Abstract
Background
The bacterium Moraxella osloensis is a mutualistic symbiont of the slug-parasitic nematode Phasmarhabditis hermaphrodita. In nature, P. hermaphrodita vectors M. osloensis into the shell cavity of the slug host Deroceras reticulatum in which the bacteria multiply and kill the slug. As M. osloensis is the main killing agent, genes expressed by M. osloensis in the slug are likely to play important roles in virulence. Studies on pathogenic interactions between bacteria and lower order hosts are few, but such studies have the potential to shed light on the evolution of bacterial virulence. Therefore, we investigated such an interaction by determining gene expression of M. osloensis in its slug host D. reticulatum by selectively capturing transcribed sequences.
Results
Thirteen M. osloensis genes were identified to be up-regulated post infection in D. reticulatum. Compared to the in vitro expressed genes in the stationary phase, we found that genes of ubiquinone synthetase (ubiS) and acyl-coA synthetase (acs) were up-regulated in both D. reticulatum and stationary phase in vitro cultures, but the remaining 11 genes were exclusively expressed in D. reticulatum and are hence infection specific. Mutational analysis on genes of protein-disulfide isomerase (dsbC) and ubiS showed that the virulence of both mutants to slugs was markedly reduced and could be complemented. Further, compared to the growth rate of wild-type M. osloensis, the dsbC and ubiS mutants showed normal and reduced growth rate in vitro, respectively.
Conclusion
We conclude that 11 out of the 13 up-regulated M. osloensis genes are infection specific. Distribution of these identified genes in various bacterial pathogens indicates that the virulence genes are conserved among different pathogen-host interactions. Mutagenesis, growth rate and virulence bioassays further confirmed that ubiS and dsbC genes play important roles in M. osloensis survival and virulence, respectively in D. reticulatum.
Background
As the dialog between a host and bacterium requires the coordinated activity of many bacterial gene products in response to the host [1], investigating the bacterial pathogenesis in a diverse set of pathogen-host interactions can contribute to our understanding of the evolution of bacterial virulence. There have been extensive studies on bacteria pathogenic to higher order hosts [2-7], particularly the zebrafish and mouse which have emerged as model animals for the study of bacterial pathogenesis [8]. These studies have provided ample information on the virulence genes essential for pathogenesis, yet we know little about the origin and evolution of these genes. Characterizing the bacterial genes expressed in hosts representing different evolutionary history may enable us to gain a better understanding of the evolution of bacterial virulence [1]. For example, the pathogenic interaction between the nematode Caenorhabditis elegans and bacterium Salmonella enterica has showed a remarkable overlap between Salmonella virulence factors required for human and nematode pathogenesis [9]. While there are several reports of bacterial illness in snakes, tortoises, and reptiles [10-13], little is known about bacteria-involved infections in a slug host [14].
As lower order invertebrates, mollusks are proven to be excellent model systems for studies in neurophysiology, behavioral ecology and population genetics [15]. The slug Deroceras reticulatum is one of the important mollusk invertebrates. Moraxella osloensis is a gram-negative, oxidase positive, aerobic bacterium within the family Moraxellaceae in the gamma subdivision of the purple bacteria. This bacterium has recently been identified as one of the natural symbionts of a bacteria-feeding nematode, Phasmarhabditis hermaphrodita (Rhabditida: Rhabditidae), which is a lethal endoparasite of slugs [16,17], including the slug D. reticulatum [18]. In nature, bacteria colonize the gut of nematode infective juveniles (IJs) which represent a specialized stage of development adapted for survival in the unfavorable environment. The IJs seek out and enter the slug's shell cavity through the posterior mantle region. Once inside the shell cavity, the bacteria are released, and the IJs resume growth, feeding on the multiplying bacteria [18-20]. The infected slugs die in 4–10 days, and the nematodes colonize the entire cadaver and produce next generation IJs which leave the cadaver to seek a new host [18].
The lethality of these nematodes to slugs has been shown to correlate with the number of M. osloensis cells carried by IJs [20]. Tan and Grewal [20] demonstrated that the 72 h old M. osloensis cultures inoculated into the shell cavity were highly pathogenic to the slug. They further reported that M. osloensis produced an endotoxin which was identified to be a rough type lipopolysaccharide (LPS) with a molecular weight of 5300 KD, and the purified M. osloensis LPS was toxic to the slug with an estimated 50% lethal dose of 48 μg when injected into the shell cavity [21]. Although these studies laid the foundation for the bacteria-slug interaction, the virulence mechanisms of M. osloensis that result in pathogenesis and slug mortality are not established. The present study was designed to determine the molecular and genetic basis of M. osloensis virulence to the slug D. reticulatum.
Several techniques have been developed to study bacterial genes that are expressed during infection or that are required for virulence in the host during infection. The most commonly used techniques include in vivo expression technology (IVET) [22], signature-tagged mutagenesis (STM) [23] and differential fluorescence induction (DFI) [24]. Recently, the selective capture of transcribed sequences (SCOTS) technique has been developed to study bacterial gene expression specifically in macrophage [25]. This technique has been used to identify bacterial virulence factors, and has been demonstrated to be capable of identifying genes expressed in specific tissues of infected animals [5,26]. More recently, microarray analysis has also been applied in monitoring bacterial gene expression during host infection [27]. As the host and bacterial cDNAs can be easily differentiated using SCOTS, and this technique does not require prior genetic information of the pathogen, we applied SCOTS to determine M. osloensis gene expression in the slug host D. reticulatum at two time points following infection. Because important changes in gene expression can occur in bacteria during transition from active growth to stationary phase [5,28], we also investigated differential gene expression of 72 h M. osloensis in the in vitro cultures (stationary phase) relative to 24 h cultures (log phase) to confirm the infection specificity of the in vivo expressed genes. Mutational analyses (virulence of mutants to the slug and in vitro growth rate of mutants) were also carried out to examine the roles of selected genes.
Results
Differentially in vivo expressed genes
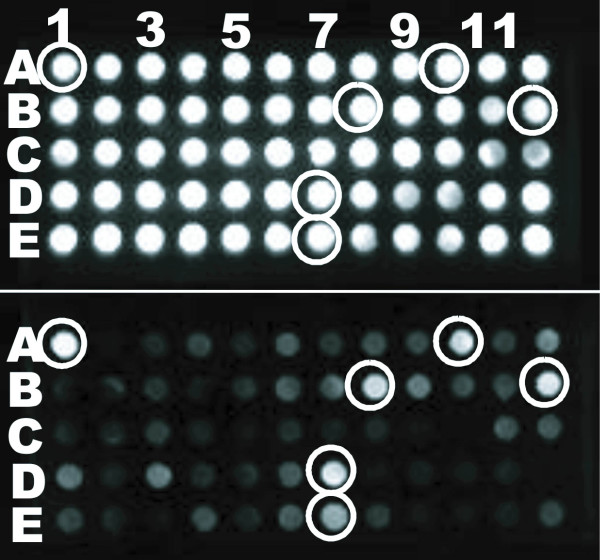
To identify M. osloensis transcripts specifically expressed in D. reticulatum, we injected slugs with 5 × 107 bacterial cells. We evaluated colonization by analyzing M. osloensis cell counts, and found that bacterial counts varied from 105 to 108 colony-forming units (CFU) per D. reticulatum 48 h or 96 h post-inoculation, indicating colonization and persistence of the injected bacteria in the slug. Transcripts expressed by M. osloensis in the slug at 48 h and 96 h post inoculation were identified by subjecting the in vivo cDNAs to three iterations of SCOTS in the presence of the transcripts expressed by 48 h late log-phase M. osloensis in vitro cultured in the Brain Heart Infusion (BHI) (Difco) medium. The enriched cDNAs were cloned into a TA vector. Each individual clone was further screened by dot blot hybridization (Fig. 1). The individual clones with signals stronger than or present on the blot hybridized to the in vivo cDNAs compared to that with signals on the blot hybridized to in vitro cDNAs were chosen for nucleotide sequencing and analysis. A total of 97 clones (27 from 48 h post inoculation, and 70 from 96 h post inoculation) were sequenced. These screened cDNAs represented the differentially expressed genes within the slug but in lower abundance or absent in the 48 h in vitro cultures. A fraction of the identified sequences was further confirmed to be M. osloensis specific by PCR amplification of M. osloensis genomic DNA with the designed primers based on identified sequences, and the results confirmed that M. osloensis transcripts were successfully isolated from the infected slugs using the SCOTS technique. The identified sequences were analyzed using the non redundant algorithms of BLAST [29] in the website of NCBI (National Center for Biotechnology Information) [30]. For each identified sequence, about two to five cDNA clones were detected in the screened library, and these clones showed the same hits in the GenBank. In this study, the identity of up-regulated sequences at 48 h and 96 h post infection was identical. We identified 13 distinct sequences (arbitrarily termed as M1, M2, M3...M13) representing 13 up-regulated genes (Table 1) which carry putative functions in cell structure integrity, energy metabolism, degradation, and translocation. Three of these sequences, M11, M12, and M13, have similarity with genes encoding hypothetical proteins, and further analysis by inverse PCR amplification and sequencing demonstrated that they were similar to the genes encoding preprotein translocase (SecA), acyl-coA synthetases (Acs), and acetyl-coA carboxylase (Acc). One of the sequences (M3) did not exhibit similarity to any genes or gene products in current databases, and is possibly novel. M1, M2 and M4 genes contain translated sequences of "Gly-Asp-Pro-Asp" repeats, and are similar to membrane proteins. Sequence M5 shows similarity with the gene encoding protein-disulfide isomerase (DsbC) in other bacteria. Sequences M6, M8, and M10 have similarity to genes that encode iron regulation related proteins in other bacteria; thus, the identification of these genes suggests that iron availability in slug host may be limited. The sequence M7 shares similarity to the gene encoding acetyltransferase (Ats) that functions in the energy metabolism and is often involved in antigen synthesis [31]. The sequence M9 has similarity with the predicted gene β-carboxymuconolactone decarboxylase (PCA) which encodes enzymes participating in the conversion of protocatechuate to succinate and acetylcoenzyme A (Acyl-CoA) [32].
Figure 1.
One of the dot blots (exposing 2 min) showing enriched cDNAs hybridized to a digoxigenin-labeled probe generated from normalized in vivo cDNAs (upper) or normalized in vitro cDNAs (lower) after three rounds of SCOTS normalization. The dots at the same position in the two arrays were loaded with the same amplicon of each individual clone from the enriched cDNA library, and the concentration of probes was standardized to be the same.
Table 1.
SCOTS identified M. osloensis genes differentially expressed in vivo
| Seq | Gene | Accession | %Identity/%Similaritya | E-value | Spanb | Possible functionc |
| M1 | sclB | DQ324274 | 50/63, Streptococcus | 9e-24 | 96 | Surface lipoprotein |
| M2 | vspC | DQ324276 | 46/56, Mycobacterium | 4e-06 | 50 | Ala and Pro rich protein |
| M11 | secA | DQ324262 | 77/88, Psychrobacter | 5e-127 | 299 | Preprotein translocase |
| M12 | acs | DQ324263 | 76/89, Psychrobacter | 0 | 442 | Acyl-CoA synthetases |
| M13 | acc | DQ324264 | 85/90, Psychrobacter | 0 | 480 | Acetyl-CoA carboxylase |
| M4 | spp | DQ324275 | 50/63, Lactobacillus | 2e-05 | 37 | Surface protein precursor |
| M5 | dsbC | DQ324260 | 49/67, Psychrobacter | 5e-70 | 257 | Disulfide isomerase |
| M6 | abp | DQ324271 | 80/86, Acinetobacter | 2e-51 | 145 | ATP-binding protein |
| M7 | ats | DQ324266 | 31/46, Psychrobacter | 7e-11 | 163 | Acetyltransferases |
| M8 | ubiS | DQ324261 | 71/83, Psychrobacter | 0 | 557 | Ubiquinone synthesis |
| M9 | pca | DQ324272 | 67/80, Psychrobacter | 3e-46 | 131 | Carboxymuconolactone decarboxylase |
| M10 | adh | DQ324273 | 90/96, Acinetobacter | 5e-55 | 114 | Aldehyde dehydrogenase |
| M3 | No similarity | Unknown |
a %Identity/%Similarity percentages apply to amino acids.
b Span means the number of amino acids over which similarity was detected.
c Genes with highest similarity to SCOTS identified sequences.
Differentially in vitro expressed genes at the stationary phase
In order to determine if the identified in vivo expressed genes are infection-specific, the differential expression of transcripts corresponding to 72 h M. osloensis cultures (stationary phase, data not shown) relative to 24 h cultures (early-log phase) was examined by SCOTS technique. Nine sequences (descriptive Mo1, Mo2...Mo9) were identified to be differentially expressed in 72 h relative to 24 h cultures (Table 2). One sequence (Mo9) did not show any similarity in current NCBI databases, and is thus possibly novel for M. osloensis. Sequences Mo2 and Mo4 have similarity to transposases and integrase, Mo6 is similar to topoisomerase, Mo1 has high similarity to Actin-like ATPase, and Mo7 is similar to ATPase involved in DNA replication. Two sequences, Mo3 and Mo8 are similar to genes of ubiS and acs that were also found to be differentially expressed under in vivo condition, suggesting that these two genes are not infection specific.
Table 2.
SCOTS identified genes exclusively expressed by 72 h in vitro cultures
| Seq | Gene | Accession | %Identity/similaritya | E-value | Spanb | Possible functionc |
| Mo1 | ala | DQ904630 | 87/94, Flavobacterium | 1e-27 | 86 | Actin-like ATPase |
| Mo2 | tra | DQ904631 | 72/78, Psychrobacter | 6e-13 | 52 | Transposase |
| Mo3 | ubiS | DQ904632 | 97/100, Moraxella | 1e-12 | 47 | Ubiquinone synthesis |
| Mo4 | int | DQ904633 | 75/84, Psychrobacter | 3e-21 | 105 | Integrase |
| Mo5 | DQ324278 | 80/90, Psychrobacter | 1e-46 | 110 | Conserved protein | |
| Mo6 | top | DQ324269 | 71/88, Psychrobacter | 2e-28 | 80 | Topoisomerase |
| Mo7 | pol | DQ904634 | 38/58, Psychrobacter | 4e-17 | 134 | DNA polymerase |
| Mo8 | acs | DQ904635 | 64/82, Acinetobacter | 3e-24 | 79 | Acyl-CoA synthetase |
| Mo9 | No similarity | Unknown |
a %Identity/%Similarity percentages apply to amino acids.
b Span means the number of amino acids over which similarity was detected.
c Genes with highest similarity to SCOTS identified sequences.
Mutational analysis of identified genes
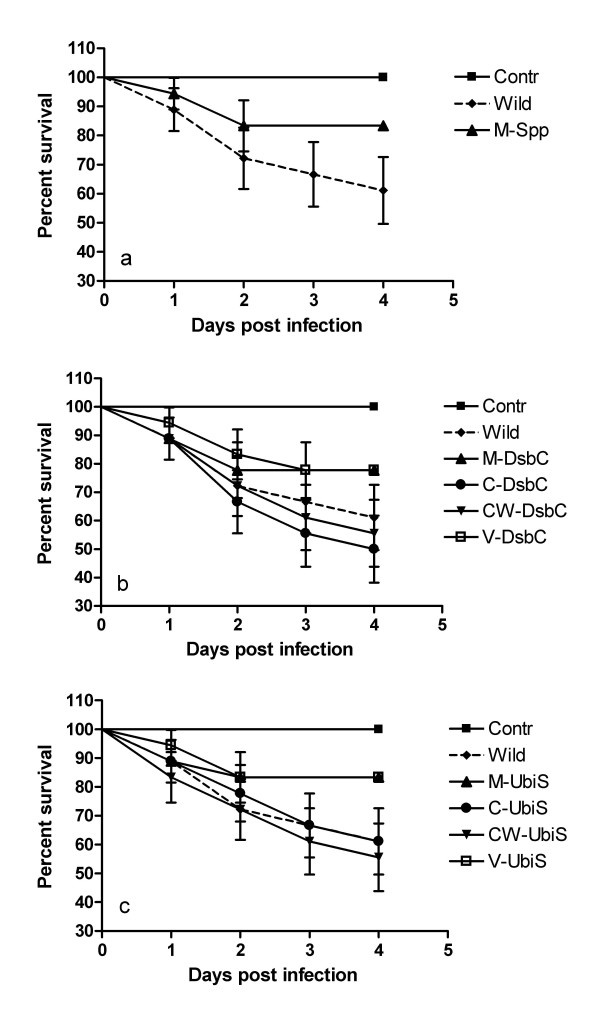
To investigate the role of up-regulated genes in M. osloensis in the slug, three mutants of selected genes were constructed using inverse PCR combined with insertional inactivation. The selected genes represent three distinct groups: cell surface protein (M4, spp) which has been previously reported as a virulence gene in pathogenic bacteria [33,34], protein-disulfide isomerase (M5, dsbC) which has been recently identified as a virulence gene in pathogenic bacteria associated with higher order hosts [35], and ubiquinone synthetase (M8, ubiS) which has been reported to be important for bacterial survival and was found to be up-regulated in vitro at the stationary phase [36,37]. As described in Materials and Methods, the inverse PCR was performed using primers with the engineered Ape I and Bma I sites. The 800-bp non-polar kanamycin resistance gene was inserted into the 50-bp internal deletion within the open reading frame (ORF) of spp, dsbC, or ubiS. Compared to the whole sequences of dsbC and ubiS amplified in this study, the insertions were located 250 bp downstream from the ATG codon of dsbC, and 300 bp downstream from the GTG start codon for ubiS. After natural transformation and double recombination, the resulting isogenic mutants were obtained in vitro after 72 h incubation, and they were termed as M-spp, M-dsbC, and M-ubiS, respectively. The virulence of wild type and mutants was determined by direct inoculation into the slug D. reticulatum and subsequent survival analysis. All three mutants demonstrated the level of virulence similar to the wild-type strain in the first two days (P > 0.05) (Fig. 2). However, at 3 days post infection, all three mutants produced significantly lower slug mortality relative to the wild-type parent strain (P = 0.036, 0.015, and 0.023 for M-spp, M-dsbC, and M-ubiS, respectively).
Figure 2.
The slug survival post M. osloensis infection was plotted with Kaplan-Meier plots. The slug mortality caused by bacterial cultures is significantly different between wild type and mutants post 2-days infection. Contr, Saline; Wild, wild-type M. osloensis; M-Spp, mutant of spp gene; M-DsbC, mutant of dsbC gene; V-DsbC, dsbC mutant containing the empty vector; C-DsbC, the complemented dsbC mutant; CW-DsbC, wild-type M. osloensis containing the plasmid borne dsbC gene (CW-dsbC serves as a control for C-dsbC); M-UbiS, mutants of ubiS gene; V-UbiS, ubiS mutant containing the empty vector; C-UbiS, the complemented ubiS mutant; CW-UbiS, wild-type M. osloensis containing the plasmid borne ubiS gene (CW-ubiS serves as a control for C-ubiS).
We further complemented the constructed mutants. The full length and promoter region of dsbC and ubiS were obtained by inverse PCR strategy with the aid of six pairs of primers (data not shown) and six restriction enzymes (Ape I, EcoR I, Hinf I, Pst I, Bma I, and Bxa I).
Amplification of the spp ORF was not successful due to the presence of repetitive sequences, and thus was not used for further analysis. The ORF of dsbC and ubiS were 843 bp and 1671 bp, and the blast searches confirmed that they were similar to protein-disulfide isomerase and ubiquinone synthesis genes, respectively. The sequence analysis revealed that dsbC gene had an N-terminal signal peptide, and contained thioredoxin-like domain with the characteristic Cys-x-x-Cys active site. The complemented mutants, termed as C-dsbC and C-ubiS, were obtained by transferring intact dsbC and ubiS genes borne in the plasmid into the respective M. osloensis mutant by electroporation. The level of virulence of complemented mutants in the slug was similar to wild-type strains (Fig. 2).
Growth rates of mutants
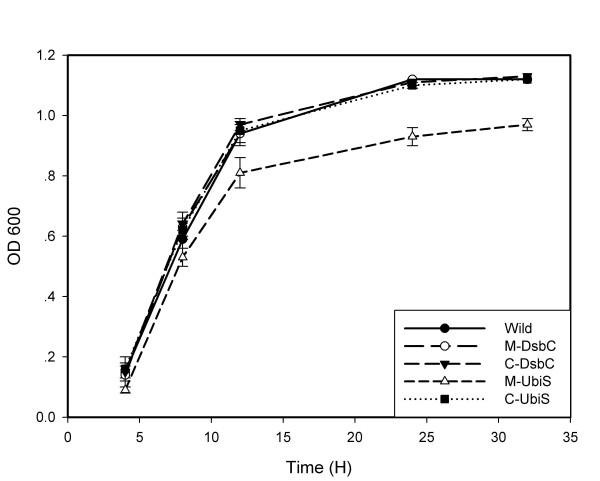
As reduced virulence of mutants may result from growth defect, the growth rates of dsbC and ubiS mutants in the BHI medium were examined. Compared to the wild-type M. osloensis, the dsbC mutant did not show any growth defect in vitro (Fig. 3). However, the ubiS mutant showed significant reduction in growth rate in vitro. The ubiS mutant grew slower than the wild type and its viability (shown as OD600) was about 12% lower relative to the wild-type strain after 12 h (Fig. 3), and the growth defect became evident at the time of the late log phase entering into the stationary phase. In addition, the growth defect phenotype was fully complemented by the trans-complementation with the gene itself.
Figure 3.
Growth curve for M. osloensis in culture derived from measurements of OD 600 over time. Wild: wild-type M. osloensis; M-DsbC: mutant of dsbC gene; C-DsbC: complemented dsbC mutant; M-UbiS: mutant of ubiS gene; C-UbiS: complemented ubiS mutant. All data are shown as the mean of three repeated experiments with standard errors.
Discussion
The current study is the first description of virulence genes expressed by a bacterium in a slug host. The SCOTS analysis demonstrates that M. osloensis expressed 13 genes in the slug host when compared with log-phase bacteria growing in vitro in the BHI medium. Comparison with differentially expressed genes by the 72 h relative to 24 h in vitro cultures revealed that 11 of the 13 genes appeared infection specific. Among these infection-specific genes, some genes likely play roles in energy metabolism, and may affect virulence indirectly by sustaining the basic cell functions during infection. For example, the pca gene identified here plays a role both in nutrition and detoxification in other bacteria [38]. Gene abp identified in this study is similar to the genes encoding ATP binding proteins that confer multiple resistance and pH adaptation. Blast search reveals that abp also shares high similarity to the polysaccharide export protein gene. In case of pathogenic bacteria, polysaccharide is often secreted as an evolutionary adaptation to adhere to host surfaces and to escape from drying and host immune defense [39,40]. Therefore, it is possible that abp gene product may play an important role in M. osloensis survival in the slug.
ubiS gene exhibits sequence similarity to the gene encoding protein that functions in ubiquinone synthesis. Ubiquinone is a central component of the electron transport chain under aerobic conditions and functions in the formation of disulfide bonds in periplasmic proteins by facilitating the reoxidation of protein-disulfide isomerase [36,37]. As ubiS gene was also up-regulated in the stationary phase in M. osloensis, this gene may be important for bacterial survival during starvation and could be induced by the stress conditions. Further examination of ubiS mutant growth in vitro indicated that the reduced virulence of this mutant may be due to the reduced growth rate.
Preproteins are exported primarily across the cytoplasmic membrane to the periplasm or the outer membrane by the sec system composed of the translocase SecYEG (a protein-conducting channel) and SecA (an ATP-dependent motor protein) proteins [41-43]. SecA interacts dynamically with the SecYEG components to drive the transmembrane movement of newly synthesized preproteins [44]. Tomkiewicz et al. [45] showed that in Escherichia coli SecA drives a constant rate of preprotein translocation consistent with a stepping mechanism of translocation. Further, the secA expression is subject to the translational control in response to the cellular activity of protein translocation [44,46]. Therefore, the up-regulation of secA gene in vivo in M. osloensis suggests that the expression of secA is regulated by the infection process.
Three of the identified genes, sclB, vspC, and spp, are similar to the genes encoding structural proteins of the outer membrane. Outer membrane proteins are known to be critical for establishment of disease in the host by causing resistance to immune killing [33,47] as demonstrated in a fish pathogenic bacterium, Edwardsiella tarda [34]. Virulence of spp mutant to the slug is significantly reduced indicating that surface membrane proteins are also important for M. osloensis infection in the slug.
Genes ats, adh, acs and acc found to be up-regulated in vivo in M. osloensis are similar to the genes reported to be important for the virulence in other bacterial pathogens. ats gene is similar to the gene for acetyltransferase which has been known as an effector protein in bacterium Yersinia [48], and plays an important role in regulating biological signaling. ald gene is similar to the gene for aldehyde dehydrogenase (AldH). In Vibrio cholerae, aldH gene has been found to be located in a pathogenicity island in epidemic and pandemic strains but absent from non-pathogenic strains [49]. Gene acs shares sequence similarity with the gene of long chain fatty acid CoA synthetase (FadD), an enzyme involved in lipid synthesis and whose expression may be important at various stages of infection [50-53]. fadD gene expression has been demonstrated to be important in virulence in a number of organisms, including Xanthomonas campestris [54] and Salmonella enterica serovar Typhimurium [55]. In this study, acs gene was identified to be up-regulated both in vivo and in vitro stationary phase. Thus, the expression of acs protein may be related to M. osloensis survival and virulence under in vivo conditions.
The gene dsbC belongs to the gene family dsb involved in disulfide bond exchange which catalyses the folding of various factors including virulence determinants such as the components of type III secretory machinery and assembly of type II secreted subunits into effector proteins in a number of bacteria [56-58]. Various gram-negative bacterial pathogens use type II or III secretion system as a basic virulence mechanism [2]. Dsb protein has been identified as a virulence factor in diverse bacterial pathogens associated with different higher order hosts. In pathogenic Shigella flexneri, dsb gene is necessary for intracellular survival and cell-to-cell spread in the host [57]. In the fish pathogenic bacterium, Flavobacterium psychrophilum, mutants of Dsb-like protein gene exhibited reduced virulence and cytotoxicity [59]. dsb gene was also identified as a virulence factor by SCOTS technique in the pig pathogenic bacterium Actinobacillus pleuropneumoniae [30]. Mutation of dsbC in Bordetella pertussis resulted in decreased toxin secretion [60]. In our study, the dsbC mutant showed significantly reduced virulence to the slug, but its normal growth rate in vitro confirms that dsbC serves as a virulence gene in M. osloensis in the slug host.
Previous studies by Tan and Grewal [21] showed that lipopolysaccharide (LPS) of M. osloensis alone was sufficient to cause slug mortality. Compared to the virulence defects of dsbC and ubiS mutant, it may be hypothesized that there is relationship between LPS and DsbC or UbiS despite the current lack of experimental evidence. The link between UbiS and Dsb has been reported in a study in Escherichia coli which demonstrated that disulfide bond formation in exported proteins is catalyzed by Dsb protein, and is directly coupled to the electron transport chain via reoxidation of Dsb protein by either ubiquinone or menaquinone [35]. In addition, most animals possess agglutinating activity to recognize self or non-self particles, and the agglutinins are believed to be lectins in invertebrates. According to Barker [15], the land slug Incilaria contains three lectins which show significant sequence similarity to other animals, having two disulphide bonds which may be related to lectin activity. Since dsb gene family in prokaryotes has the ability to rearrange non-native disulfides to their native configuration [61], it is proposed that dsbC functioning as a virulence factor in M. osloensis may be related to the host self/non-self recognition.
Conclusion
In this study, no differences in M. osloensis gene expression between 48 h and 96 h post inoculation were observed indicating that the infection was established in 48 h. Thirteen M. osloensis genes were up-regulated in the slug. Most of these genes have been previously identified in other bacterial pathogens, suggesting the conserved nature of bacterial virulence genes. Among the 13 up-regulated genes, acs and ubiS were also expressed in the bacterial stationary phase in vitro, and ubiS was further confirmed to be important for M. osloensis survival in the slug. The remaining 11 genes were infection specific. One of these genes, dsbC, belonging to dsb gene family, was confirmed to be a virulence gene in M. osloensis based on the mutagenesis study, growth rate and virulence bioassays. To our knowledge this is the first report on bacterial gene expression in a slug host interface.
Methods
Bacteria, slugs, and culture conditions
A type strain of Moraxella osloensis acquired from the American Type Culture Collection (ATCC) was used in this study. The bacteria were cultured in BHI (Difco) broth at 25°C, and confirmed to be M. osloensis by 16S rDNA sequencing. The association between this bacterial strain and the nematode P. hermaphrodita had been previously evaluated by assessing the recovery of infective juveniles in the bacterial cultures. The growth rate of M. osloensis was determined by turbidity measurements according to Tortora et al. [28]. Slug D. reticulatum adults were collected from the field and fed on pieces of fresh carrots at 18°C for at least 12 days. All the slugs were fed on sterile water for three days to wash the intestine before bacterial infection. Only healthy actively moving adult slugs were used in experiments.
General techniques
Bacterial genomic DNA was prepared using standard method for Gram negative bacteria [62]. Biotinylation of bacterial genomic DNA was obtained with EZ-Link Psoralen-PEO-Biotin (Pierce) according to the manufacturer's instructions. The total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. RNA samples were treated with RNase-free DNase I (Ambion, Austin, TX) according to the manufacturer's guidelines, and were concentrated by spectrophotometer and gel electrophoresis. Total RNA was converted to first-strand cDNA using Superscript II reverse transcriptase (Invitrogen RT-PCR kit) according to the manufacturer's instructions. First strand cDNA was made double-stranded with Klenow fragment (NEB, Beverly, MA) as described by Froussard [63]. Restriction endonucleases and ligase enzymes (Promega) were used according to the manufacturer's guidelines. The DNA samples were sequenced at the Biotechnology Center, Madison, WI, USA; and McGill University and Genome Quebec Innovation Centre, Montreal, Canada. Sequence analysis was carried out using BLAST algorithms (blastx) in GenBank NCBI.
SCOTS analysis
We extended SCOTS technique to determine in vivo gene expression of M. osloensis within the slug at two different time points post infection. The SCOTS technique for this study involves the capture of in vivo transcribed sequences by using biotinylated M. osloensis genomic DNA coupled to streptavidin-coated paramagnetic beads and a PCR-based subtractive hybridization with transcripts from the in vitro cultured bacteria.
Briefly, for the control, 5 μg total RNA samples obtained from 48 h M. osloensis cultures (OD 600 = 0.8) were converted to cDNAs and then made double-stranded. The primer (SCOT09) with a defined sequence (SCOT0) at the 5' end and random sequence at the 3' end was used for both first and second strand cDNA synthesis (Table 3). cDNAs were then PCR amplified using the defined primer (SCOT0) for 30 cycles. Bacterial cDNA normalization was done according to Graham and Clark-Curtiss with some modifications [25]. In this study, M. osloensis ribosomal operon (rDNA) was amplified using primers Mor-F and Mor-R (Table 3). A plasmid containing the amplified operon was constructed using TOPO XL PCR Cloning kit (Invitrogen), and the constructed plasmid was used to block the abundant rDNA sequence in order to effectively capture the cDNA molecules representing mRNA transcripts. The rDNA operon was added to biotinylated genomic DNA at a ratio of 10:1. The genomic DNA-rDNA mixture was sonicated to a size range of 1 to 5 kb. The sonicated, biotinylated genomic DNA-rDNA mixture containing 6 μg rDNA and 0.6 μg genomic DNA was denatured and hybridized for 30 min at 67°C. PCR amplified cDNAs (6 μg) from 48 h cultures were denatured and added to the genomic DNA-rDNA prehybridized mixture, and hybridized at 67°C for 24 h. Streptavidin magnesphere paramagnetic particles (Promega) were used to capture the bacterial cDNA that hybridized to biotinylated genomic DNA according to the manufacturer's instructions. Captured cDNAs were then eluted, precipitated, and amplified by PCR with the defined primers for additional two successive rounds of SCOTS. The finally amplified cDNAs were considered to be normalized in vitro cDNAs.
Table 3.
Primers used in SCOTS analysis
| Primers | Sequences |
| Mor-F | 5-GAACGCTGGCGGCAGGCTTAACACATGC |
| Mor-R | 5-GCTGGCGATGACTTACTCTCACATGGCTAACGCC |
| SCOT0 | 5-ATCCACCTATCCCAGTAGGAG |
| SCOT09 | 5-ATCCACCTATCCCAGTAGGAGNNNNNNNNN-3 |
| SCOT18 | 5-GACAGATTCGCACTTAACCCT |
| SCOT189 | 5-GACAGATTCGCACTTAACCCTNNNNNNNNN-3 |
| SCOT110 | 5-ATGCGAATCCAGACTGTAAGA |
| SCOT1109 | 5-ATGCGAATCCAGACTGTAAGANNNNNNNNN-3 |
For the treatment, 50 μl suspension of 48 h M. osloensis cultures (OD 600 = 0.8) containing 5 × 107 cells according to the bioassay test [24] was inoculated into the shell cavity of slug D. reticulatum. Inoculation into the shell cavity is mimicking the natural infection of the slug by nematode-bacteria complex. Before RNA isolation, a parallel experiment was performed to evaluate the number of viable M. osloensis to confirm the successful bacterial infection in the slug post inoculation. In brief, the infected slug was surface sterilized by immersing in 0.1% thimerosal for 3 h, washed by sterile water, and mortared. The slug homogenate was serially (10-fold) diluted and plated on Mueller-Hinton II agar (MHAII; BBL Becton Dickinson, Cockeysville, Md). The number of CFU was then counted based on the colony color and oxidase reaction [64]. After that, the total RNA was isolated from survived slugs. As about 50% slug mortality occurred within 96 h post bacterial injection or nematode infection, two time point, 48 and 96 h post bacterial injection were selected for total RNA isolation. Three 5 μg total RNA samples obtained from three infected slugs were pooled and made double-stranded by the random primer (Primer SCOT189 was used for 48 h post infection, and SCOT1109 was used for 96 h post infection). Double-stranded cDNAs were then PCR amplified for 30 cycles by the defined primer (Primer SCOT18 was used for post 48 h infection, and SCOT110 was used for post 96 h infection). Amplified cDNAs from 48 h or 94 h post infection were added to the genomic DNA-rDNA prehybridized mixture to hybridize at 67°C for 24 h. The bacterial cDNAs were then captured by streptavidin-coated magnetic particles. The captured cDNAs were eluted and PCR amplified. In the first round of SCOTS, three separate samples of cDNAs were captured by hybridization to biotinylated genomic DNA in parallel reactions. After the first round of SCOTS, the three amplified cDNA preparations for 48 h or 96 h post infection were combined, denatured, and again hybridized to genomic DNA-rDNA mixture for two successive rounds of SCOTS. The finally amplified cDNAs were considered to be normalized in vivo cDNAs.
To enrich the normalized in vivo cDNAs, the normalized cDNAs from 48 h or 96 h post infection were hybridized to biotinylated genomic DNA that has been prehybridized with both rDNA and normalized in vitro cDNAs. After hybridization, the bacterial cDNAs were captured and PCR amplified for next round of enrichment. Finally, the enriched bacterial cDNAs were cloned into an original TA cloning kit (Invitrogen). Each individual clone was PCR amplified, and the concentration of the amplicon was standardized to 0.3 μg/μl. The 80 μl individual amplicon was transferred to each well of the dot-blot containing the positively charged nylon membrane using the low vacuum. The nylon membrane with immobilized individual cDNA was transferred into a hybridization bottle for hybridization using Dig easy hyb granules (Roche) according to the manufacturer's instruction. Digoxigenin-labeled probes were made from the normalized in vitro or in vivo cDNAs using PCR Dig Probe Synthesis kit from Roche Molecular Biochemicals (Indianapolis, Ind.) in accordance with the manufacturer's instructions. The concentration of the generated probes was standardized to be same. Individual cDNA was then screened by dot blot hybridization with the generated probe. The hybridization between the probe and the individual cDNA was performed at 65°C for 24 h. The dilution of anti-digoxigenin-HRP conjugate (1: 800) and Amersham ECL Plus western blotting detection reagents (GE healthcare Bio-Sciences Corp, Piscataway, NJ) were used to detect the hybridization between individual cDNA and the probe. The individual clones with signals stronger or present on the blot hybridized to the normalized in vivo cDNAs compared to that with signals on the blot hybridized to the normalized in vitro cDNAs were chosen for nucleotide sequencing and analysis.
Furthermore, in order to determine that the identified in vivo expressed genes were specific to infection or suppressive condition, we examined the differential in vitro gene expression of 72 h M. osloensis cultures (stationary phase) relative to 24 h cultures (early-log phase) using the similar procedure.
Inactivation of SCOTS identified genes
Several M. osloensis mutants of SCOTS identified genes were constructed by insertion-deletion strategy using inverse PCR according to Furano and Campagnar [65]. Briefly, the SCOTS identified cDNA molecule was cloned into a PGEM-T vector (Promega). The primers with engineered restriction sites were used in the inverse PCR to amplify the cDNA fragment. This resulted in a deletion of about 50 bp nucleotides internal to SCOTS identified sequence. The nonpolar kanamycin resistance gene was amplified from a TA vector with the engineered primers. Inverse PCR product and kanamycin resistance gene were subjected to restriction digestion and ligation, resulting in the plasmid containing the cDNA sequence and kanamycin resistance gene. Sequence analysis was performed to confirm the proper insertion of the cassette. The resulting plasmid was PCR amplified, and product was used to naturally transform wild-type M. osloensis [65,66]. Insertion of the kanamycin gene through a double recombination resulted in the disruption of the wild-type gene. In brief, a 100 μl aliquot of an early-log phase M. osloensis culture was plated onto BHI agar, and 20 ng of the purified mutagenesis constructed sequence from above was spotted onto a portion of the bacterial lawn. After 6 h incubation under standard growth conditions, the area of the bacterial lawn that had been inoculated with the constructed sequence was swabbed onto BHI agar plates containing kanamycin or added to BHI broth (broth method) containing kanamycin and slug homogenate. Samples from broth method were incubated for 12 h, and then different dilutions were spot inoculated on the selective plates containing kanamycin to select the mutants. The mutant chromosomal DNA was isolated and subjected to PCR analysis as well as sequence analysis to confirm the integration of the inactivated genes into the genome. The stability of these mutants was verified by growing the mutants in medium lacking kanamycin and the frequency of kanamycin resistance, and the mutants with stable gene disruptions were selected for the virulence test and complementation study.
Due to the limited genetic information for M. osloensis, we used inverse PCR strategy to amplify the full sequence of SCOTS captured sequences. The amplified sequences were further analyzed by the bio-software such as SignalP 3.0 [67]. The cloned intact genes were cloned into pCR2.1 using the TA Cloning kit (Invitrogen) according to the manufacturer's instructions. The plasmid containing the intact genes was then transferred into the responded mutant and wild-type backgrounds by electroporation. The complemented mutants were further confirmed before virulence test. The virulence of wild type, mutants, and complemented mutants of M. osloensis cultures were determined by quantifying slug mortality following the injection of 72 h bacterial culture into the shell cavity as described below.
Virulence of M. osloensis to the slug
The virulence of M. osloensis cultures was determined by quantifying slug mortality following the injection of bacteria into the shell cavity. The different bacterial cells were precipitated and washed several times using a sterile saline solution (0.85% NaCl), and the total numbers of bacteria in each suspension were measured with a spectrophotometer at a wavelength 600 nm. Different M. osloensis cultures were injected into the slug shell cavity with the same dose used in the SCOTS analysis, and which is consistent with the dose used by Tan and Grewal [20]. Slugs injected with the saline solution served as controls. Six slugs were tested in each treatment, and three replicas were used for each treatment. Slug mortality was recorded 4 days after inoculation at 18°C. The survival analysis was plotted as Kaplan-Meier plots by using the statistical software GraphPad Prism 4 (GraphPad software Inc.), and the Chi-square value for significance at each time point was calculated with significant difference tests at P= 0.05.
Growth rate of mutants in the media
M. osloensis cells of wild type, mutants, and complemented mutants were streaked on BHI agar plates and incubated at 25°C for 72 h. Single colonies from the cultures were picked and inoculated into 3 ml BHI broth, and cultured at 25°Cfor 72 h. To obtain growth rate, 100 ml BHI broth was inoculated with 1 ml of 72 h cultured bacterial cells, and the OD 600 value was recorded until the wild-type bacteria reached the stationary phase.
List of abbreviations
IJs, infective juveniles; SCOTS, selective capture of transcribed sequences
Authors' contributions
RA performed all the experiments, analyzed the data, and wrote the first draft. Both SS and PSG designed the study, wrote the grant proposal and obtained the funding. PSG initiated the project and helped in writing the manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We appreciate the helpful reviews of the manuscript by Dr. Michael Sadowsky (Department of Soil, Water, and Climate, University of Minnesota, St. Paul, MN), Dr. David Denlinger (Department of Entomology, The Ohio State University, Columbus, OH), and Dr. Man-Wah Tan (Genetics and Microbiology and Immunology Departments, Stanford University, Palo Alto, CA). We thank Ms. Judy A. Smith for help with slug collection and Ms. Megan Strother for technical assistance. This research was supported by an interdisciplinary grant from the Ohio Agricultural Research and Development Center, Wooster, Ohio.
Contributor Information
Ruisheng An, Email: an.48@osu.edu.
Srinand Sreevatsan, Email: sreev001@umn.edu.
Parwinder S Grewal, Email: grewal.4@osu.edu.
References
- Chiang SL, Mekalanos JJ, Holden DW. In vivo genetic analysis of bacterial virulence. Annu Rev Microbiol. 1999;53:129–154. doi: 10.1146/annurev.micro.53.1.129. [DOI] [PubMed] [Google Scholar]
- Hueck CJ. Type III secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–343. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodgen KA, Roth JA, Stanton TB, Bolin CA, Minion FC, Wannemuehler MJ. Virulence mechanisms of bacterial pathogens. ASM Press, Washington, DC; 2000. [Google Scholar]
- Donnenberg MS. Pathogenic strategies of enteric bacteria. Nature. 2000;406:768–774. doi: 10.1038/35021212. [DOI] [PubMed] [Google Scholar]
- Dozois CM, Daigle F, Curtiss R., III Identification of pathogen-specific and conserved genes expressed in vivo by an avian pathogenic Escherichia coli strain. PNAS. 2003;100:247–252. doi: 10.1073/pnas.232686799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coady AM, Murray AL, Elliott DG, Rhodes LD. Both msa genes in Renibacterium salmoninarum are needed for full virulence in bacterial kidney disease. Appl Environ Microbiol. 2006;72:2672–2678. doi: 10.1128/AEM.72.4.2672-2678.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer JL, Hedrick RP. Piscirichettsia salmonis: a gram-negative intracellular bacterial pathogen of fish. J Fish Dis. 2003;26:251–262. doi: 10.1046/j.1365-2761.2003.00460.x. [DOI] [PubMed] [Google Scholar]
- Pradel E, Ewbank JJ. Genetic models in pathogenesis. Annu Rev Genet. 2004;38:347–363. doi: 10.1146/annurev.genet.38.072902.092528. [DOI] [PubMed] [Google Scholar]
- Tenor JL, McCormick BA, Ausubel FM, Aballay A. Caenorhabditis elegans-based screen identifies Salmonella virulence factors required for conserved host-pathogen interactions. Curr Biol. 2004;14:1018–1024. doi: 10.1016/j.cub.2004.05.050. [DOI] [PubMed] [Google Scholar]
- Tu ZC, Eisner W, Kreiswirth BN, Blaser MJ. Genetic divergence of Campylobacter fetus strains of mammal and reptile origins. J Clin Microbiol. 2005;43:3334–3340. doi: 10.1128/JCM.43.7.3334-3340.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant M, Mills K, Williams S, Kleckley F, Sims A, Elsey RM, Bushnell J. Effects of bacterial lipopolysachharide on peripheral leukocytes in the American alligator (Alligator mississippiensis) Vet Immnunol Immunopathol. 2006;111:315–320. doi: 10.1016/j.vetimm.2006.01.018. [DOI] [PubMed] [Google Scholar]
- Dickinson VM, Duck T, Schwalbe CR, Jarchow JL, Trueblood MH. Nasal and cloacal bacteria in free-ranging desert tortoises from the western United States. J Wildl Dis. 2001;37:252–257. doi: 10.7589/0090-3558-37.2.252. [DOI] [PubMed] [Google Scholar]
- Isaza R, Garner M, Jacobson F. Proliferative osteoarthritis and osteoarthrosis in 15 snakes. J Zoo Wildl Med. 2000;31:20–27. doi: 10.1638/1042-7260(2000)031[0020:POAOIS]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Paillard C, Roux FL, Borrego JL. Bacterial disease in marine bivalves, a review of recent studies: trands and evolution. Aquat Living Resour. 2004;17:477–498. doi: 10.1051/alr:2004054. [DOI] [Google Scholar]
- Barker GM. The Biology of Terrestrial Molluscs. CABI publishing, New York, NY; 2001. [Google Scholar]
- Wilson MJ, Glen DM, Pearce JD, Rodgers PB. Monoxenic culture of the slug parasite Phasmarhabditis hermaphrodita (Nematoda: Rhabditidae) with different bacteria in liquid and solid phase. Fundam Appl Nematol. 1995;18:159–166. [Google Scholar]
- Wilson MJ, Grewal PS. Biology, production and formulation of slug-parasitic nematodes. In: Grewal PS, Ehlers RU, Shapiro-Ilan D, editor. Nematodes As Biological Control Agents. CABI Publishing, Cambridge, MA; 2005. pp. 421–430. [Google Scholar]
- Wilson MJ, Glen DM, George SK. The Rhabditid nematode Phasmarhabditis hermaphrodita as a potential biological control agent for slugs. Biocontr Sci Technol. 1993;3:503–511. [Google Scholar]
- Tan L, Grewal PS. Infection behavior of the rhabditid nematode Phasmarhabditis hermaphrodita to the grey garden slug Deroceras reticulatum. J Parasitol. 2001;87:1349–1354. doi: 10.1645/0022-3395(2001)087[1349:IBOTRN]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Tan L, Grewal PS. Pathogenicity of Moraxella osloensis, a bacterium associated with the nematode Phasmarhabditis hermaphrodita, to the slug Deroceras reticulatum. Appl Environ Microbiol. 2001;67:5010–5016. doi: 10.1128/AEM.67.11.5010-5016.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Grewal PS. Endotoxin activity of Moraxella osloensis against the grey garden slug Deroceras reticulatum. Appl Environ Microbiol. 2002;68:3943–3947. doi: 10.1128/AEM.68.8.3943-3947.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahan MJ, Slauch JM, Mckalanos JJ. Selection of bacterial virulence genes that are specifically induced in host tissues. Science. 1993;259:686–688. doi: 10.1126/science.8430319. [DOI] [PubMed] [Google Scholar]
- Hensel M, Shea JE, Gleeson C, Jones MD, Dalton E, Holden DW. Simultaneous identification of bacterial virulence genes by negative selection. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- Valdivia RH, Falkow S. Fluorescence-based isolation of bacterial genes expressed within host cells. Science. 1997;277:2007–2011. doi: 10.1126/science.277.5334.2007. [DOI] [PubMed] [Google Scholar]
- Graham JE, Clark-Curtiss JE. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS) Proc Natl Acad Sci USA. 1999;96:11554–11559. doi: 10.1073/pnas.96.20.11554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltes N, Gerlach GF. Identification of genes transcribed by Actinobacillus pleuropneumoniae in necrotic porcine lung tissue by using selective capture of transcribed sequences. Infect Immun. 2004;72:6711–6716. doi: 10.1128/IAI.72.11.6711-6716.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinton JC, Hautefort I, Eriksson S, Thompson A, Rhen M. Benefits and pitfalls of using microarrays to monitor bacterial gene expression during infection. Curr Opin Microbiol. 2004;7:277–282. doi: 10.1016/j.mib.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Tortora GJ, Funke BR, Case CL. Microbial growth. In: Tortora GJ, Funke BR, Case CL, editor. Microbiology: an introduction. Benjamin Cummings, San Francisco, CA; 2004. pp. 156–183. [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BLAST http://www.ncbi.nlm.nih.gov/BLAST/
- Liu D, Lindqvist L, Reeves PR. Transferases of O-antigen biosynthesis in Salmonella enterica: dideoxyhexosyltransferases of groups B and C2 and acetyltransferase of group C2. J Bacteriol. 1995;177:4084–4088. doi: 10.1128/jb.177.14.4084-4088.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorite MJ, Sanjuan J, Velasco L, Olivares J, Bedmar EJ. Characterization of Bradyrhizobium japonicum pcaBDC genes involved in 4-hydroxybenzoate degradation. Biochim Biophys Acta. 1998;1397:257–261. doi: 10.1016/s0167-4781(98)00048-7. [DOI] [PubMed] [Google Scholar]
- Whatmore AM. Streptococcus pyogenes sclB encodes a putative hypervariable surface protein with a collagen-like repetitive structure. Microbiology. 2001;147:419–429. doi: 10.1099/00221287-147-2-419. [DOI] [PubMed] [Google Scholar]
- Rasmussen M, Bjorck L. Unique regulation of SclB – a novel collagen-like surface protein of Streptococcus pyogenes. Mol Microbiol. 2001;40:1427–1438. doi: 10.1046/j.1365-2958.2001.02493.x. [DOI] [PubMed] [Google Scholar]
- Stenson TH, AA W. DsbA and DsbC are required for secretion of pertussis toxin by Bordetella pertussis. Infect Immun. 2002;70:2297–2303. doi: 10.1128/IAI.70.5.2297-2303.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander K, Young LG. Alternative hydroxylases for the aerobic and anaerobic biosynthesis of ubiquinone in Escherichia coli. Biochemistry. 1978;17:4750–4755. doi: 10.1021/bi00615a024. [DOI] [PubMed] [Google Scholar]
- Sheehan KB, Fagg JA, Ferris MJ, Henson JM. Identification of Actinobacillus pleuropneumoniae genes important for survival during infection in its natural host. Appl Environ Microbiol. 2003;69:5914–5918. doi: 10.1128/AEM.69.10.5914-5918.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orville AM, Lipscomb JD, Ohlendorf DH. Crystal structures of substrate and substrate analog complexes of protocatechuate 3,4-dioxygenase: endogenous Fe3+ ligand displacement in response to substrate binding. Biochemistry. 1997;36:10052–10066. doi: 10.1021/bi970469f. [DOI] [PubMed] [Google Scholar]
- Boulnois GJ, Jann K. Bacterial polysaccharide capsule synthesis, export and evolution of structural diversity. Mol Microbiol. 1989;3:1819–1823. doi: 10.1111/j.1365-2958.1989.tb00168.x. [DOI] [PubMed] [Google Scholar]
- Moxon ER, Kroll JS. The role of bacterial polysaccharide capsules as virulence factors. Curr Top Microbiol Immunol. 1990;150:65–85. doi: 10.1007/978-3-642-74694-9_4. [DOI] [PubMed] [Google Scholar]
- Economou A. Bacterial preprotein translocase: mechanism and conformational dynamics of a processive enzyme. Mol Microbiol. 1998;27:511–518. doi: 10.1046/j.1365-2958.1998.00713.x. [DOI] [PubMed] [Google Scholar]
- Vrontou E, Economou A. Structure and function of SecA, the preprotein translocase nanomotor. Biochim Biophys Acta. 2004;1694:67–80. doi: 10.1016/j.bbamcr.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Wickner W, Schekman R. Protein translocation across biological membranes. Science. 2005;310:1452–1456. doi: 10.1126/science.1113752. [DOI] [PubMed] [Google Scholar]
- Mori H, Ito K. The Sec protein-translocation pathway. Trend Microbiol. 2001;9:494–500. doi: 10.1016/S0966-842X(01)02174-6. [DOI] [PubMed] [Google Scholar]
- Tomkiewicz D, Nouwen N, van Leeuwen R, Tans S, Driessen AJ. SecA supports a constant rate of preprotein translocation. J Biol Chem. 2006;281:15709–15713. doi: 10.1074/jbc.M600205200. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Ito K. Secretion monitor, SecM, undergoes self-translation arrest in the cytosol. Mol Cell. 2001;7:185–192. doi: 10.1016/S1097-2765(01)00166-6. [DOI] [PubMed] [Google Scholar]
- Rao PSS, Lim TM, Leung KY. Functional genomics approach to the identification of virulence genes involved in Edwardsiella tarda pathogenesis. Infect Immun. 2003;71:1343–1351. doi: 10.1128/IAI.71.3.1343-1351.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Keitany G, Li Y, Wang Y, Ball HL, Goldsmith EJ, Orth K. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science. 2006;312:1211–1214. doi: 10.1126/science.1126867. [DOI] [PubMed] [Google Scholar]
- Karaolis DK, Johnson JA, bailey CC, Boedeker EC, Kaper JB, Reeves PR. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic stranis. Proc Natl Acad Sci USA. 1998;95:3134–3139. doi: 10.1073/pnas.95.6.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black PN, Faergeman NJ, Dirusso CC. Long-chain acyl-CoA-dependent regulation of gene expression in bacteria, yeast and mammals. J Nutr. 2000;130:305–309. doi: 10.1093/jn/130.2.305S. [DOI] [PubMed] [Google Scholar]
- Gargiulo CE, Stuhlsatz-Krouper SM, Schaffer JE. Localization of adipocyte long-chain fatty acyl-CoA synthetase at the plasma membrane. J Lipid Res. 1999;40:881–892. [PubMed] [Google Scholar]
- Greenway DLA, Silbert DF. Altered acyltransferase activity in Escherichia coli associated with mutations in acyl coenzyme-a synthetase. J Biol Chem. 1983;258:3034–3042. [PubMed] [Google Scholar]
- Hill CE, Metcalf DS, Macinnes JI. A search for virulence genes of Haemophilus parasuis using differential display RT-PCR. Vet Microbiol. 2003;96:189–202. doi: 10.1016/S0378-1135(03)00212-8. [DOI] [PubMed] [Google Scholar]
- Soto MJ, Fernandez-Pascual M, Sanjuan J, Olivares J. A fadD mutant of Sinorhizobium meliloti shows multicellular swarming migration and is impaired in nodulation efficiency on alfalfa roots. Mol Microbiol. 2002;43:371–382. doi: 10.1046/j.1365-2958.2002.02749.x. [DOI] [PubMed] [Google Scholar]
- Utley M, Franklin DP, Krogfelt KA, Lauz DC, Cohen PS. A Salmonella typhimurium mutant unable to utilize fatty acids and citrate is avirulent and immunogenic in mice. FEMS Microbiol Lett. 1998;163:129–134. doi: 10.1111/j.1574-6968.1998.tb13036.x. [DOI] [PubMed] [Google Scholar]
- Yu J. Inactivation of DsbA, but Not DsbC and DsbD, affects the intracellular survival and virulence of Shigella flexneri. Infect Immun. 1998;66:3909–3917. doi: 10.1128/iai.66.8.3909-3917.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Kroll JS. DsbA: a protein-folding catalyst contributing to bacterial virulence. Microbes Infect. 1999;1:1221–1228. doi: 10.1016/S1286-4579(99)00239-7. [DOI] [PubMed] [Google Scholar]
- Sandkvist M. Type II secretion and pathogenesis. Infect Immun. 2001;69:3523–3535. doi: 10.1128/IAI.69.6.3523-3535.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez B, Secades P, Prieto M, McBride MJ, Guijarro JA. A mutation in Flavobacterium psychrophilum tlpB inhibits gliding motility and induces biofilm formation. Appl Environ Microbiol. 2006;72:4044–4053. doi: 10.1128/AEM.00128-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadokura H, Bader M, Tian H, Bardwell JC, Beckwith J. Roles of a conserved arginine residue of DsbB in linking protein disulfide-bond-formation pathway to the respiratory chain of Escherichia coli. Proc Natl Acad Sci USA. 2000;97:10884–10889. doi: 10.1073/pnas.97.20.10884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamoto H, Bardwell CA. Catalysis of disulfide bond formation and isomerization in the Escherichia coli periplasm. BBA. 2004;1694:111–119. doi: 10.1016/j.bbamcr.2004.02.012. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW, Russell D. Plasmids and their usefulness in molecular cloning. In: Sambrook J, Russell DW, Russell D, editor. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press, New York, NY; 2000. pp. 7–13. [Google Scholar]
- Froussard P. A random-PCR method (rPCR) to construct whole cDNA library from low amounts of RNA. Nucl Acids Res. 1992;20:2900. doi: 10.1093/nar/20.11.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaneechoutte M, Claeys G, Steyaert S, Baere TD, Peleman R, Verschraegen G. Isolation of Moraxella canis from an ulcerated metastatic lymph node. J Clin Microbiol. 2000;38:3870–3871. doi: 10.1128/jcm.38.10.3870-3871.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furano K, Campagnar AA. Inactivation of the Moraxella catarrhalis 7169 ferric uptake regulator increases susceptibility to the bactericidal activity of normal human sera. Infect Immun. 2003;71:1843–1848. doi: 10.1128/IAI.71.4.1843-1848.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juni E. Simple genetic transformation assay for rapid diagnosis of Moraxella osloensis. Appl Microbiol. 1974;27:16–24. doi: 10.1128/am.27.1.16-24.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, Heijne GV, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]