344 ± 82 (n = 10) | 332 ± 56 (n = 20) |
| MOR122-1 | 0 (n = 6) | 30 ± 16 (n = 6) | 78 ± 24 (n = 9) | 138 ± 38 (n = 7) | 211 ± 28 (n = 10) | 319 ± 45 (n = 18) | 331 ± 37 (n = 18) |
| MOR160-5 | 116 ± 32 (n = 6) | 125 ± 43 (n = 8) | 198 ± 30 (n = 12) | 188 ± 25 (n = 10) | 460 ± 66 (n = 12) | 310 ± 48 (n = 28) | 327 ± 40 (n = 20) |
| MOR267-16 | 46 ± 22 (n = 7) | 37 ± 19 (n = 6) | 88 ± 28 (n = 7) | 134 ± 23 (n = 7) | 290 ± 39 (n = 10) | 292 ± 44 (n = 19) | 297 ± 33 (n = 20) |
The averaged cell density per cross-sectional area of the septal organ is denoted as cell number ± standard error/mm2 with n = the number of septal organ sections (thickness = 20 μm).
Asynchronous Onsets and Differential Developments of OSNs Expressing Different ORs
We then investigated development of the expression patterns of these nine ORs in the septal organ at different stages (see Fig. 3). We started at E16 because before this stage, the septal organ cannot be positively identified and there are too few OR positive cells for quantification. At E16, the OSNs expressing five of the nine ORs were detectable (Table 1), among which MOR236-1 and MOR256-17 cells had significantly higher densities than others (p < 0.05 when compared with MOR256-3 and MOR160-5 cells, and p < 0.001 when compared with others). The mRNAs of the other four ORs, including MOR235-1 (the closest counterpart of MOR236-1), were not found. Because the septal organ was not completely separated from the MOE at this stage, the presumptive septal organ region could only be approximately outlined based on the thickening of the olfactory-like epithelium and the fact that the dorsal boundary often starts at a dent in the septal epithelium [Fig. 3(A,D,G)]. Adjacent sections were labeled by different OR probe to minimize the effects caused by potential inaccuracy in the cross-sectional area measurement (Materials and Methods Section). At E16, the number of OR positive cells usually exceeds the number of OMP positive cells. The cell density of OMP positive cells was 194 ± 34/mm2 (n = 8 septal organ sections), much lower than the sum density of the five ORs present at E16 (690/mm2, Table 1), indicating that some immature cells express ORs (Supplemental data, sFig. 1).
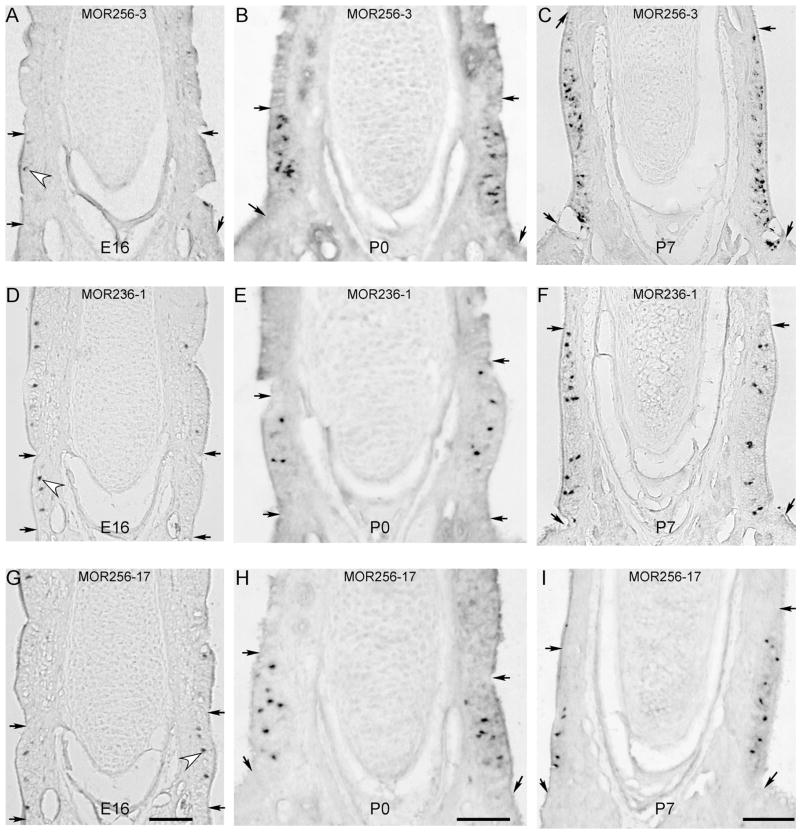
Figure 3.
The septal organ neurons expressing different ORs show distinct growth rates. A–C: Coronal sections from E16 (A), P0 (B) and P7 (C) animals were hybridized with the MOR256-3 antisense RNA probe. D–F: Coronal sections from E16 (D), P0 (E) and P7 (F) animals were hybridized with the MOR236-1 probe. G–I: Coronal sections from E16 (G), P0 (H) and P7 (I) animals were hybridized with the MOR256-17 probe. The images at E16, P0 and P7 were taken from the regions within the rectangles shown in Figure 1(H,I,K), respectively. Paired arrows mark the dorsal and ventral boundaries of the septal organ. Hollow arrowheads mark examples of labeled cells at E16. Each scale bar applies to the whole column and all scale bars = 0.1 mm. During the course of the study, we changed our picture taken system from an upright microscope (Olympus BX61) to a dissecting microscope (Olympus SZX12). We always focused on the labeled cells, even though the images taken under a dissecting microscope (B, E, H and I) appeared slightly different than the others.
At E18, the OSNs expressing three other ORs (MOR244-2, MOR122-1, and MOR235-1) appeared and MOR256-3 cells became as abundant as MOR236-1 and MOR256-17 cells (Table 1). At P0, OSNs expressing all nine ORs were evident and MOR256-3 cells became the most dominant type followed by MOR236-1 and MOR256-17 cells [Fig. 3(B,E,H)]. At P3 and P7, MOR244-3 cells became the second most abundant type after MOR256-3 cells (Table 1). The results indicate that the septal organ OSNs expressing different ORs have asynchronous onsets.
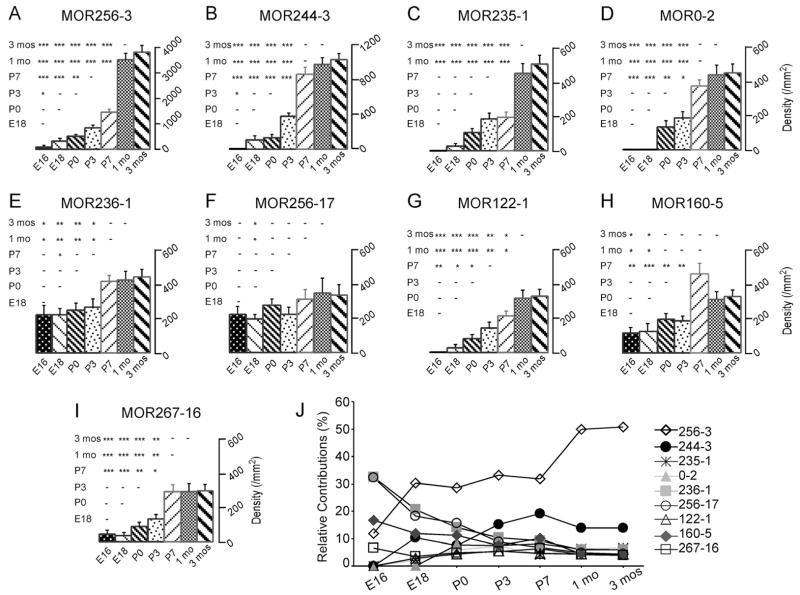
To quantify the growth rates of OSNs expressing different ORs after the onsets, we compared the cell densities for each OR at different time points [Fig. 4(A–I)]. The cell densities for three ORs (MOR256-3, MOR235-1, and MOR122-1) continued to increase after their initial appearance. Their densities reached a maximum at about 1 month. It is possible that the cell densities for some ORs (e.g. MOR256-3 and MOR235-1) would keep rising after this point, because their densities showed a trend of increment from 1 to 3 months (the last point examined), although the change was not significant. The OSNs expressing five other ORs (MOR244-3, MOR0-2, MOR236-1, MOR160-5, and MOR267-16) achieved their highest densities at P7. The density of MOR256-17 cells did not increase significantly from E16 to 3 months. The data suggest that different types of OR cells have differential developmental trajectories.
Figure 4.
The cell densities for individual odorant receptors reach a maximum at different stages in the septal organ. A–I: The averaged cell densities per cross-sectional area (cell number/mm2) are plotted for the nine ORs: MOR256-3 (A), MOR244-3 (B), MOR235-1 (C), MOR0-2 (D), MOR236-1 (E), MOR256-17 (F), MOR122-1 (G), MOR160-5 (H) and MOR267-16 (I). Error bars = standard errors. 1 mo = 1 month. 3 mos = 3 months. Pairwise comparison at all time points is obtained by ANOVA post-hoc tests and shown in the triangle matrix. The P value range is marked as *** (p < 0.001), ** (0.001 < p < 0.01), * (0.01 < p < 0.05), or – (a dash, p > 0.05). (J) Relative contributions of the septal organ cells expressing different ORs change during development. At each time point, the averaged cell density of each OR is normalized to the sum of the cell densities from all nine ORs.
Attributed to the differential onsets and developmental courses, the relative contributions of different OR cells altered during development. At any time point, the cell density for each OR was normalized to the sum density of all nine ORs, making the total contributions from these nine OR cell types as 100% [Fig. 4(J)]. MOR236-1 and MOR256-17 cells were dominant at E16 with each contributing to 33% of the cells and their cell densities slightly increased (for MOR236-1) or stayed constant (for MOR256-17) from E16 to 3 months [Fig. 4(E,F)]. However, their relative contributions continued to decline during development and reached ~5% in adults, presumably due to greater additions of other OR cells. Notably, MOR235-1 (the closest counterpart of MOR236-1) cells did not appear until E18, but displayed a similar density as MOR236-1 cells in adults [Figs. 2(C,E),4(J)]. MOR256-3 cells became dominant at P0, but its relative contribution stayed at 30% from E18 to P7 and did not reach 50% until postnatal 1 month. This finding resulted from a drastic increase of MOR256-3 cells from P7 to 1 month, but only moderate changes in other OR cell types (e.g. MOR244-3 and MOR0-2) during this period. These data suggest that early onset does not correspond with high abundance at later stages and the composition of the septal organ neurons expressing different ORs is dynamic during development.
DISCUSSION
Using in situ hybridization, we have analyzed the cell densities of nine highly expressed ORs in the septal organ from E16 to 3 months. The OSNs expressing different ORs (even closely-related ones) show asynchronous temporal onsets and differential developmental courses, and early onset does not correlate with high abundance in adult. Consequently, the relative contributions of OSNs expressing different ORs are dynamic during development.
Development of the Septal Organ
The OMP-positive OSNs start to cluster in the presumptive septal organ region at about E16 in the mouse, consistent with previous studies in the rat (Giannetti et al., 1995; Oikawa et al., 2001). The septal organ then progressively separates from the MOE with gradual disappearance of the OSNs in the transitional zone (see Fig. 1). The fact that all nine predominant septal organ ORs are also found in the most ventral zone of the MOE raises the possibility that the septal organ is a displaced patch of the ventral zone of the MOE. However, because the septal organ primarily covers only a small fraction of the ORs expressed in the ventral MOE and their relative abundance in the septal organ does not resemble that in the MOE (Tian and Ma, 2004), it is possible that the septal organ is a distinct chemosensory system with its own developmental course as previously suggested (Giannetti et al., 1995; Oikawa et al., 2001). This is also evident during postnatal development, i.e., the rat septal organ reaches a maximal size at 2–3 months while the MOE continues to expand beyond 10 months (Weiler and Farbman, 2003). Because of significant development during the postnatal days, the septal organ may be subject to modification by sensory experience.
Asynchronous Onsets of OSNs Expressing Different ORs
The OSNs expressing different ORs have asynchronous onsets in the septal organ. The cells expressing five of the nine ORs (MOR256-17, MOR236-1, MOR160-5, MOR256-3, and MOR267-16) are already present at E16, while the cells expressing the other four ORs are only evident at E18 (MOR244-3, MOR122-1, and MOR235-1) or P0 (MOR0-2) (Table 1). The temporal difference in the expression onset of these ORs is potentially more significant in the MOE. The first appearance of OR124 cells (the rat counterpart of MOR256-3) is between E12 and E14 (Strotmann et al., 1995), and the onset for MOR256-17 expression is as early as E12 (Schwarzenbacher et al., 2006). The four ORs with relatively late onsets in the septal organ (MOR244-3, MOR122-1, MOR235-1, and MOR0-2) have similar onsets in the MOE (data not shown). From the limited number of ORs examined, the expression onset of different ORs can differ by at least 6 days in the mouse olfactory epithelium (MOR256-17 at E12 and MOR0-2 after E18). Most studies including the current one investigate the expression onset by detecting the mRNAs of individual OR genes. There is no evidence suggesting independent transcriptional and translational regulation of OR expression, but it remains to be determined if the asynchronous onset holds true for protein expression of different ORs.
Notably, the asynchronous onset is observed for very similar ORs. MOR236-1 and MOR235-1, the closest counterparts in the mouse genome, are encoded by two genes located next to each other on Chromosome 2, sharing 91% similarity in the amino acid sequence (Tsuboi et al., 1999). Although their expression patterns are comparable in adults (Table 1 and Fig. 2), MOR236-1 is abundantly expressed at E16, while MOR235-1 cells do not appear until E18 (Table 1). The results suggest that the expression patterns of different ORs in the adult olfactory epithelium are not correlated with their expression onsets.
Differential Developments of OSNs Expressing Different ORs
After the initial onset, the OSNs expressing different ORs exhibit distinct developmental courses in the septal organ. For different ORs, the maximum cell densities are achieved at different time points, ranging from E16 (MOR256-17) to P7 (MOR244-3, MOR0-2, MOR236-1, MOR160-5, and MOR267-16) and 1 month (MOR256-3, MOR235-1, and MOR122-1) (see Fig. 4). The differential developmental courses may reflect the distinct proliferation and/or apoptotic rates for different OR identities (see Introduction Section). The results are consistent with a previous study using a high-throughput DNA micro-array to examine the expression levels of mouse OR genes at different developmental stages. There is a steady increase in the number of OR genes detected in the MOE during early (pre- and postnatal) development (Zhang et al., 2004b). These findings may partly explain why the number of glomeruli in the olfactory bulb gradually increases during postnatal development (Pomeroy et al., 1990). Although apoptosis continuously occurs in the olfactory epithelium, none of the nine ORs examined here shows a significant decrease in the cell density from E16 to 3 months. Loss of the entire cell population of an OR in the olfactory epithelium may be rare, consistent with the fact that elimination of existing glomeruli is never observed in a vital imaging study (LaMantia et al., 1992). However, the same microarray study mentioned earlier demonstrates that the expression levels of some OR genes fall below the detection thresholds during postnatal development (Zhang et al., 2004b). An in situ examination of more ORs in the MOE is required to address whether loss of certain OSN populations occurs.
Because the OSNs with different OR identities have asynchronous onsets and differential growth rates, the relative contributions of different types of OSNs change with developmental stages. In the septal organ, MOR256-17 and MOR236-1 cells dominate at E16 with each type contributing to ~33% of the total labeled cells by all nine OR probes (Table 1, Fig. 3). However, their relative contributions drop to ~5% in adults (see Fig. 4). On the other hand, the density of MOR235-1 cells keeps increasing after the onset at E18 and reaches about the same level as MOR236-1 cells in adults (Figs. 2, 4). The results indicate that similar expression abundance in the adult olfactory epithelium can be achieved by very different developmental trajectories.
This is also true for MOR256-3 cells, the most prevalent OSNs in the adult septal organ. MOR256-3 cells are detectable at E16, but with a significantly lower density than MOR256-17 and MOR236-1 cells (see Fig. 3). The density of MOR256-3 cells continues to increase, but its relative contribution stays at ~30% from E18 to P7 and reaches 50% at 1 month, depending on the growth rates of all nine types of OSNs (see Fig. 4). Such calculation is only meaningful if a single cell expresses only one receptor type. Extensive double in situ labeling with different combinations of these nine OR probes indicates that coexpression of any two ORs in single OSNs happens at a very low frequency in the septal organ, about 2% at P0 and 0.2% at 1 month (our unpublished data). This factor is not corrected in the above quantification.
We cannot rule out that some abundant receptors in the septal organ are missing in the analysis, but it is unlikely. OR-positive cells labeled by a mixture of the nine probes account for >90% of the OMP positive cells in the adult septal organ (see Fig. 2). At E16, the number of OR-positive cells exceeds that of OMP-GFP cells (Supplementary data, sFig. 1), suggesting OR expression precedes OMP expression in some cells, consistent with the finding that some immature, OMP-negative cells express ORs (Iwema and Schwob, 2003). Even though in the case that we did not analyze the complete set of ORs in the septal organ, the relative contributions of these nine receptors still hold true. Because developing OSNs can migrate tangentially in the olfactory epithelium (Caggiano et al., 1994; Schwob et al., 1994; Huard et al., 1998), the number of septal organ fated cells could be potentially underestimated during embryonic days such as E16.
The current study suggests that the onset and development of OR expression is not random, which may depend on the intrinsic properties of the progenitor cells and/or their microenvironment. Similar regulation may also occur during regeneration after lesion to the olfactory epithelium, because the recovered OR expression pattern closely resembles that in the control animals (Iwema et al., 2004). The developmental trajectories of the OR expression patterns may be modified by the postnatal (or even prenatal) chemical environment.
Acknowledgments
This article contains supplementary material available via the Internet at http://www.mrw.interscience.wiley.com/suppmat/1932-8451/suppmat/
Contract grant sponsor: NIDCD/NIH; contract grant number: R01 DC006213.
Contract grant sponsors: Whitehall foundation, Institute on Aging at the University of Pennsylvania (a pilot grant).
References
- Beites CL, Kawauchi S, Crocker CE, Calof AL. Identification and molecular regulation of neural stem cells in the olfactory epithelium. Exp Cell Res. 2005;306:309–316. doi: 10.1016/j.yexcr.2005.03.027. [DOI] [PubMed] [Google Scholar]
- Caggiano M, Kauer JS, Hunter DD. Globose basal cells are neuronal progenitors in the olfactory epithelium: A lineage analysis using a replication-incompetent retrovirus. Neuron. 1994;13:339–352. doi: 10.1016/0896-6273(94)90351-4. [DOI] [PubMed] [Google Scholar]
- Carr VM, Farbman AI. The dynamics of cell death in the olfactory epithelium. Exp Neurol. 1993;124:308–314. doi: 10.1006/exnr.1993.1201. [DOI] [PubMed] [Google Scholar]
- Cowan CM, Roskams AJ. Caspase-3 and caspase-9 mediate developmental apoptosis in the mouse olfactory system. J Comp Neurol. 2004;474:136–148. doi: 10.1002/cne.20120. [DOI] [PubMed] [Google Scholar]
- Fung KM, Peringa J, Venkatachalam S, Lee VM, Trojanowski JQ. Coordinate reduction in cell proliferation and cell death in mouse olfactory epithelium from birth to maturity. Brain Res. 1997;761:347–351. doi: 10.1016/s0006-8993(97)00467-8. [DOI] [PubMed] [Google Scholar]
- Fuss SH, Omura M, Mombaerts P. Local and cis effects of the H element on expression of odorant receptor genes in mouse. Cell. 2007;130:373–384. doi: 10.1016/j.cell.2007.06.023. [DOI] [PubMed] [Google Scholar]
- Giannetti N, Pellier V, Oestreicher AB, Astic L. Immunocytochemical study of the differentiation process of the septal organ of Masera in developing rats. Brain Res Dev Brain Res. 1995;84:287–293. doi: 10.1016/0165-3806(94)00195-6. [DOI] [PubMed] [Google Scholar]
- Guillery RW. On counting and counting errors. J Comp Neurol. 2002;447:1–7. doi: 10.1002/cne.10221. [DOI] [PubMed] [Google Scholar]
- Huard JM, Youngentob SL, Goldstein BJ, Luskin MB, Schwob JE. Adult olfactory epithelium contains multipotent progenitors that give rise to neurons and non-neural cells. J Comp Neurol. 1998;400:469–486. [PubMed] [Google Scholar]
- Ishii T, Omura M, Mombaerts P. Protocols for two-and three-color fluorescent RNA in situ hybridization of the main and accessory olfactory epithelia in mouse. J Neurocytol. 2004;33:657–669. doi: 10.1007/s11068-005-3334-y. [DOI] [PubMed] [Google Scholar]
- Iwema CL, Fang H, Kurtz DB, Youngentob SL, Schwob JE. Odorant receptor expression patterns are restored in lesion-recovered rat olfactory epithelium. J Neurosci. 2004;24:356–369. doi: 10.1523/JNEUROSCI.1219-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwema CL, Schwob JE. Odorant receptor expression as a function of neuronal maturity in the adult rodent olfactory system. J Comp Neurol. 2003;459:209–222. doi: 10.1002/cne.10583. [DOI] [PubMed] [Google Scholar]
- Kaluza JF, Gussing F, Bohm S, Breer H, Strotmann J. Olfactory receptors in the mouse septal organ. J Neurosci Res. 2004;76:442–452. doi: 10.1002/jnr.20083. [DOI] [PubMed] [Google Scholar]
- LaMantia AS, Pomeroy SL, Purves D. Vital imaging of glomeruli in the mouse olfactory bulb. J Neurosi. 1992;12:976–988. doi: 10.1523/JNEUROSCI.12-03-00976.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewcock JW, Reed RR. A feedback mechanism regulates monoallelic odorant receptor expression. Proc Natl Acad Sci USA. 2004;101:1069–1074. doi: 10.1073/pnas.0307986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- Mahalik TJ. Apparent apoptotic cell death in the olfactory epithelium of adult rodents: Death occurs at different developmental stages. J Comp Neurol. 1996;372:457–464. doi: 10.1002/(SICI)1096-9861(19960826)372:3<457::AID-CNE8>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Mombaerts P. Axonal wiring in the mouse olfactory system. Annu Rev Cell Dev Biol. 2006;22:713–737. doi: 10.1146/annurev.cellbio.21.012804.093915. [DOI] [PubMed] [Google Scholar]
- Oikawa T, Saito H, Taniguchi K, Taniguchi K. Immunohistochemical studies on the differential maturation of three types of olfactory organs in the rats. J Vet Med Sci. 2001;63:759–765. doi: 10.1292/jvms.63.759. [DOI] [PubMed] [Google Scholar]
- Pomeroy SL, LaMantia AS, Purves D. Postnatal construction of neural circuitry in the mouse olfactory bulb. J Neurosci. 1990;10:1952–1966. doi: 10.1523/JNEUROSCI.10-06-01952.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter SM, Zheng C, Koos DS, Feinstein P, Fraser SE, Mombaerts P. Structure and emergence of specific olfactory glomeruli in the mouse. J Neurosci. 2001;21:9713–9723. doi: 10.1523/JNEUROSCI.21-24-09713.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H, Mimmack ML, Keverne EB, Kishimoto J, Emson PC. Expression of olfactory receptors. G-proteins and Ax CAMs during the development and maturation of olfactory sensory neurons in the mouse. Brain Res Mol Brain Res. 1998;110:69–81. doi: 10.1016/s0165-3806(98)00096-0. [DOI] [PubMed] [Google Scholar]
- Schwarzenbacher K, Fleischer J, Breer H. Odorant receptor proteins in olfactory axons and in cells of the cribriform mesenchyme may contribute to fasciculation and sorting of nerve fibers. Cell Tissue Res. 2006;323:211–219. doi: 10.1007/s00441-005-0073-4. [DOI] [PubMed] [Google Scholar]
- Schwob JE, Huard JM, Luskin MB, Youngentob SL. Retroviral lineage studies of the rat olfactory epithelium. Chem Senses. 1994;19:671–682. doi: 10.1093/chemse/19.6.671. [DOI] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Nakatani H, Suzuki M, Saito M, Yoshihara Y, Sakano H. Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science. 2003;302:2088–2094. doi: 10.1126/science.1089122. [DOI] [PubMed] [Google Scholar]
- Shykind BM, Rohani SC, O’Donnell S, Nemes A, Mendelsohn M, Sun Y, Axel R, et al. Gene switching and the stability of odorant receptor gene choice. Cell. 2004;117:801–815. doi: 10.1016/j.cell.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Strotmann J, Wanner I, Helfrich T, Breer H. Receptor expression in olfactory neurons during rat development: in situ hybridization studies. Eur J Neurosci. 1995;7:492–500. doi: 10.1111/j.1460-9568.1995.tb00345.x. [DOI] [PubMed] [Google Scholar]
- Sullivan SL, Bohm S, Ressler KJ, Horowitz LF, Buck LB. Target-independent pattern specification in the olfactory epithelium. Neuron. 1995;15:779–789. doi: 10.1016/0896-6273(95)90170-1. [DOI] [PubMed] [Google Scholar]
- Tian H, Ma M. Molecular organization of the olfactory septal organ. J Neurosci. 2004;24:8383–8390. doi: 10.1523/JNEUROSCI.2222-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treloar HB, Feinstein P, Mombaerts P, Greer CA. Specificity of glomerular targeting by olfactory sensory axons. J Neurosci. 2002;22:2469–2477. doi: 10.1523/JNEUROSCI.22-07-02469.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi A, Yoshihara S, Yamazaki N, Kasai H, Asai-Tsuboi H, Komatsu M, Serizawa S, et al. Olfactory neurons expressing closely linked and homologous odorant receptor genes tend to project their axons to neighboring glomeruli on the olfactory bulb. J Neurosci. 1999;19:8409–8418. doi: 10.1523/JNEUROSCI.19-19-08409.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voyron S, Giacobini P, Tarozzo G, Cappello P, Perroteau I, Fasolo A. Apoptosis in the development of the mouse olfactory epithelium. Brain Res Dev Brain Res. 1999;115:49–55. doi: 10.1016/s0165-3806(99)00055-3. [DOI] [PubMed] [Google Scholar]
- Wang F, Nemes A, Mendelsohn M, Axel R. Odorant receptors govern the formation of a precise topographic map. Cell. 1998;93:47–60. doi: 10.1016/s0092-8674(00)81145-9. [DOI] [PubMed] [Google Scholar]
- Weiler E, Farbman AI. The septal organ of the rat during postnatal development. Chem Senses. 2003;28:581–593. doi: 10.1093/chemse/bjg047. [DOI] [PubMed] [Google Scholar]
- Young JM, Friedman C, Williams EM, Ross JA, Tonnes-Priddy L, Trask BJ. Different evolutionary processes shaped the mouse and human olfactory receptor gene families. Hum Mol Genet. 2002;11:1683. doi: 10.1093/hmg/11.5.535. [DOI] [PubMed] [Google Scholar]
- Young JM, Shykind BM, Lane RP, Tonnes-Priddy L, Ross JA, Walker M, Williams EM, et al. Odorant receptor expressed sequence tags demonstrate olfactory expression of over 400 genes, extensive alternate splicing and unequal expression levels. Genome Biol. 2003;4:R71. doi: 10.1186/gb-2003-4-11-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Firestein S. The olfactory receptor gene superfamily of the mouse. Nat Neurosci. 2002;5:124–133. doi: 10.1038/nn800. [DOI] [PubMed] [Google Scholar]
- Zhang X, Rodriguez I, Mombaerts P, Firestein S. Odorant and vomeronasal receptor genes in two mouse genome assemblies. Genomics. 2004a;83:802–811. doi: 10.1016/j.ygeno.2003.10.009. [DOI] [PubMed] [Google Scholar]
- Zhang X, Rogers M, Tian H, Zou DJ, Liu J, Ma M, Shepherd GM, et al. High-throughput microarray detection of olfactory receptor gene expression in the mouse. Proc Natl Acad Sci USA. 2004b;101:14168–14173. doi: 10.1073/pnas.0405350101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou DJ, Feinstein P, Rivers AL, Mathews GA, Kim A, Greer CA, Mombaerts P, et al. Postnatal refinement of peripheral olfactory projections. Science. 2004;304:1976–1979. doi: 10.1126/science.1093468. [DOI] [PubMed] [Google Scholar]