Abstract
Genetic data from humans and mice reveal that the formation of cementum is sensitive to intra- and extracellular phosphate/pyrophosphate distribution. The intracellular molecular pathways whereby altered levels of extracellular phosphate concentration may affect cementum formation have not been elucidated. To initiate inquiry, we have studied the temporal effects of extracellular phosphate on global patterns of gene expression in a line of immortalized mouse cementoblasts. Total RNA from cultured cementoblasts treated with 5 mM inorganic phosphate over a designated time period, from 1–48 hrs, was analyzed for global patterns of gene expression by means of DNA microarrays representing the complete mouse genome. Analyses of significant hybridization signals indicated that 5 mM extracellular phosphate alters the expression of genes comprising several gene ontology (GO) groups, including transcription factor activity and Wnt signaling.
Keywords: cementoblasts, phosphate, global gene expression
INTRODUCTION
Experimental and clinical demonstrations that periodontal regeneration is possible beg a better understanding of the cellular and molecular mechanisms that regulate the formation of these tissues during development and regeneration. In this regard, our laboratory has been using a murine model of tooth development to identify the temporospatial expression of molecules considered to be regulators of cementogenesis. Our group, as well as others, has demonstrated that genes linked to phosphate regulation may prove to have a vital role in modulating cementoblast behavior and subsequent cementogenesis.
Genetic defects in several cell membrane proteins that are known to regulate extracellular pyrophosphate (ePPi) levels have been implicated in the regulation of cementum formation (Beertsen et al., 1999). These include PC-1 (NPP1, a nucleoside triphosphate pyrophosphohydrolase), TNAP (tissue non-specific alkaline phosphatase), and ANK (ankylosis) (Terkeltaub, 2001). TNAP disruption leads to hypophosphatemia and premature exfoliation of primary teeth. TNAP-/- mice fail to form cementum. PC-1 and ANK mutant mice develop more than 10 times as much cementum, while dentin, the periodontal ligament, and alveolar bone appear to be structurally and functionally normal (Nociti et al., 2002).
The addition of inorganic or β-glycerol phosphate to cultures of cells involved in the formation of mineralized tissue is essential for mineral formation in vitro. We have determined that 5 mM extracellular inorganic phosphate (ePi) increased transcripts for Dmp1, Opn, Ank, PC-1, and Pit 1 and decreased expression of Bsp, Ocn, Col 1, and Tnap, and that these changes were significantly reduced by foscarnet, an inhibitor of ePi uptake (Foster et al., 2006).
Analysis of these and other data (Harmey et al., 2004) suggests that the modulation of extracellular levels of pyrophosphate/inorganic phosphate (PPi/Pi) is important in the regulation of mineralized tissue formation. The molecular pathway(s) whereby ePi regulates mineralized tissue formation has/have not been fully defined. The purpose of these studies was to test the hypothesis that Pi is a signaling molecule for cementoblasts that induces changes in the expression of currently unidentified groups of functionally related genes critical to cementogenesis. To systematically identify the mechanisms by which Pi alters cell function and, specifically, gene expression over time, we examined the effects of 5 mM extracellular Pi (ePi), over a designated time period, on global patterns of gene expression in an immortalized, cloned line of cementoblasts in vitro.
MATERIALS & METHODS
Cell Culture
Immortalized murine cementoblasts were maintained in culture as previously described (Foster et al., 2006) in a protocol approved by the University Committee on Use and Care of Animals and in compliance with state and Federal laws.
Gene Expression Experiments
Our immortalized clones of murine cementoblasts were plated in 60-mm dishes at a concentration of 2.6 x 104 cells/cm2 and maintained in Dulbecco’s Modified Eagle Medium (DMEM) with 10% fetal bovine serum (FBS) supplemented with penicillin, streptomycin, and glutamine. Upon reaching confluence, media were changed to DMEM with 5% FBS, and experimental treatments were added. Phosphate was added to media at a dose of 5 mM. The selection of this dose was based on our earlier data (Foster et al., 2006). A stock solution of 100 mM Pi was made in DMEM media at a pH of 7.4, and filter-sterilized. Total RNA was isolated with Trizol® Reagent (Invitrogen/GIBCO/BRL, Carlsbad, CA, USA) at the indicated times after the initial addition of 5 mM Pi. The Agilent Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA, USA) was used to ensure RNA quality (unpublished observations).
Real-time RT-PCR
Selected genes were confirmed by real-time RT-PCR as described (Foster et al., 2006). For real-time RT-PCR analysis, RNA was converted to cDNA, and 2.0 μL of the resulting cDNA product was used per 20-μL reaction in a real-time PCR Roche LightCycler system (Roche Diagnostics GmbH, Mannheim, Germany). PCR reactions were carried out with the DNA Master SYBR Green I kit (Roche Diagnostic Co., Indianapolis, IN, USA), with a total volume of 20 μL. Primers were designed by LightCycler probe design software (Roche GmbH, Mannheim, Germany). A BLAST search of GenBank was performed on the primer sequences to ensure specificity, and melting curve analysis of products was additionally performed to ensure specificity. Expression was analyzed for genes of interest with GAPDH serving as a housekeeping/reference gene for normalization. The amplification profile used on the LightCycler was: 95/0, 55/7, 72/20 (temperature [°C]/time [sec]), and 35–40 cycles. Primer sequences used are listed in the Table. All primers were used at a concentration of 0.5 μM (except DMP1, used at 0.25 μM to optimize PCR conditions), in 3 mM MgCl2.
Table.
GO Groups with Cementoblast Genes Regulated by Pi
| Gene Ontology (GO) Term | Percentage of Nested Genes Meeting Criterion (fold change ≥± 2) |
|---|---|
| Transcription | 12* |
| Cell growth and maintenance | 7.5 |
| Development | 20 |
| Cell-matrix adhesion | 50 |
| Ossification | 50 |
| Wnt receptor signaling | 24 |
| Protein amino acid dephosphorylation | 10.5 |
Values are the percent of cementoblast genes meeting our pre-set criterion nested in the GO terms as calculated by MAPPFinder 2 after 24 hrs of Pi treatment.
Relative quantification of PCR products was achieved by use of the LightCycler Relative Quantification Software, version 1.0 (Roche Diagnostics GmbH, Mannheim, Germany), to compare amplification of the target gene of interest with that of GAPDH as a reference gene, with calibrator normalization and amplification efficiency correction.
Microarray Scanning
We obtained fluorescent-intensity raw data by scanning hybridized arrays (CodelinkTM) using an Axon GenePix 4000B fluorescent scanner and the GenePix Pro imaging software. Fluorescent intensity of each spot in the image was determined with ImaGeneTM 5 software (Biodiscovery, Marina del Rey, CA, USA) for spot-finding and quantitative analysis. Statistical analysis and data normalization were carried out with Bioconductor software. The spot-quantified data files were input into Bioconductor for further processing. Recent reports in statistical analysis of the background correction have pointed out the disadvantages of background correction due to large increases in variance. Therefore, foreground spot intensities from all Codelink Bioarrays were normalized as a group, by quantile normalization as described (Bolstad et al., 2003).
We estimated reproducibility by calculating the coefficient of variation, CV, for the biologically replicated (triplicate) arrays. For an estimate of overall variability (biological plus technical), we used 3 arrays with RNA from 3 different “no treatment” cultures at 24 hrs to calculate an overall median CV of 0.044, 3 arrays with RNA from 3 different “no treatment” cultures at 48 hrs to calculate an overall median CV of 0.038, 3 arrays with RNA from 3 different ± 5 mM phosphate cultures at 24 hrs to calculate an overall median CV of 0.041, and 3 arrays with RNA from 3 different ± 5 mM Pi cultures at 48 hrs to calculate an overall median CV of 0.041. These CV values are within the manufacturer’s quality specifications for technical variability, and are similar to values reported elsewhere (Ramakrishnan et al., 2002).
Differentially expressed genes between the no-treatment groups and the ± 5 mM Pi groups at each time interval were determined by a modified t test followed by a correction for false-discovery rate, FDR (FDR = 0.05), as described elsewhere (Storey and Tibshirani, 2003). These significant gene lists were subsequently analyzed by GenMAPP and MAPPFinder (Doniger et al., 2003), with ≥ ± two-fold as the selection criteria. Experiments, data management, analyses, and storage complied with MIAME standards (Brazma et al., 2001).
RESULTS
Initial studies demonstrated that ePi influences the expression patterns of genes known to be involved in extracellular matrix formation in cementoblasts (Foster et al., 2006). To expand these findings, we determined the effects of culturing cementoblasts in the presence or absence of 5 mM Pi over time on global changes in gene expression. To anchor these data to previous results, we confirmed, by quantitative RT-PCR, ePi-related positive (Ank, Tnap, Dmp 1, Bsp, Ocn, Opn, Timp 1, and Mmp 3 and 23) and negative (S/GK 1, Omd, and BMP 2 and 3) changes in the gene expression patterns of cementoblast cultures over time. The total numbers of significant genes after ePi treatment were: 27 at 1 hr, 299 at 3 hrs, 431 at 6 hrs, 877 at 12 hrs, 4671 at 24 hrs, and 488 at 48 hrs.
Statistically validated gene lists from the array experiments were analyzed by GenMAPP2 and MAPPFinder2 (Doniger et al., 2003), as an initial step for elucidating the biological processes and functions served by ePi-altered genes. Using ≥ ± two-fold change as the limiting criterion, we identified several GO (Gene Ontology) groups of interest (Table, Appendix) for possible further hypothesis formulation and testing. Based on this information, we have chosen to focus initially on transcription factors and Wnt signaling as possible early mediators of ePi signaling.
Alterations in transcription factor genes were among the earliest detected changes. Egr (early growth response) 1 and 2 were elevated at 1 hr and declined to undetectable levels by 3 and 6 hrs, respectively (Fig. 1). Foxc2 (forkhead box c2) was elevated by 3 hrs and sustained through 24 hrs of ePi treatment (Fig. 1). The expression of osterix (Nakashima et al., 2002) and of the transcriptional/developmental regulator Id2 (Peng et al., 2004) was strongly depressed for up to 24 and 48 hrs, respectively. In total, the expression of 18 GO-listed transcription factors met our pre-set limiting criterion of ≥± two-fold in 5 mM ePi-treated cementoblasts (Appendix).
Figure 1.
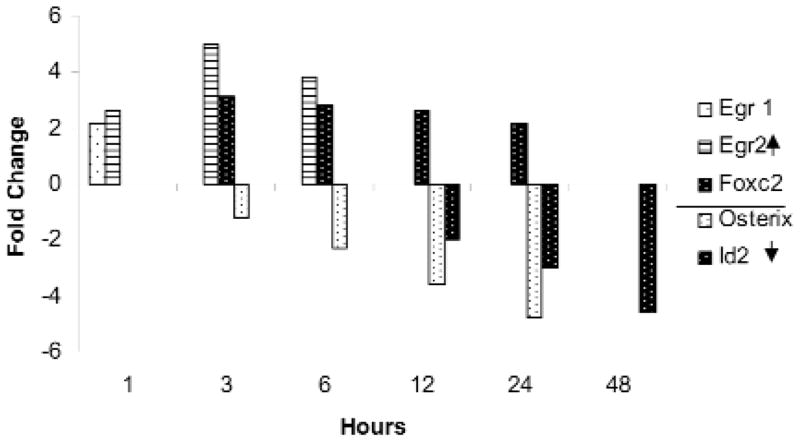
The fold change in expression of selected transcription factors from the total transcription factor GO Group altered by exposure of cementoblasts to 5 mM ePi is displayed. Values are the average of triplicate arrays and differ from controls, p ≤ 0.05.
Of the 58 cementoblast-expressed genes listed in the GO group, “Developmental Processes”, 6 were increased and 12 decreased by 48 hrs of ePi exposure (Appendix). A subset involved in Wnt signaling is depicted in Fig. 2. The regulated genes include: Ppap2b, up by 6 hrs; the signaling Wnts 4 and 10b, down by 3 hrs; Wif (Wnt inhibitory factor )1 and Dkk (Dickkopf) 3, a Wnt receptor-bound inhibitor, down at 12 hrs; as well as Sfrp (secreted frizzed related peptide) 4, another inhibitor of Wnt signaling, up after 12 hrs.
Figure 2.
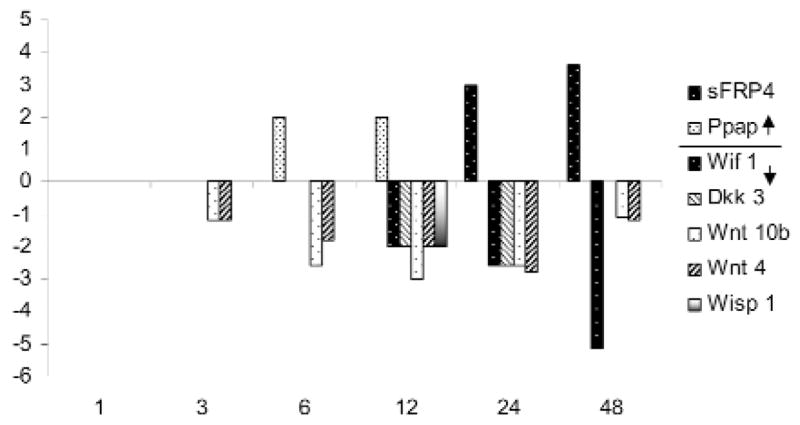
The fold change in expression of selected transcription factors from the total Wnt signaling GO Group altered by exposure of cementoblasts to 5 mM ePi is displayed. Values are the average of triplicate arrays and differ from controls, p ≤ 0.05.
DISCUSSION
Genetic and laboratory data have implicated ePi in the regulation of cementogenesis, yet the relevant metabolic pathways are unknown. Here we demonstrate the utility of global gene expression profiling to discover and further explore signaling and other pathways likely involved in this regulation. Public domain software, GenMAPP2 and MAPPFinder 2 (Doniger et al., 2003), rather than mathematical clustering, was used for initial searching of our validated gene expression datasets for biological relevance. One example of the utility of this method is confirmed by the fact that the data revealed simultaneously enhanced and diminished expression of genes serving common biological functions, such as transcription factor activity. Mathematical clustering groups only genes with identical directional changes in gene expression patterns over time, independent of function.
Transcription Factors
Our hypothesis holds that if ePi is a signaling molecule, then ePi-induced alterations in the expression of transcription factor genes are likely an early event in the regulation of cementogenesis. In this regard, the enhanced expression of Foxc2 and the simultaneous decline in the levels of both osterix and Id2 (Fig. 1) are of particular interest. Foxc2 (that is, Mfh-1), implicated in the proliferation and differentiation of neural-crest-derived mesenchymal cells, has been localized to the perichondrium of developing endochondral bones (Nifuji et al., 2001). The null mutants have craniofacial and other skeletal defects (Nifuji et al., 2001). The zinc finger transcription factor Egr 1, enhanced by 1 hr, is expressed in developing teeth (Karavanova et al., 1992) and joints (Storm and Kingsley, 1999). Interestingly, an Egr 2 binding site is a glucocorticoid-sensitive enhancer of Ocn in osteoblasts (Leclerc et al., 2005), and Egr2-/- mice develop osteoporosis (Levi et al., 1996).
Osterix and Runx2 regulate the differentiation of osteoblasts in intramembranous as well as endochrondral bones (Zelzer and Olsen, 2003), possibly by distinct pathways (Celil et al., 2005). The expression of Id2—one of a family of genes that inhibit basic helix-loop-helix transcription factors and thereby regulate transcription, development, and cell differentiation (Kreider et al., 1992)—must be decreased for terminal differentiation of osteoblasts (Peng et al., 2004). Cementoblast expression of BMP4, which stimulates the early expression of Id2 in osteoblasts, is repressed by ePi (Appendix), while TOB1, an inhibitor of BMP signaling, is enhanced after 24 hrs (Appendix). These intriguing findings, suggesting that ePi diminishes genes associated with bone formation while enhancing a transcription factor (Foxc2) promoting osteoblast differentiation, support the need for functional studies on the role of these genes in cementoblasts and cementogenesis.
Wnt Signaling
The analysis of existing data demonstrates a role for Wnt signaling in bone formation or remodeling (Glass et al., 2005) and tooth development (Pispa and Thesleff, 2003). In this regard, the ability of ePi to alter the expression pattern of genes involved in Wnt signaling is of interest. The expression of one secreted blocker of canonical Wnt signaling, Sfrp 4, was enhanced, while that of another, Wif 1, was depressed. Two Wnt signaling genes, Wnt 10b and Wnt 4, were diminished, as was the level of the membrane-bound inhibitor Dkk 3 (Dickkoff) (Fig. 2). Sfrp 4 (Leimeister et al., 1998) and Wnt 10b (Thesleff et al., 2001) are expressed in developing teeth, and Sfrp 4 has been identified as a potential phosphatonin (Quarles, 2003), a putative circulating regulator of phosphate concentration. Interestingly, Id2 expression, elevated in C3H10T1/2 and C2C12 cells by BMPs 2, 6, and 9 (Peng et al., 2004), is a target of β-catenin (Rockman et al., 2001). BMP4 was depressed in cementoblasts by ePi (Appendix), while BMP 6 and 9 were not detected. Analysis of these data suggests a complex interaction between phosphate regulation and Wnt signaling in cementogenesis (Fig. 3). The regulation of Wnt pathways is also complex, with several pathways leading to β-catenin stabilization (Jones and Jomary, 2002). It is possible that the Pi-related decrease in Wnt signaling results in decreased levels of β-catenin and Id2 expression (Fig. 3). A vital role for Wnt signaling in the regulation of mineralized tissue formation is emerging (Glass et al., 2005). Ppap2b (that is, type 1 collagen-inducible protein, VCIP) increased by 12 hrs and may have a role in cell-cell interactions (Humtsoe et al., 2003).
Figure 3.
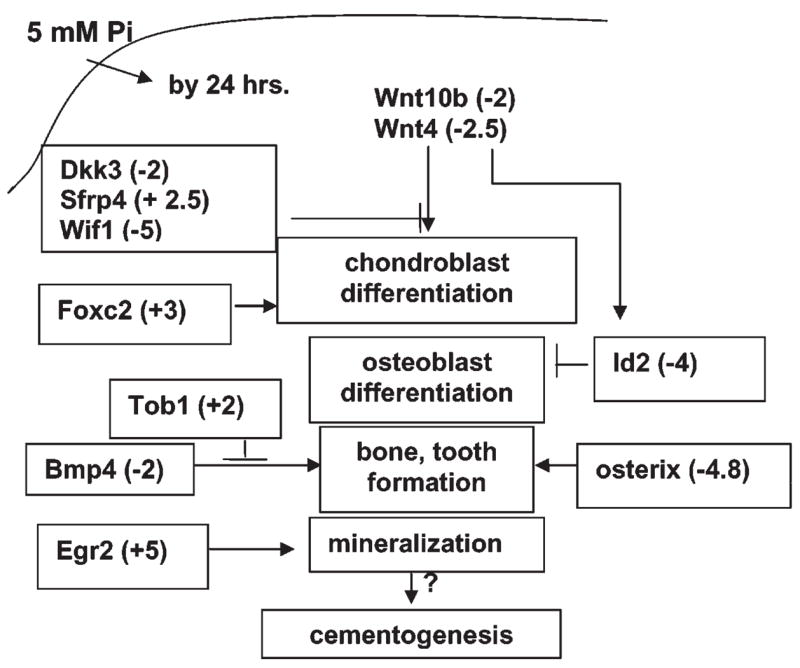
Speculative schematic depicting some known functions of genes whose expression was affected by ePi by 24 hrs. ePi resulted in differential patterns of expression of genes mediating Wnt signaling, suggesting a complex role for these genes in ePi signaling. The expression of Bmp4 and osterix (Sp7), implicated in bone and tooth development, as well as an inhibitor of Bmp Tob1, appears to be co-regulated, so that BMP signaling is diminished. The transcription factors Foxc2, a stimulator of chondroblast differentiation, and Egr2, a promoter of mineralization, may play a similar role in cementoblasts.
Other Genes of Interest
Perusal of ePi-induced changes in GO-grouped genes (Table, Appendix) revealed some unexpected findings. For example, by 24 hrs of ePi treatment, only 10.5% of genes listed under the GO term “protein amino acid dephosphorylation” (Table) were identified. However, these included DUSP (dual-specificity phosphatase, also known as MAP kinase phosphatase) 1, 4, and 6, proposed to be involved in the regulation of mitogenic signals by dephosphorylation of ERK 1 and 2. The expression of genes comprising the MAP kinase GO groups is not significantly altered by ePi, but the activity of cellular functions regulated by these enzymes, such as proliferation, may be diminished. Id2 may influence cell proliferation (Norton et al., 1998). ePi, which has been implicated in osteoblast apoptosis (Meleti et al., 2000), may also have a role in the regulation of cementoblast proliferation.
It appears that some pathways known to be related to mineralized tissue formation are inhibited by ePi, while others are stimulated in cementoblasts. Precise temporospatial regulation of these pathways may be required for maintaining functional boundaries between mineralized and non-mineralized extracellular matrices, such as the cementum-periodontal ligament junction. Speculative ideas regarding possible interactions of these pathways in the regulation of cementogenesis are depicted in Fig. 3. The functional significance of the effects of these signals on the regulation of key molecular pathways and the specific pathways in the transduction of ePi signals is under study.
Supplementary Material
A supplemental appendix to this article is published electronically only at http://www.dentalresearch.org.
Acknowledgments
This project was supported by NIH/NIDCR grant DE15109 and by the UW NIEHS-sponsored Center for Ecogenetics and Environmental Health, Grant #NIEHS P30ES07033.
References
- Beertsen W, VandenBos T, Everts V. Root development in mice lacking functional tissue non-specific alkaline phosphatase gene: inhibition of acellular cementum formation. J Dent Res. 1999;78:1221–1229. doi: 10.1177/00220345990780060501. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TD. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, et al. Minimum information about a microarray experiment (MIAME) - toward standards for microarray data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- Celil AB, Hollinger JO, Campbell PG. Osx transcriptional regulation is mediated by additional pathways to BMP2/Smad signaling. J Cell Biochem. 2005;95:518–528. doi: 10.1002/jcb.20429. [DOI] [PubMed] [Google Scholar]
- Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4(1):R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster BL, Nociti FH, Jr, Swanson EC, Matsa-Dunn D, Berry JE, Cupp CJ, et al. Regulation of cementoblast gene expression by inorganic phosphate in vitro. Calcif Tissue Int. 2006;78:103–112. doi: 10.1007/s00223-005-0184-7. [DOI] [PubMed] [Google Scholar]
- Glass DA, 2nd, Bialek P, Ahn JD, Starbuck M, Patel MS, Clevers H, et al. Canonical Wnt signaling in differentiated osteoblasts controls osteoclast differentiation. Dev Cell. 2005;8:751–764. doi: 10.1016/j.devcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Harmey D, Hessle L, Narisawa S, Johnson KA, Terkeltaub R, Millan JL. Concerted regulation of inorganic pyrophosphate and osteopontin by akp2, enpp1, and ank: an integrated model of the pathogenesis of mineralization disorders. Am J Pathol. 2004;164:1199–1209. doi: 10.1016/S0002-9440(10)63208-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humtsoe JO, Feng S, Thakker GD, Yang J, Hong J, Wary KK. Regulation of cell-cell interactions by phosphatidic acid phosphatase 2b/VCIP. EMBO J. 2003;22:1539–1554. doi: 10.1093/emboj/cdg165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SE, Jomary C. Secreted Frizzled-related proteins: searching for relationships and patterns. Bioessays. 2002;24:811–820. doi: 10.1002/bies.10136. [DOI] [PubMed] [Google Scholar]
- Karavanova I, Vainio S, Thesleff I. Transient and recurrent expression of the Egr-1 gene in epithelial and mesenchymal cells during tooth morphogenesis suggests involvement in tissue interactions and in determination of cell fate. Mech Dev. 1992;39:41–50. doi: 10.1016/0925-4773(92)90024-e. [DOI] [PubMed] [Google Scholar]
- Kreider BL, Benezra R, Rovera G, Kadesch T. Inhibition of myeloid differentiation by the helix-loop-helix protein Id. Science. 1992;255:1700–1702. doi: 10.1126/science.1372755. [DOI] [PubMed] [Google Scholar]
- Leclerc N, Noh T, Khokhar A, Smith E, Frenkel B. Glucocorticoids inhibit osteocalcin transcription in osteoblasts by suppressing Egr2/Krox20-binding enhancer. Arthritis Rheum. 2005;52:929–939. doi: 10.1002/art.20872. [DOI] [PubMed] [Google Scholar]
- Leimeister C, Bach A, Gessler M. Developmental expression patterns of mouse sFRP genes encoding members of the secreted frizzled related protein family. Mech Dev. 1998;75:29–42. doi: 10.1016/s0925-4773(98)00072-0. [DOI] [PubMed] [Google Scholar]
- Levi G, Topilko P, Schneider-Maunoury S, Lasagna M, Mantero S, Cancedda R, et al. Defective bone formation in Krox-20 mutant mice. Development. 1996;122:113–120. doi: 10.1242/dev.122.1.113. [DOI] [PubMed] [Google Scholar]
- Meleti Z, Shapiro IM, Adams CS. Inorganic phosphate induces apoptosis of osteoblast-like cells in culture. Bone. 2000;27:359–366. doi: 10.1016/s8756-3282(00)00346-x. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, et al. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- Nifuji A, Miura M, Kato N, Kellermann O, Noda M. Bone morphogenetic protein regulation of forkhead/winged helix transcription factor Foxc2 (Mfh1) in a murine mesodermal cell line C1 and in skeletal precursor cells. J Bone Miner Res. 2001;16:1765–1771. doi: 10.1359/jbmr.2001.16.10.1765. [DOI] [PubMed] [Google Scholar]
- Nociti FH, Jr, Berry JE, Foster BL, Gurley KA, Kingsley DM, Takata T, et al. Cementum: a phosphate-sensitive tissue. J Dent Res. 2002;81:817–821. doi: 10.1177/154405910208101204. [DOI] [PubMed] [Google Scholar]
- Norton JD, Deed RW, Craggs G, Sablitzky F. Id helix-loop-helix proteins in cell growth and differentiation. Trends Cell Biol. 1998;8:58–65. [PubMed] [Google Scholar]
- Peng Y, Kang Q, Luo Q, Jiang W, Si W, Liu BA, et al. Inhibitor of DNA binding/differentiation helix-loop-helix proteins mediate bone morphogenetic protein-induced osteoblast differentiation of mesenchymal stem cells. J Biol Chem. 2004;279:32941–32949. doi: 10.1074/jbc.M403344200. [DOI] [PubMed] [Google Scholar]
- Pispa J, Thesleff I. Mechanisms of ectodermal organogenesis. Dev Biol. 2003;262:195–205. doi: 10.1016/s0012-1606(03)00325-7. [DOI] [PubMed] [Google Scholar]
- Quarles LD. Evidence for a bone-kidney axis regulating phosphate homeostasis. J Clin Invest. 2003;112:642–646. doi: 10.1172/JCI19687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan R, Dorris D, Lublinsky A, Nguyen A, Domanus M, Prokhorova A, et al. An assessment of Motorola CodeLink microarray performance for gene expression profiling applications. Nucleic Acids Res. 2002;30:e30. doi: 10.1093/nar/30.7.e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman SP, Currie SA, Ciavarella M, Vincan E, Dow C, Thomas RJ, et al. Id2 is a target of the beta-catenin/T cell factor pathway in colon carcinoma. J Biol Chem. 2001;276:45113–45119. doi: 10.1074/jbc.M107742200. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm EE, Kingsley DM. GDF5 coordinates bone and joint formation during digit development. Dev Biol. 1999;209:11–27. doi: 10.1006/dbio.1999.9241. [DOI] [PubMed] [Google Scholar]
- Terkeltaub RA. Inorganic pyrophosphate generation and disposition in pathophysiology. Am J Physiol Cell Physiol. 2001;281:C1–11. doi: 10.1152/ajpcell.2001.281.1.C1. [DOI] [PubMed] [Google Scholar]
- Thesleff I, Keranen S, Jernvall J. Enamel knots as signaling centers linking tooth morphogenesis and odontoblast differentiation. Adv Dent Res. 2001;15:14–18. doi: 10.1177/08959374010150010401. [DOI] [PubMed] [Google Scholar]
- Zelzer E, Olsen BR. The genetic basis for skeletal diseases. Nature. 2003;423:343–348. doi: 10.1038/nature01659. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A supplemental appendix to this article is published electronically only at http://www.dentalresearch.org.


