Abstract
One of the top-selling medicinal products worldwide is Hypericum perforatum (St. John's Wort). Despite its cosmopolitan distribution and utilization, little is known regarding the relationship of the bioactive compounds in H. perforatum to the plants from which they are purportedly derived. In this study, amplified fragment length polymorphism (AFLP) analysis of 56 Hypericum accessions, representing 11 species, was conducted to gain a better understanding of diversity within Hypericum species, especially within cultivated accessions of H. perforatum, and to establish a molecular methodology that will provide breeders and regulators with a simple, affordable, and accurate tool with which to identify purported H. perforatum material. Utilizing four primer combinations, a total of 298 polymorphic markers were generated, of which 17 were present in all H. perforatum accessions and 2 were specific to only H. perforatum. This study demonstrates that AFLP can be utilized not only to determine the relationships of closely related Hypericum accessions, but as a tool to authenticate material in herbal remedies through the use of genetic fingerprinting.
Keywords: Hypericum, Hypericaceae, marker analysis, genetic diversity, St. John's wort
Introduction
The genus Hypericum L., family Hypericaceae, is composed of approximately 450 species of trees, shrubs, and herbs widely distributed in temperate regions across the globe [1]. Originally native to southern Europe, H. perforatum is commonly found throughout temperate regions of both the northern and southern hemispheres [2]. Classified within the second largest section (Hypericum) of the genus, H. perforatum, commonly known as St. John's wort, is the best known species of the family. Hypericum perforatum has been suggested to have originated from the ancient hybridization and subsequent polyploidization of two diploids (2n = 2× = 16), H. maculatum subsp. maculatum Crantz and H. attenuatum Choisy [3]. It is a facultative apomict, as both sexual and aposporic processes can take place on the same plant [4]. While most H. perforatum individuals generated through apomixis are tetraploid (2n = 4× = 32) there are hexaploid (2n = 6× = 48), diploid (2n = 2× = 16), and aneuploid individuals as well [5], [6], [7].
Hypericum perforatum's biological extracts are widely recognized as valuable phytopharmaceutical agents with antiviral capabilities [8], and the potential to treat maladies such as depression, skin wounds, and burns [9]. Hypericum perforatum contains at least ten classes of biologically active compounds [10], of which two of the more important bioactive compounds, hypericin and hyperforin, are broadly variable in biological activity in humans [11]. Research indicates that these compounds vary in concentration and or constituency depending on species origin, tissue type, genetics, and environmental factors [11]. In addition, concentrations of these compounds can vary widely between accessions derived from the same species [12], [13].
Quality control involved with the production and distribution of phytopharmaceutical medicines has not been highly regulated with respect to species of plants being used in the preparation of commercial products and the concentration of bioactive compounds. Moreover, the technology available for identification of H. perforatum plant material in commercially available products is not standardized and thus variation between products is an issue [14]. Because of the importance of H. perforatum to the phytopharmaceutical industry, it is important to develop a reliable marker system that can be used to affordably and accurately identify plant material purported to be H. perforatum in order to aid producers while protecting consumers from potentially adulterated products.
Studies conducted by Arnholdt-Schmidt [15] and Mayo et al. [7] demonstrated that techniques such as RAPD (random amplification of polymorphic DNA) and AFLP (amplified fragment length polymorphism) analysis, would enable the elucidation of genetic diversity in wild populations of Hypericum spp. In this study, AFLP analysis was used to describe patterns of genetic variation and distribution within and among wild and commercially cultivated accessions of H. perforatum, and additionally, to develop a suite of species-specific markers that can be used to identify H. perforatum plant material. AFLP analysis is a whole-genome approach that has broad applicability in determining genetic variability within and among plant populations [17], crop origins [18], and relationships among cultivars [9]. AFLP markers are highly repeatable [19], provide broad genomic coverage and a virtually limitless number of genetic markers. Using AFLP technology, we identify two monomorphic and 28 polymorphic species-specific markers that can be used to accurately identify plant material purported to be H. perforatum.
Materials and Methods
Hypericum spp. were obtained from the North Central Regional Plant Introduction Station in Ames, Iowa (Table 1). Fifty-six Hypericum accessions from three different continents were studied, including 11 different species, 38 wild-collected and four cultivated accessions of H. perforatum, and two accessions of the outgroup Triadenum walteri [20]. The taxonomic identities of these accessions follow the systematic treatment used in the Germplasm Resources Information Network database, http://www.ars-grin.gov/npgs, except that accessions of H. perforatum were identified to subspecies on the basis of available herbarium vouchers, digital images, living plants, and geographic origin by following Robson's (2002) key [21]. Leaf material was obtained from three individual plants per accession, flash frozen in liquid nitrogen, and stored at −80°C prior to DNA extraction.
Table 1.
Accessions of Hypericum spp. studied
| NCRPIS number | Species | Original location |
|---|---|---|
| Ames 26862 | H. perforatum L. perforatum | Coimbra, Portugal 2 |
| Ames 27342 | H. perforatum L. veronense | Gegark ‘unik’, Armenia 3 |
| Ames 27343 | H. perforatum L. veronense | Ararat, Armenia 3 |
| Ames 27427 | H. perforatum L. perforatum | East Bohemia, Czech Republic 2 |
| Ames 27428 | H. perforatum L. perforatum | Germany 2 |
| Ames 27429 | H. perforatum L. perforatum | East Bohemia, Czech Republic 2 |
| Ames 27443 | H. perforatum L. chinense | China 3 |
| Ames 27452 | H. perforatum L. perforatum ‘Elixir’ | Denmark 1 |
| Ames 27453 | H. perforatum L. perforatum ‘Helos’ | Denmark 1 |
| Ames 27454 | H. perforatum L. perforatum ‘New Stem’ | Germany 1 |
| Ames 27455 | H. perforatum L. perforatum ‘Topas’ | Germany 1 |
| Ames 27490 | H. perforatum L. perforatum | California, United States 4 |
| Ames 27491 | H. perforatum L. perforatum | Kansas, United States 4 |
| Ames 27493 | H. perforatum L. perforatum | Kansas, United States 4 |
| Ames 27510 | H. perforatum L. perforatum | Lithuania 2 |
| Ames 27511 | H. perforatum L. perforatum | Lithuania 2 |
| Ames 27512 | H. perforatum L. perforatum | Lithuania 2 |
| Ames 27513 | H. perforatum L. perforatum | Lithuania 2 |
| Ames 27515 | H. perforatum L. perforatum | Lithuania 2 |
| Ames 27516 | H. perforatum L. perforatum | Lithuania 2 |
| Ames 27517 | H. perforatum L. perforatum | Lithuania 2 |
| Ames 27518 | H. perforatum L. perforatum | Lithuania 2 |
| Ames 27519 | H. perforatum L. perforatum | Lithuania 2 |
| Ames 27520 | H. perforatum L. perforatum | Lithuania 2 |
| Ames 27700 | H. perforatum L. perforatum | South Bohemia, Czech Republic2 |
| Ames 27701 | H. perforatum L. perforatum | South Moravia, Czech Republic 2 |
| Ames 27702 | H. perforatum L. perforatum | South Moravia, Czech Republic 2 |
| Ames 27703 | H. perforatum L. perforatum | South Moravia, Czech Republic 2 |
| Ames 27705 | H. perforatum L. perforatum | South Moravia, Czech Republic 2 |
| Ames 27706 | H. perforatum L. perforatum | South Moravia, Czech Republic 2 |
| Ames 27708 | H. perforatum L. perforatum | South Moravia, Czech Republic 2 |
| Ames 27710 | H. perforatum L. perforatum | East Bohemia, Czech Republic 2 |
| Ames 27711 | H. perforatum L. perforatum | East Bohemia, Czech Republic 2 |
| Ames 27712 | H. perforatum L. perforatum | East Bohemia, Czech Republic 2 |
| Ames 27713 | H. perforatum L. perforatum | North Moravia, Czech Republic 2 |
| Ames 27714 | H. perforatum L. perforatum | North Moravia, Czech Republic 2 |
| Ames 27716 | H. perforatum L. perforatum | West Bohemia, Czech Republic 2 |
| Ames 27736 | H. perforatum L. perforatum | Missouri, United States 4 |
| Ames 27753 | H. perforatum L. songaricum | Uzbekistan 3 |
| Ames 27756 | H. perforatum L. songaricum | Uzbekistan 3 |
| Ames 27757 | H. perforatum L. songaricum | Uzbekistan 3 |
| PI 325351 | H. perforatum L. perforatum | Stavropol Region, Russia 3 |
| Ames 27430 | H. tetrapterum Fr. | East Bohemia, Czech Republic |
| PI 636398 | H. undulatum Schousb. | Coimbra, Portugal |
| Ames 27737 | H. punctatum Lam. | Missouri, United States |
| Ames 27744 | H. punctatum Lam. | Arkansas, United States |
| Ames 27747 | H. punctatum Lam. | Missouri, United States |
| Ames 27424 | H. hirsutum L. | Central Bohemia, Czech Rep. |
| Ames 27426 | H. humifusum L. | Central Bohemia, Czech Rep. |
| Ames 27061 | H. densiflorum Pursh | Tennessee, United States |
| Unknown | H. adpressum W. P. C. Barton | Unknown |
| Ames 27440 | H. ascyron subsp. pyramidatum N. Robson | Unknown |
| Ames 27470 | H. ascyron subsp. pyramidatum N. Robson | Iowa, United States |
| Ames 27593 | H. ascyron subsp. pyramidatum N. Robson | Illinois, United States |
| Ames 26858 | H. androsaemum L. | Coimbra, Portugal |
| Ames 27480 | H. gentianoides L. | Florida, United States |
| Ames 27751 | Triadenum walteri (J. G. Gmel.) | Arkansas, United States |
| Ames 27752 | Triadenum walteri (J. G. Gmel.) | Arkansas, United States |
Accession collection locations of Hypericum spp. are designated as follows: 1 = domesticated, 2 = Europe, 3 = East Europe/Asia, 4 = United States of America.
Total genomic DNA was extracted from leaf tissue using the DNeasy Plant Mini kit (Qiagen Inc.; Valencia, CA, USA) in accordance with the supplied protocol and quantified using a Nanodrop (Nanodrop Technologies; Wilmington, DE, USA) spectrophotometer. Amplified fragment length polymorphism (AFLP) analysis was run on each sample and its technical replicate in accordance to Vos et al. [16], with modifications to include slight differences in adapter and primer sequences (Table 2). Digestion, ligation, pre-selective and selective amplifications were performed as in Hawkins et al. [22]. Following amplifications, samples were submitted to the DNA facility of the Iowa State University and run on an ABI 3100 Genetic Analyzer (Applied Biosystems; Foster City, CA, USA).
Table 2.
AFLP primer and adapter sequences
| Adapters | 5′-Sequence-3′ |
|---|---|
| EcoRI forward adapter | CTC GTA TAC TGC GTA CC |
| EcoRI reverse adapter | AAT TGG TAC GCA GTA |
| Msel forward adapter | GAC GAT GAG TCC TGA G |
| Msel reverse adapter | TAC TCA GGA CTC ATC |
| + 1 Pre-selective primers | |
| EcoRI + A | TAC TGC GTA CCA ATT C – A |
| Msel + C | GAC GAT GAG TCC TGA GTA A – C |
| + 3 Selective primers | |
| Msel + CAA (I) | GAC GAT GAG TCC TGA GTA A – CAA |
| Msel + CAC (III, IV) | GAC GAT GAG TCC TGA GTA A – CAC |
| EcoRI + AGC (A) | (FAM) - TAC TGC GTA CCA ATT C – AGC |
| EcoRI + ACG (B) | (HEX) - TAC TGC GTA CCA ATT C – ACG |
| EcoRI + AAC (C) | (HEX) - TAC TGC GTA CCA ATT C – AAC |
Four + 3 selective amplifications, designated IA, IIIA, IIIB, and IVC, were performed. Roman numerals represent non-labeled selective primers and the letters “A, B, or C” represent the 5′ FAM or 5′ HEX labeled primers with bold-type indicating selective nucleotides.
AFLP banding patterns were visualized with Genographer 1.6.0 [23]. For analytical purposes, bands of the same size were considered homologous, even though it is possible that some bands of the same size may actually represent non-homologous genomic fragments. Visual comparisons between three biological replicates, as well as two technical replicates, were used to determine reproducibility. Bands absent from two of the three biological replicates and their corresponding technical replicates were excluded from the study. Homologous bands were scored for presence (1) or absence (0).
To visualize relationships among accessions, Neighbor-joining analysis was conducted in Paup* version 4.0 [24], using the 56 accessions of Hypericum spp. and rooting with two accessions of Triadenum walteri. Default settings were employed, except “Break ties” was set to “randomly” and distances were calculated using Nei's [25] restriction-site distances. Branch support was assessed through the implementation of 5000 bootstrap replicates. Principal coordinate analysis (PCO) was performed with NTSYS-pc [26] to obtain an additional visual representation of patterns of genetic variation in the wild and cultivated material and to explore possible relationships with geography. Genetic diversity within H. perforatum was hierarchically partitioned using analysis of molecular variance (AMOVA) [27] in the GenAlEx program [28].
Results
AFLP markers were generated for 56 accessions of Hypericum spp. and 2 accessions of Triadenum walteri. Four AFLP primer combinations produced a total of 298 easily scored and reproducible markers. Within the 42 H. perforatum accessions, 221 markers were generated, of which 204 (92%) are polymorphic and 17 (8%) are present (monomorphic) in all accessions. Of the 17 monomorphic markers, only two (IVC134 and IVC335) were specific to H. perforatum, while the other 15 were present in accessions outside of H. perforatum. However, AFLP analysis generated 28 polymorphic H. perforatum-specific markers (Table 3). Of these, 10 were present at a frequency of 50% or more, 9 were present in 20–49% of the accessions, and 11 were present in less than 20% of the H. perforatum accessions.
Table 3.
Markers detected in H. perforatum as revealed by AFLP analysis
| Marker | % present | Accessions positive for markers |
|---|---|---|
| Markers present only in H. perforatum (30) | ||
| IA258 | Polymorphic (40 %) | 27427, 27429, 27453, 27454, 27455, 27510, 27511, 27512, 27516, 27517, 27518, 27519, 27700, 27701, 27703, 27705, 27708 |
| IA320 | Polymorphic (2 %) | 27515 |
| IA321 | #Polymorphic (31 %) | 27427, 27428, 27455, 27490, 27515, 27700, 27701, 27702, 27708, 27710, 27712, 27713, 27714 |
| IA323 | Polymorphic (7 %) | 27452, 27511, 27512 |
| IA360 | Polymorphic (21 %) | 27443, 27452, 27491, 27493, 27706, 27716, 27736, 27756, 27757 |
| IA409 | Polymorphic (62 %) | All accessions except: 26862, 27429, 27443, 27452, 27453, 27454, 27491, 27493, 27516, 27517, 27518, 27519, 27706, 27711, 27713, 27753 |
| IA415 | Polymorphic (36 %) | 27427, 27453, 27454, 27455, 27511, 27512, 27700, 27701, 27703, 27705, 27708, 27716, 27736, 27757, 325351 |
| IA430 | Polymorphic (7 %) | 27517, 27519, 27713 |
| IIIA321 | Polymorphic (2 %) | 27443 |
| IIIA358 | Polymorphic (33 %) | 26862, 27428, 27490, 27491, 27493, 27513, 27515, 27520, 27702, 27710, 27711, 27712, 27714, 27736 |
| IIIA378 | Polymorphic (74 %) | All accessions except: 26862, 27429, 27491, 27493, 27516, 27517, 27518, 27519, 27520, 27736, 27756 |
| IIIA398 | Polymorphic (62 %) | All accessions except: 27342, 27427, 27443, 27452, 27513, 27520, 27700, 27701, 27702, 27706, 27716, 27736, 27753, 27756, 27757, 325351 |
| IIIA467 | Polymorphic (14 %) | 27342, 27428, 27453, 27454, 27706, 27712 |
| IIIB200 | Polymorphic (83 %) | All accessions except: 26862, 27343, 27513, 27736, 27753, 27756, 27757 |
| IIB245 | Polymorphic (71 %) | All accessions except: 26862, 27429, 27452, 27491, 27493, 27510, 27516, 27517, 27518, 27519, 27704, 27711, 27757 |
| IIIB286 | Polymorphic (21 %) | 27455, 27516, 27518, 27519, 27700, 27701, 27708, 27713, 325351 |
| IIIB288 | Polymorphic (36 %) | 27427, 27428, 27429, 27453, 27454, 27490, |
| IIIB320 | Polymorphic (14 %) | 27427, 27428, 27490, 27702, 27710, 27712 |
| IIIB389 | Polymorphic (33 %) | 27343, 27427, 27428, 27452, 27490, 27491, 27493, 27510, 27515, 27702, 27710, 27712, 27714, 27753 |
| IIIB390 | Polymorphic (52 %) | 27427, 27428, 27452, 27453, 27454, 27455, 27490, 27513, 27515, 27700, 27701, 27702, 27703, 27705, 27706, 27708, 27710, 27712, 27713, 27714, 27716, 325351 |
| IIIB404 | Polymorphic (2 %) | 26862 |
| IIIB473 | Polymorphic (2 %) | 27490 |
| IVC134 | Monomorphic (100 %) | All accessions |
| IVC253 | Polymorphic (29 %) | 27427, 27453, 27454, 27455, 27511, 27512, 27700, 27701, 27703, 27705, 27708, 27716 |
| IVC254 | Polymorphic (5 %) | 27511, 27512 |
| IVC259 | Polymorphic (16 %) | 27427, 27455, 27700, 27701, 27708, 27736 |
| IVC335 | Monomorphic (100 %) | All accessions |
| IVC355 | Polymorphic (52 %) | 27427, 27428, 27455, 27490, 27491, 27493, 27510, 27512, 27513, 27515, 27519, 27700, 27701, 27702, 27706, 27708, 27710, 27712, 27713, 27714, 27716, 27736 |
| IVC383 | Polymorphic (10 %) | 27453, 27454, 27736, 325351 |
| IVC412 | Polymorphic (50 %) | 27342, 27343, 27428, 27453, 27454, 27490, 27491, 27493, 27513, 27515, 27701, 27702, 27703, 27705, 27706, 27710, 27712, 27714, 27716, 27736, 27753 |
Markers present in all H. perforatum accessions (17)
IA119, IA127, IA185, IA222, IA290, IIIA106, IIIA121, IIIA222, IIIA249, IIIB271, IVC114, IVC118, IVC134, IVC202, IVC217, IVC310, IVC335
Neighbor-joining analysis (Fig.1) revealed a monophyletic H. perforatum clade supported by a bootstrap value of 80. Within the H. perforatum clade there is a basal monophyletic group (clade 3) composed primarily of accessions from Lithuania and supported by a bootstrap value of 99. The remainder of the H. perforatum accessions are sister to this basal Lithuanian group and are divided into two additional major clades (clades 1 and 2) and one minor paraphyletic group (“paraphyletic accessions”). The larger clade (clade 2) predominately contains accessions from the Czech Republic and appears to be divided into 3 groups, each with bootstrap support of 100. Three of the four domesticated H. perforatum accessions studied (Ames 27453–27455) are located within this clade. The remaining major clade (clade 1) is comprised of accessions representing all 4 of the sub-species found in H. perforatum. As expected, the H. perforatum clade and its sister group, the Hypericum spp. clade, are composed of species sharing the characteristic dark leaf glands, these containing hypericin, pseudohypericin and hyperforin.
Fig. 1.
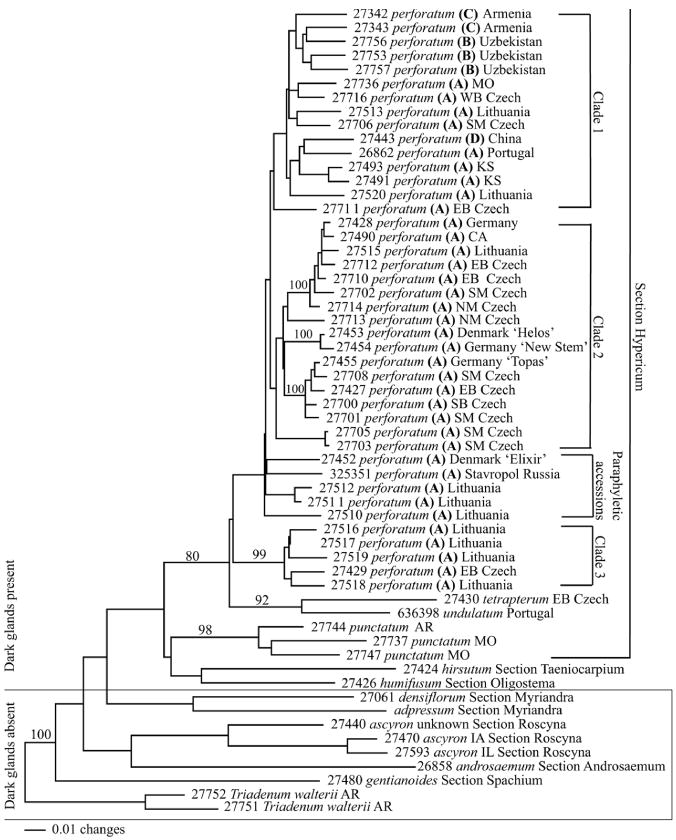
Neighbor-joining analysis of Hypericum spp. Bold letters represent different sub-species A = perforatum, B = songaricum, C = veronense, and D = chinense. Accessions in single quotes indicate cultivated accessions. Taxa outside of box contain dark glands. Numbers along branches denote bootstrap support.
PCO analysis on AFLP data derived from all accessions show a clear delineation between H. perforatum and all other accessions (Fig. 2A). Congruent with the neighbor-joining analysis, the H. perforatum accessions appear in a tight cluster most closely associated with other Hypericum spp. that produce dark glands. When only H. perforatum accessions are included in the PCO analysis, three separate clusters are apparent, consistent with the three major clades recovered in the neighbor-joining analysis (Fig. 2B). Additionally, 5 accessions (Ames 27452, 27510, 27511, and 27512, and PI 325351) occupy an intermediate position in the PCO outside of the three major clusters, indicative of possible introgression. These five accessions are basal to the larger two H. perforatum clades in the neighbor-joining tree.
Fig. 2.
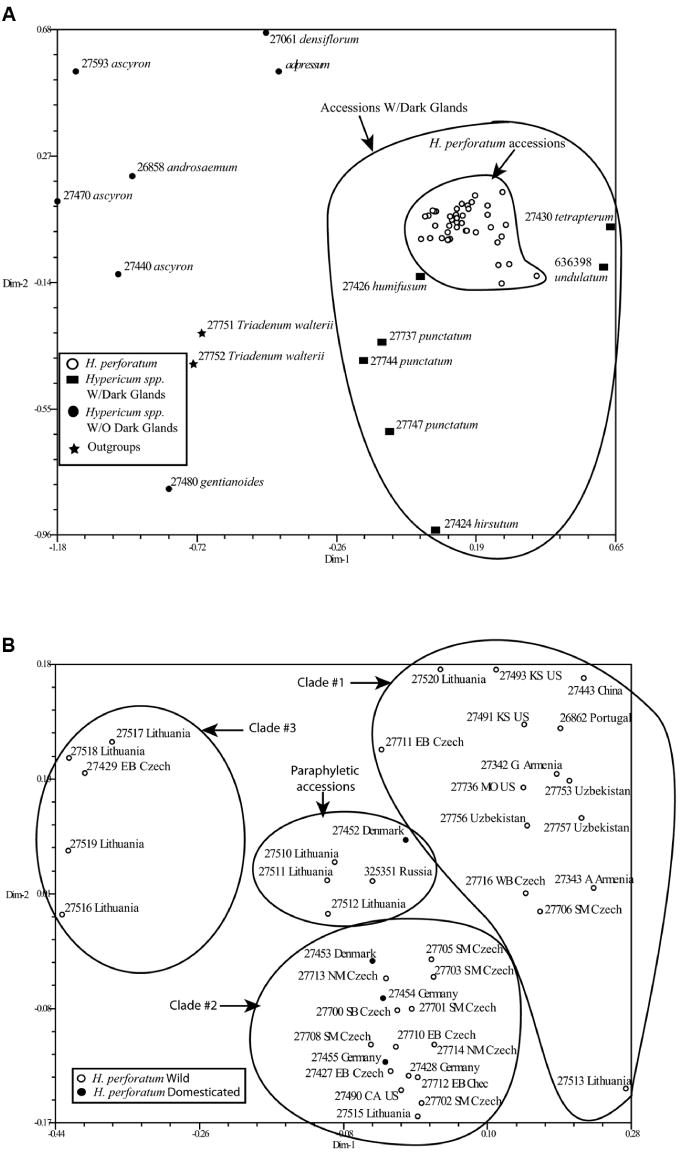
A PCO illustrating the relationship of all Hypericum spp. and the outgroup Triadenum walteri. Inner circle is composed of only H. perforatum accessions and outer circle contains all accessions with dark, hypericin-containing glands. B PCO illustrating the relationship of domesticated and non-domesticated accessions of Hypericum perforatum. Circles delineate accessions based upon clade designation from the neighbor-joining analysis.
The distribution of genetic diversity within and between Hypericum spp. populations was explored using AMOVA. Accessions were grouped together based on region of origin and/or domestication, only polymorphic markers were employed, and the analysis examined both the global and locus-by-locus partitioning of genetic diversity (Table 4). First, AMOVA was used to evaluate partitioning of genetic diversity within and among accessions from different geographic areas. Results indicate that the majority of variation (64%) present in the H. spp. used for this study can be attributed to within-population differences, while 36% of the variation can be ascribed to among-population genetic variation. When only H. perforatum accessions are included in the AMOVA, 88% of the variation can be attributed to within-population differences due to geographic collection locations, while only 12% of the variation can explain among-populations differences. Second, if the populations are segregated by domestication, the measure of variation within-populations increased to 94%, while variation among-populations decreased to 6%. A closer look at the PCO indicates that there is a domesticated accession (Ames 27452 Elixir; Richters, Goodward, Canada) central to the 3 main groups. This could be a result of hybridization and introgression of genetic material from the other primary clusters. With this in mind, the same comparison was made while excluding Elixir. The resulting analysis indicated that the within-population variance is 11% and the among-population variance is 89%. A fourth AMOVA, consisting of only H. perforatum accessions and segregated into 4 populations based on relatedness as indicated by neighbor-joining analysis (accessions in clade 1 = population 1 etc.), was conducted. The among-population variance is 33% and the within-population variance is 67%.
Table 4.
Analysis of molecular variance (AMOVA) for Hypericum spp.
| Source of variation | d.f. | Sum of Squares | Variance components | % Total variance | Probability |
|---|---|---|---|---|---|
| Comparison between H. perforatum and other Hypericum species | |||||
| Among-population | 1 | 454.19 | 19.95 | 36% | 0.001 |
| Within-population | 54 | 1905.67 | 35.29 | 64% | |
| Total | 55 | 2359.86 | |||
| Comparison of H. perforatum accessions by geographic collection locations (Domesticated, Europe, East Europe/Asia, and US) | |||||
| Among-populations | 3 | 178.97 | 4.00 | 12% | 0.002 |
| Within-populations | 38 | 1117.70 | 29.41 | 88% | |
| Total | 41 | 1296.67 | |||
| Comparison of H. perforatum accessions by domestication state (Domesticated, Wild) | |||||
| Among-populations | 1 | 46.17 | 2.06 | 6% | 0.073 |
| Within-populations | 40 | 1250.50 | 31.26 | 94% | |
| Total | 41 | 1296.67 | |||
| Comparison of H. perforatum accessions by domestication excluding 27452 ‘Elixir’ (Domesticated, Wild) | |||||
| Among-populations | 1 | 52.73 | 3.90 | 11% | 0.034 |
| Within-populations | 39 | 1210.00 | 31.03 | 89% | |
| Total | 40 | 1262.73 | |||
| Comparison of H. perforatum accessions by distribution within neighbor-joining analysis | |||||
| Among-populations | 3 | 403.40 | 11.65 | 33% | 0.001 |
| Within-populations | 38 | 893.27 | 23.51 | 67% | |
| Total | 41 | 1296.67 | |||
Genetic distance within H. perforatum was measured using Nei's unbiased measure of genetic distance. The highest estimate of genetic distance (0.187) was between domesticated accessions and accessions from the Eastern Europe/Asia region. While the lowest estimate of genetic distance (0.077) was between domesticated accessions and European accessions.
Discussion
One obstacle facing breeders, horticulturists, researchers, and oversight agencies working with medicinals is the inability to genetically determine the source of plant material. Markers generated in this study may aid in overcoming this obstacle. Of 298 polymorphic markers generated, 17 markers are present in all Hypericum accessions and 30 markers are present in only H. perforatum. Two markers (IVC134, IVC335), are specific for H. perforatum, and present in all accessions studied; these may prove particularly useful for identification of H. perforatum plant material. Collectively, the 30 unique H. perforatum markers may aid breeders in determining genetic identity and source, can be employed as a tool by producers to accurately diagnose the identity of individual plant lots, and could be useful to agencies or consumer groups as a means to evaluate end-user H. perforatum “St. John's wort” preparations. Additionally, this molecular marker study provides the foundation for future work focused on developing species-specific primers that could be used to identify material purported to be H. perforatum with a single PCR reaction.
Neighbor-joining analysis supports the delineation of Hypericum spp. that either have or lack hypericin-containing dark glands. It is also evident from both neighbor-joining and PCO analysis that H. perforatum clusters tightly and separately from other species of Hypericum. Within H. perforatum, three distinct clades and one minor paraphyletic group are observed. Three of the four domesticated accessions belong to the same clade (clade 2), and two of those accessions are phylogenetically sister to one another and share boot-strap support of 100%. Interestingly, clade 2 is comprised entirely of subspecies perforatum, suggesting that the domesticated accessions originate from within this group. None of the domesticated accessions in our study belong to clades 1 or 3, which contain members of subspecies perforatum (clade 3) and a mixture of subspecies perforatum, songaricum, and veronense (clade 1). It is within these clades that breeders may choose to look in order to identify new traits or increase genetic diversity within the domesticated accessions.
Accessions from regional geographic areas tend to be more closely related. However, there are multiple instances where H. perforatum accessions from one location are more closely associated with those from different locations. Additionally, only 12% of the total amount of genetic diversity observed can be attributed to among-population difference, indicative of high levels of gene flow between populations. For example, the presence of the California accession Ames 27490 in clade 2; given that wild populations from California are naturalized from foreign introductions, it is not surprising that this accession groups with the European, domesticated species in clade 2. Observations such as this are consistent with results previously shown in H. perforatum illustrating that populations from different geographic areas can and often times are more closely related [29].
Analysis of molecular variance between H. perforatum accessions and Hypericum accessions from other species indicates that there is a high level of among-population variation (36%). This indicates an abundance of variation at the genus level, which the phylogenetic and clustering analyses readily partitioned into distinctive groupings. When comparing only H. perforatum accessions by geographic region of collection, 88% of the variance occurs within populations. The variance due to geographic distribution was similar to variance attributed to domestication if the cultivated accession Elixir was excluded from the analysis. Elixir, which is centrally located in the PCO, is responsible for 5% of the among-population variance within the domesticated varieties. The high within-population variation exhibited in the analysis when the populations are distinguished by either geographic location or domestication, along with the findings of Maron et al. [25], encouraged us to re-analyze the data with the populations segregated in accordance with the neighbor-joining analysis. When analyzed under these conditions, the level of among-population variation is substantially increased. These findings imply dispersal of plant material outside of their original range, most likely with human assistance.
Genetic distance analysis of the AFLP data revealed that the cultivated populations studied share higher genetic identity with the Western and Central European populations (0.925) than with populations from East Europe and Asia (0.828). This could be attributed to the fact that the cultivated varieties used in this study were developed in Germany and Denmark. Additional studies involving a larger sampling of domesticated material will help distinguish these possibilities, and may shed additional light on the source(s) and number of times that H. perforatum has been domesticated.
While other studies have utilized a molecular approach to place H. perforatum within a phylogenetic framework [14], [20], [29], [30], this is the first study placing an emphasis on the relationships and diversity between both wild and cultivated accessions of H perforatum, within the overall phylogenetic framework of the genus Hypericum. This study demonstrates that there is a great deal of genetic diversity among Hypericum species as well as within H. perforatum, and that this diversity is structured phylogenetically and geographically. These data provide the foundation for future work characterizing the evolutionary history, genetic relationships, and recent domestication of St. John's wort and its closely related species.
Acknowledgments
We would like to thank Nathan Johnson for his technical assistance. Mention of commercial brand names does not constitute an endorsement of any product by the U.S. Department of Agriculture or cooperating agencies. This publication was made possible by grant number P01ES012020 from the National Institute of Environmental Health Sciences (NIEHS) and the Office of Dietary Supplements (ODS), NIH. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIEHS, NIH.
References
- 1.Robson N. Hypericum botany. In: Ernst E, editor. The genus Hypericum. London: Taylor and Francis; 2003. pp. 1–22. [Google Scholar]
- 2.Robson NKB. Studies in the genus Hypericum L. (Guttiferae) 1. Infrageneric classification. Bull Brit Mus (Natural History, Botany series) 1977:291–355. [Google Scholar]
- 3.Campbell MH, Delfosse ES. The biology of Australian weeds. 13. Hypericum perforatum L. J Aust I Agr Sci. 1984;50:63–73. [Google Scholar]
- 4.Martonfi P, Brutovska R, Cellarova E, Repcak M. Apomixis and hybridity in Hypericum perforatum. Folia Geobot Phytotaxon. 1996;31:389–96. [Google Scholar]
- 5.Matzk F, Meister A, Brutovska R, Schubert I. Reconstruction of reproductive diversity in Hypericum perforatum L. opens novel strategies to manage apomixis. Plant J. 2001;26:275–82. doi: 10.1046/j.1365-313x.2001.01026.x. [DOI] [PubMed] [Google Scholar]
- 6.Barcaccia G, Arzenton F, Sharbel TF, Varotto S, Parrini P, Lucchin M. Genetic diversity and reproductive biology in ecotypes of the facultative apomict Hypericum perforatum L. Heredity. 2006;96:322–34. doi: 10.1038/sj.hdy.6800808. [DOI] [PubMed] [Google Scholar]
- 7.Mayo GM. Modes of reproduction in Australian populations of Hypericum perforatum L. (St. John's wort) revealed by DNA fingerprinting and cytological methods. Genome. 2003;46:573–9. doi: 10.1139/g03-038. [DOI] [PubMed] [Google Scholar]
- 8.Axarlis S, Mentis A, Demetzos C, Mitaku S, Skaltsounis AL, Marselos M, et al. Antiviral in vitro activity of Hypericum perforatum L. extract on the human cytomegalovirus (HCMV) Phytother Res. 1998;12:507–11. [Google Scholar]
- 9.Barnes J, Anderson LA, Phillipson JD. St John's wort (Hypericum perforatum L.): a review of its chemistry, pharmacology and clinical properties. J Pharm Pharmacol. 2001;53:583–600. doi: 10.1211/0022357011775910. [DOI] [PubMed] [Google Scholar]
- 10.Greeson JM, Sanford B, Monti DA. St. John's wort (Hypericum perforatum): a review of the current pharmacological, toxicological, and clinical literature. Psychopharmacology. 2001;153:402–14. doi: 10.1007/s002130000625. [DOI] [PubMed] [Google Scholar]
- 11.Nahrstedt A, Butterweck V. Biologically active and other chemical constituents of the herb of Hypericum perforatum L. Pharmacopsychiatry. 1997;30:129–34. doi: 10.1055/s-2007-979533. [DOI] [PubMed] [Google Scholar]
- 12.Campbell MH, May CE, Southwell IA, Tomlinson JD, Michael PW. Variation and varietal determination in Hypericum perforatum L. (St. John's wort) in New South Wales. Plant Prot Q. 1992;7:43–5. [Google Scholar]
- 13.Southwell IA, Campbell MH. Hypericin content variation in Hypericum perforatum in Australia. Phytochemistry. 1991;30:475–8. [Google Scholar]
- 14.Aziz N, Sauve RJ, Long D, Cherry M. Genetic and phytochemical diversity assessment among eleven Hypericum accessions via AFLP and HPLC analyses. J Herbs Spices Med Plants. 2006;12:97–105. [Google Scholar]
- 15.Arnholdt-Schmitt B. RAPD analysis: a method to investigate aspects of the reproductive biology of Hypericum perforatum L. Theor Appl Genet. 2000;100:906–11. [Google Scholar]
- 16.Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–14. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaudeul M, Taberlet P, Till-Bottraud I. Genetic diversity in an endangered alpine plant, Eryngium alpinum L. (Apiaceae), inferred from amplified fragment length polymorphism markers. Mol Ecol. 2000;9:1625–37. doi: 10.1046/j.1365-294x.2000.01063.x. [DOI] [PubMed] [Google Scholar]
- 18.Soleimani VD, Baum BR, Johnson DA. AFLP and pedigree-based genetic diversity estimates in modern cultivars of durum wheat [Triticum turgidum L. subsp. durum (Desf.) Husn.] Theor Appl Genet. 2002;104:350–7. doi: 10.1007/s001220100714. [DOI] [PubMed] [Google Scholar]
- 19.Blears MJ, Grandis SA, Lee H, Trevors JT. Amplified fragment length polymorphism (AFLP): a review of the procedure and its applications. J Ind Microbiol Biotechnol. 1998;21:99–114. [Google Scholar]
- 20.Gustafsson MHG, Bittrich V, Stevens PF. Phylogeny of Clusiaceae based on rbcL sequences. Int J Plant Sci. 2002;163:1045–54. [Google Scholar]
- 21.Robson N. Studies in the genus Hypericum L. (Guttiferae) 4(2). Section 9. Hypericum sensu lato (part 2): subsection 1. Hypericum series 1. Hypericum. Bull Brit Mus (Natural History, Botany series) 2002;32:61–123. [Google Scholar]
- 22.Hawkins JS, Pleasants J, Wendel JF. Identification of AFLP markers that discriminate between cultivated cotton and the Hawaiian island endemic, Gossypium tomentosum Nuttall ex Seeman. Genet Resour Crop Evol. 2005;52:1069–78. [Google Scholar]
- 23.Benham J, Jeung JU, Jasieniuk M, Kanazin V, Blake T. Genographer: a graphical tool for automated fluorescent AFLP and microsatellite analysis. J Agric Genomics. 1999:1–3. [Google Scholar]
- 24.Swafford DL. Phylogenetic analysis using parsimony (*and other methods), version 4. Sunderland: Sinauer; 2002. [Google Scholar]
- 25.Nei M. Genetic distance between populations. Am Nat. 1972;106:283–92. [Google Scholar]
- 26.Rohlf FJ. NTSYS-pc. Numerical taxonomy and multivariate analysis system, Version 2.10. New York: Exeter Software; 2002. [Google Scholar]
- 27.Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–91. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peakall R, Smouse PE. GENEALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6:288–95. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maron JL, Vila M, Bommarco R, Elmendorf S, Beardsley P. Rapid evolution of an invasive plant. Ecol Monogr. 2004;74:261–80. [Google Scholar]
- 30.Crockett SL, Douglas AW, Scheffler BE, Khan IA. Genetic profiling of Hypericum (St. John's wort) species by nuclear ribosomal ITS sequence analysis. Planta Med. 2004;70:929–35. doi: 10.1055/s-2004-832619. [DOI] [PubMed] [Google Scholar]


