Abstract
To get a better understanding of mutagenic mechanisms in humans, we have cloned and sequenced the human homolog of the Saccharomyces cerevisiae REV3 gene. The yeast gene encodes the catalytic subunit of DNA polymerase ζ, a nonessential enzyme that is thought to carry out translesion replication and is responsible for virtually all DNA damage-induced mutagenesis and the majority of spontaneous mutagenesis. The human gene encodes an expected protein of 3,130 residues, about twice the size of the yeast protein (1,504 aa). The two proteins are 29% identical in an amino-terminal region of ≈340 residues, 39% identical in a carboxyl-terminal region of ≈850 residues, and 29% identical in a 55-residue region in the middle of the two genes. The sequence of the expected protein strongly predicts that it is the catalytic subunit of a DNA polymerase of the pol ζ type; the carboxyl-terminal domain possesses, in the right order, the six motifs characteristic of eukaryotic DNA polymerases, most closely resembles yeast pol ζ among all polymerases in the GenBank database, and is different from the human α, δ, and ɛ enzymes. Human cells expressing high levels of an hsREV3 antisense RNA fragment grow normally, but show little or no UV-induced mutagenesis and are slightly more sensitive to killing by UV. The human gene therefore appears to carry out a function similar to that of its yeast counterpart.
Keywords: UV mutagenesis, yeast REV3, 5′ rapid amplification of cDNA ends
In budding yeast, Saccharomyces cerevisiae, virtually all DNA damage-induced mutagenesis and a major fraction of spontaneous mutagenesis depends on the activity of the REV3 (scREV3) gene (1–4). The scREV3 locus encodes the catalytic subunit of DNA polymerase ζ, which contains at least one other subunit, the product of the REV7 gene (5). Pol ζ, together with the REV1 gene product and possibly other proteins, is thought to carry out translesion replication; that is, replication past DNA lesions that block chain elongation by the normal replicases (2, 5). Translesion replication is regulated, in a way that is still poorly understood, by the RAD6 and RAD18 genes, which encode an E2 ubiquitin conjugase and a DNA binding protein with which the conjugase associates, respectively (6).
Although budding yeast is often a good model for other eukaryotes, it is not yet known whether similar proteins and mechanisms exist in other members of this group. Information of this kind from humans might be particularly useful, in view of its possible relevance to understanding genotoxic hazards and cancer. We have therefore conducted a search for human homologs of the scREV genes, as a preliminary step to investigating the enzymological basis of translesion replication in Homo sapiens. In this paper we report the isolation and sequence of a human homolog of scREV3 and evidence from human cells expressing an hsREV3 antisense mRNA fragment that this gene performs a function similar to that of its yeast counterpart.
MATERIALS AND METHODS
cDNA Synthesis, 5′ Rapid Amplification of cDNA Ends (RACE), and Cloning.
Unless otherwise noted, the materials used were obtained from Life Technologies (Gaithersburg, MD) and were used according to the manufacturer’s specifications. cDNAs were synthesized by using Superscript II reverse transcriptase and human fetal brain total RNA (Stratagene) with primers based on the known human REV3 (hsREV3) sequence. The cDNA was purified on Glassmax columns and subsequently tailed with oligo(dC) using terminal transferase. The tailed cDNA was then selectively amplified by using a nested hsREV3-specific primer and a commercial primer whose 3′ end was complementary to the oligo(dC) tail. In some experiments, amplification used Taq polymerase for 35 cycles of 94°C for 30 sec, 50°C for 30 sec, and 72°C for 3 min, using a Hybaid thermal cycler. Where higher melting temperature primers were used, the annealing temperature was increased to 55°C. Products of this reaction were further amplified by using two dU-containing primers compatible with the cloneAMP plasmid cloning system. In later experiments, the initial PCR used non-dU-containing primers and elongase for 35 cycles of 94°C for 20 sec, 50°C for 15 sec, and 68°C for 4 min, and a second amplification using dU-containing primers and Taq polymerase. PCR products to be cloned were separated on agarose gels, purified by using Wizard PCR Prep spin columns (Promega) and treated with uracil glycosylase in the presence of the pAMP-1 vector. The resulting annealed DNAs were used to transform Escherichia coli DH5α cells. Additionally, 3′-sequence was confirmed and extended by using four plasmids isolated from a human cDNA library by the Genetrapper method.
DNA Purification and Sequence Analysis.
Plasmid DNAs were isolated by alkaline lysis (7) and those with large inserts purified by using midi columns (Qiagen, Chatsworth, CA) for sequence analysis. In some experiments, plasmids were screened for suitable hsREV3 inserts using dideoxy sequencing from the 3′-end of the insert with Klenow fragment pol I. Some clones were sequenced entirely with Sequenase II (United States Biochemical). In most instances, however, thermal cycling with one of three dye terminator chemistries was used (PE Applied Biosystems). Primers for sequencing and PCR were developed as needed, based on known sequence as it became available. In all, 77 different primers were used. Both strands of the cDNA were sequenced in almost all cases, and at least three independent clones sequenced for each region of the gene. The sequences obtained from each reaction were aligned by using the fasta algorithm (8), and continuous sequence assembled from the observed overlaps. When sequence variants occurred, the majority sequence was taken to be the correct one. A physically continuous sequence, lacking 45 nt in the 5′-untranslated region, but which otherwise exactly agreed with the consensus, was assembled from several smaller clones, including a 5′-amplification product with one primer containing a site for EcoRI and cloned between the EcoRI and NotI sites of pBluescript KS (+) (Stratagene) to yield the clone pPGR3–3.
The sequences of human and S. cerevisiae DNA polymerases α, δ, ɛ, and ζ were aligned over the region containing the DNA polymerase motifs, by using the program multalin (9). The resulting alignment was then used to assess the genetic distance between these sequences, assuming that protein sequence evolution approximates a gamma function (10). These distances were then used to calculate a phylogenetic tree by using the neighbor-joining method of Saitou and Nei (11). Distance calculations and tree building used the program package mega (12).
Northern Blotting Analysis.
Total human brain RNA was electrophoresed in a formaldehyde agarose gel and blotted onto a nylon membrane (7). The blot was hybridized to a digoxigenin-labeled probe prepared from a clone representing the 3′ 4,240 bp of the mRNA, using the Genius labeling kit (Boehringer Mannheim). The blot was washed under high stringency conditions and hybridization detected with anti-digoxigenin alkaline phosphatase antibody conjugate and Lumiphos (Boehringer Mannheim).
Preparation of Cells with Regulatable Expression of Antisense hsREV3.
A plasmid designated pTet-Puro was prepared by excising a fragment from the vector pTet-Splice using XhoI and NotI and replacing it with a fragment carrying the gene for puromycin resistance, the bacterial origin of replication, and the gene for ampicillin resistance. A second plasmid, designated pKS-2, was made by inserting a 1,506-bp SalI fragment of the hsREV3 cDNA into pTet-Puro in the antisense orientation, under the control of the TetP promoter (13, 14). This fragment includes 48 bp of 5′-untranslated sequence, 1,415 bp of coding sequence, and polylinker sequences at each end. Plasmid pKS-2 was transfected into MSU-1.2–10A cells, an infinite lifespan, near diploid nontumorigenic human fibroblast cell line (15), into which plasmid pTet-tTak had been stably integrated. Plasmid pTet-tTak contains the tetracycline (tet) transactivator (tTA) gene under the control of the tet promoter (tetP) (13, 14). Strain MSU-1.2–10A exhibits high expression of tTA and efficient reduction of expression on addition of tet to the medium. Stable transfectants of pKS-2 were selected for puromycin resistance. Transfectants were scanned for high expression of the antisense hsREV3 fragment in the absence of tet and reduction of expression upon addition of tet to the medium. Cell strain MSU-1.2–10A-42 was chosen as suitable for studies of the effect of hsREV3 antisense on the induction of mutations by UV.
Determination of the Frequency of UV-Induced Mutants.
MSU-1.2–10A cells and their derivative cell strain MSU-1.2–10A-42, which carry an integrated fragment of hsREV3 cDNA in an antisense orientation, were cultured in the presence or absence of tet (4 μg/ml) for 7 days in Eagle’s minimal medium supplemented as described (16). Cells (21 × 106 from each population) were plated at a density of 3.4 × 104 cells per cm2 into a series of 150-mm-diameter dishes and allowed to attach. At the same time, cells from each population were plated in appropriate medium into a series of 100-mm-diameter dishes at cloning densities to be used to assay survival after UV exposure, as described (17). In addition, ≈3 × 107 cells from each population of MSU-1.2–10A-42 cells were plated for Northern blotting analysis of the level of expression of hsREV3 antisense. Approximately 6 hr after plating, when the cells to be irradiated had attached and flattened, 10 × 106 cells from each population were rinsed with PBS and irradiated with 8 J/m2 or 11 J/m2 of 254 nm UV as described (17). Mock-irradiated cells (1 × 106 from each group) were used as controls. The cells plated at cloning densities were similarly irradiated. The surviving cells in the populations plated at high densities were allowed to replicate in the presence or absence of tet (4 μg/ml) as appropriate. After 4 days, the cells in each population were dislodged with trypsin, pooled, and approximately 4 × 106 cells were plated into four 150-mm-diameter dishes and allowed to continue replicating in the presence or absence of tet as appropriate. The rest were stored frozen in liquid N2. After an 8-day expression period, the indicated number of cells from each population was assayed as described (16) for the frequency of 6-thioguanine-resistant cells.
RESULTS AND DISCUSSION
The dBEST database of the National Center for Biotechnology Information was searched by using the blast algorithm (18) for expressed sequence tags encoding polypeptides homologous to scRev3p, and a single human expressed sequence tag identified (GenBank accession no. T08184) among ≈52,000 sequences listed at that time. The corresponding clone pHIBBA34, from a human infant brain library (19), was purchased from the American Type Culture Collection and the cDNA insert of this clone sequenced in its entirety. The 2.16-kb cDNA insert was found to contain 1.16 kb of an ORF showing 40% protein sequence identity with the carboxyl-terminal sequence of S. cerevisiae rev3p and 984 bp of 3′-untranslated sequence, together with 20 bp of poly(A). A typical polyadenylation signal, AAUAAA, is separated by 17 nt from this poly(A) tract.
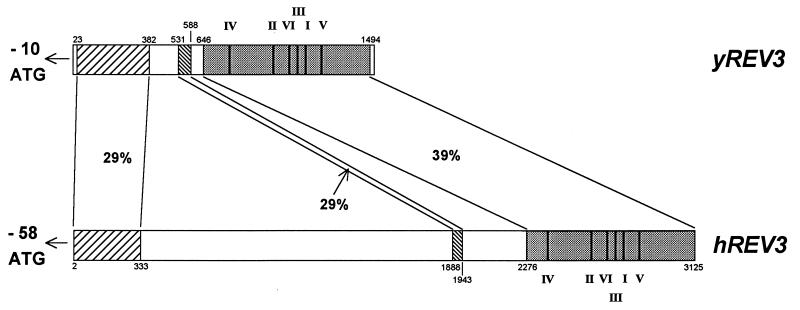
The sequence was extended toward the 5′ end by the method of 5′ RACE (20), with the products of PCR being cloned into pAMP-1. Both strands of the inserts of at least three independent clones from each round of 5′-RACE were sequenced to identify errors arising during reverse transcription. Additionally, a cDNA clone carrying 5.2 kb of 3′-terminal sequence, and three clones with shorter insets, were isolated using the Genetrapper method. Nine sequential rounds of 5′-RACE were required to reach the initiation codon, and a tenth round used to extend the 5′-untranslated sequence. The gene is very likely to be of human origin because more than 26 expressed sequence tags for this gene currently exist in the data base, and the message has been detected in RNA samples from a variety of human tissues. The complete sequence of 10,459 nt (GenBank accession no. AF058701) consists of 85 nt of 5′ untranslated sequence, 9,390 nt of ORF, and 984 nt of 3′ untranslated region. This is in good agreement with an estimate of 10–11 kb from a Northern blot of total brain RNA. The predicted human protein of 3,130 residues (Fig. 1) is therefore a little over twice the length of the yeast protein (1,504 residues) and has an expected mass of ≈353 kDa. A comparison between the expected amino acid sequence of the human protein and the sequence of the yeast polypeptide (Fig. 2) shows significant homology in a region of ≈340 residues at the amino terminus of both proteins (29% identity) in a region of ≈850 residues at their carboxyl termini (39% identity), and in a 55-residue region in the middle of both proteins (29% identity), but little similarity in the intervening regions.
Figure 1.
Amino acid sequence encoded by the 9,030-nt ORF of hsREV3. Underlined regions labeled I–VI are the conserved sequence motifs (21) characteristic of type B (22) DNA polymerases.
Figure 2.
Schematic comparison of the scREV3 and hsREV3 protein sequences. Related regions are shaded and percent identity indicated.
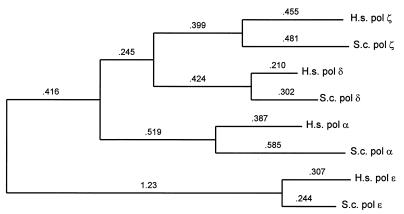
The function of the first two homologous regions, and of the species-specific regions lacking significant identity, is not known. The region of homology at the carboxyl terminus, however, is clearly the DNA polymerase domain. It contains, in the right order, the six conserved sequence motifs characteristic of DNA polymerases (21), including the canonical hexapeptide motifs within regions 1 and 2, YGDTDS and SLYPSI, respectively, found jointly only in type B DNA polymerases, as classified by Braithwaite and Ito (22). The hsREV3 gene product is therefore strongly predicted to be a DNA polymerase, and moreover to be a polymerase of the pol ζ type. The predicted protein sequence is different from that of the human α, δ, and ɛ DNA polymerases and exhibits 34% identity with the yeast Rev3 protein in the carboxyl-terminal region even when the six conserved sequences are excluded from the comparison; this is a much higher level of identity than is found in similar comparisons between different types of DNA polymerase. Among all comparable yeast and human proteins it is most closely related to yeast pol ζ (Fig. 3). The homology of human and yeast Rev3p sequences was confirmed by bootstrap analysis using 500 replications; all trees placed them on the same internal branch.
Figure 3.
Unrooted phylogenetic tree showing the relatedness of yeast and human DNA polymerases. Only the regions spanning motifs IV–V were aligned and used to generate a tree by the neighbor–joining method (11), with distances (see Materials and Methods) between nodes as indicated.
An appreciable proportion of the cDNAs analyzed contained an insertion of 128 bp between nucleotides +139 and +140 (Fig. 4). The insertion variants were detected in PCR products using primers in the 5′-untranslated region and in the region 3′ to the insertion site. Estimates based on relative band intensities on ethidium bromide-stained gels of the PCR product suggest that ≈40% of the cDNA molecules were of this type. Sequence analysis of the longer product shows that insertion leads to an almost immediate termination of the main reading frame (Fig. 4). The origin of this sequence is not yet clear, but might arise from an alternative splicing event and function to limit the amount of hsRev3p synthesized. In addition (Fig. 4), the human gene possesses an out-of-frame ATG codon at nucleotide position −58 within the 5′-untranslated region, which initiates an ORF of 106 codons that terminates within the hREV3 ORF. For the cDNA insertion variant, this reading frame terminates within the insertion (Fig. 4). An upstream out-of-frame ATG with an ORF that terminates within the main ORF is also found in the yeast gene (2), suggesting that it may be evolutionarily conserved in all REV3 genes. In both species, the sequence context of the upstream ATG is at least as good for promoting translation as that of the main ORF ATG, indicating that translational efficiency of the hsREV3 message is likely to be very low (23). High levels of pol ζ may therefore be deleterious to cells, although no evidence for this has been found thus far in yeast.
Figure 4.
Sequence at the 5′ end of the hsREV3 cDNA. Nucleotides shown in uppercase are the first 352 nt of the major mRNA species, whereas the nucleotides in lowercase are the 128-nt insertion found in the variant. The ORF for hsREV3 starts at the ATG labeled In2, and this reading frame possesses an upstream termination codon, labeled T1. The ATG labeled In1 is upstream and out of frame with respect to In2 and could direct the synthesis of a 106 amino acid polypeptide, terminating at T4. For the 128-nt-insertion variant, the ORF starting at In1 would be terminated at T3, encoding a 77 amino acid polypeptide, whereas translation initiated at In2 would terminate at T2, yielding a 47-aa polypeptide.
In yeast, the great majority of base pair substitutions and frameshift mutations induced by DNA damage occur as a result of pol ζ-dependent translesion replication. UV-induced reversion frequencies in REV3+ and null rev3 mutant strains, using a set of nine defined test alleles that included amber, ochre, initiation, and proline missense mutations, indicate that 98% of induced base pair substitutions depend on REV3 function. In addition, related experiments with a set of eight frameshift alleles indicate that pol ζ-dependent translesion replication is responsible for more than 90% of frameshift mutations induced by UV in yeast (24, 25). These results are likely to be typical for a variety of types of DNA damage, because similar data were obtained with γ-irradiated strains (26). At the same time, yeast pol ζ is involved in the production of the majority of spontaneous mutations (3, 4), including those associated with transcription (27) and double-strand break-stimulated recombination (28). However, yeast pol ζ does not appear to be involved in recombination itself (2, 28, 29), or indeed in any replication, recovery, or repair process other than translesion replication.
Evidence from experiments investigating UV-induced mutagenesis and cell killing in cultured human cells expressing an hsREV3 antisense RNA fragment indicate that the human gene performs a similar function to that of its yeast counterpart. To investigate this issue, the frequency of 6-thioguanine-resistant mutant cells induced by UV, as well as cell survival, was measured in the human fibroblast cell line MSU-1.2–10A and its derivative strain MSU-1.2–10A-42. The latter cells express a 5′ 1,506-bp hsREV3 RNA fragment in the antisense orientation from a tetracycline modulatable promoter (13, 14). Northern blotting analysis showed that in the absence of tetracycline MSU-1.2–10A-42 cells express high levels of the hsREV3 antisense RNA, and that in the presence of tetracycline such activation is suppressed ≈2-fold (data not shown).
As shown in Table 1, parental cell line MSU-1.2–10A exhibits a UV fluence-dependent increase in the frequency of 6-thioguanine-resistant cells. In contrast, no induced mutagenesis was seen in MSU-1.2–10A-42 cells grown in the absence of tetracycline, which express high levels of hsREV3 antisense RNA. In those same cells the UV sensitivity was slightly increased. This phenotype is similar to that of a yeast rev3 mutant. A 2-fold reduction in the level of antisense RNA, resulting from growth of MSU-1.2–10A-42 cells in the presence of tetracycline, appeared to increase survival to the level of normal human cells, but had no significant effect on induced mutagenesis (data not shown), presumably because even the reduced levels of antisense RNA are effective at suppressing gene function. Although the mechanisms used by higher eukaryotic cells to bypass DNA damage are poorly understood, most UV-induced mutations are thought to occur during translesion replication on templates containing residual, unrepaired photoproducts (30), a process in which hsREV3p is presumably involved. hsREV3p does not however appear to be involved in normal DNA replication, because the MSU-1.2–10A-42 cells that express high levels of hsREV3 antisense RNA in the absence of tet were found to replicate at the normal rate (data not shown).
Table 1.
Effect of hsREV3 antisense RNA level on cell killing and the frequency of 6-thioguanine-resistant mutants induced in human fibroblasts by UV
| Cell line | Tet | Antisense level | UV, J/m2 | Survival, % | No. of mutants | No. of cells selected × 10−6 | Mutants per 106 cells selected* |
|---|---|---|---|---|---|---|---|
| 10A | No | None | 0 | 100 | 0 | 2.3 | <3 |
| 8 | 49 ± 8 | 45 | 6.0 | 38 | |||
| 11 | 20 ± 10 | 88 | 6.0 | 64 | |||
| 10A-42 | No | High | 0 | 100 | 2 | 2.3 | 3 |
| 8 | 38 ± 9 | 12 | 6.0 | 6 | |||
| 11 | 22 ± 10 | 18 | 6.0 | 9 |
Data are from two independent experiments.
*Corrected for cloning efficiency.
The existence of a much larger nonhomologous or species-specific region in the expected human protein suggests that pol ζ may perform a wider range of functions in the higher eukaryotes. Such a conclusion is possibly indicated by the observation that REV3 mRNA levels are increased in cultured mouse fetal cerebral cortical cells treated with the drug pentylenetetrazol. This drug, which does not appear to damage DNA, induces epileptic-like seizures in mice. Differential screening of a cDNA library constructed from such cells led to the isolation of a partial cDNA clone (SEZ4) that contained an ORF encoding 755 residues at the carboxyl terminus of the REV3 gene (31). This sequence is 40% identical with the comparable region of the yeast gene and 96% identical with the human gene. Kajiwara et al. (31) conclude that mouse pol ζ may therefore be involved in “emergency” or stress responses.
Examination of the enzymatic properties of hsREV3p, and other hsREV gene products, is likely to lead to a better understanding of mutagenic mechanisms in humans, which in turn may aid in detection and estimation of genotoxic hazards. Better information about mutagenic mechanisms may also be relevant to cancer studies (1). The human Rev proteins may be attractive targets for therapeutic intervention, with the aim of reducing mutagenesis, and therefore perhaps cancer risks, in humans. The possibility of therapeutic benefit from reduced mutagenesis is clearly speculative at present, however, and continued work will be needed to examine its potential.
Acknowledgments
We thank George Kampo and Jack Maniloff for synthesizing oligonucleotides and sequence analysis, Kristen Sweat for help in antisense plasmid construction, Hong Zhang for preparing pTet-Puro and the MSU-1.2–10A cell strain, and Chauncey Spooner for screening the antisense transformants for expression. This work was supported by U.S. Public Health Service Grants GM21858, CA56796, and CA73984 from the National Institutes of Health.
ABBREVIATION
- RACE
rapid amplification of cDNA ends
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF058701).
References
- 1.Lawrence C W, Hinkle D C. Cancer Surv. 1996;28:21–31. [PubMed] [Google Scholar]
- 2.Morrison A, Christensen R B, Alley J A, Beck A K, Bernstine E G, Lemontt J F, Lawrence C W. J Bacteriol. 1989;171:5659–5667. doi: 10.1128/jb.171.10.5659-5667.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quah S-K, von Borstel R C, Hastings P J. Genetics. 1980;96:819–839. doi: 10.1093/genetics/96.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roche H, Gietz R D, Kunz B A. Genetics. 1994;137:637–646. doi: 10.1093/genetics/137.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nelson J R, Lawrence C W, Hinkle D C. Science. 1996;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- 6.Bailly V, Lamb J, Sung P, Prakash S, Prakash L. Genes Dev. 1994;8:811–820. doi: 10.1101/gad.8.7.811. [DOI] [PubMed] [Google Scholar]
- 7.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley; 1998. pp. 1.7.1–1.7.16. [Google Scholar]
- 8.Pearson W R, Lipman D J. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corpet F M. Nucleic Acids Res. 1989;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nei M, Chakraborty R, Fuerst P A. Proc Natl Acad Sci USA. 1976;73:4164–4168. doi: 10.1073/pnas.73.11.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 12.Kumar S, Tamura K, Nei M. mega: Molecular Evolutionary Genetics Analysis, Version 1.0. University Park, PA: Pennsylvania State Univ.; 1993. [Google Scholar]
- 13.Gossen M, Bujard H. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shockett P, Difilippantonio M, Hellman N, Schatz D G. Proc Natl Acad Sci USA. 1995;92:6522–6526. doi: 10.1073/pnas.92.14.6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin C C, Wang Q, Maher V M, McCormick J J. Cell Growth Differ. 1994;5:1381–1387. [PubMed] [Google Scholar]
- 16.Maher V M, McCormick J J. In: Technologies for Detection of DNA Damage and Mutations. Pfiefer G P, editor. New York: Plenum; 1996. pp. 381–390. [Google Scholar]
- 17.Patton J D, Rowan L A, Mendrala A L, Howell J N, Maher V M, McCormick J J. Photochem Photobiol. 1984;39:37–42. doi: 10.1111/j.1751-1097.1984.tb03401.x. [DOI] [PubMed] [Google Scholar]
- 18.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.Adams M D, Soares M B, Kerlavage A R, Fields C, Venter J C. Nat Genet. 1993;4:373–380. doi: 10.1038/ng0893-373. [DOI] [PubMed] [Google Scholar]
- 20.Frohman M A, Dush M K, Martin G R. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong S W, Wahl A F, Yuan P-M, Arai N A, Pearson B E, Arai K-I, Korn D, Hunkapiller M W, Wang T S-F. EMBO J. 1988;7:37–47. doi: 10.1002/j.1460-2075.1988.tb02781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Braithwaite D K, Ito J. Nucleic Acids Res. 1993;21:787–802. doi: 10.1093/nar/21.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozak M. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 24.Lawrence C W, Christensen R B. Genetics. 1979;92:397–408. doi: 10.1093/genetics/92.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawrence C W, O’Brien T, Bond J. Mol Gen Genet. 1984;195:487–490. doi: 10.1007/BF00341451. [DOI] [PubMed] [Google Scholar]
- 26.McKee R, Lawrence C W. Genetics. 1979;93:375–381. doi: 10.1093/genetics/93.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Datta A, Jinks-Robertson S. Science. 1995;268:1616–1619. doi: 10.1126/science.7777859. [DOI] [PubMed] [Google Scholar]
- 28.Strathern J N, Shafer B K, McGill C B. Genetics. 1995;140:965–972. doi: 10.1093/genetics/140.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lemontt J F. Mutat Res. 1971;13:319–326. doi: 10.1016/0027-5107(71)90042-x. [DOI] [PubMed] [Google Scholar]
- 30.McGregor W G, Chen R-H, Lukash L, Maher V M, McCormick J J. Mol Cell Biol. 1991;11:1927–1934. doi: 10.1128/mcb.11.4.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kajiwara K, Nagawawa H, Shimizu-Nishikawa K, Ookura T, Kimura M, Sugaya E. Biochem Biophys Res Commun. 1996;219:795–799. doi: 10.1006/bbrc.1996.0313. [DOI] [PubMed] [Google Scholar]