Abstract
Context: The LHX4 LIM-homeodomain transcription factor has essential roles in pituitary gland and nervous system development. Heterozygous mutations in LHX4 are associated with combined pituitary hormone deficiency.
Objectives: Our objectives were to determine the nature and frequency of LHX4 mutations in patients with pituitary hormone deficiency and to examine the functional outcomes of observed mutations.
Design: The LHX4 gene sequence was determined from patient DNA. The biochemical and gene regulatory properties of aberrant LHX4 proteins were characterized using structural predictions, pituitary gene transcription assays, and DNA binding experiments.
Patients: A total of 253 patients from 245 pedigrees with GH deficiency and deficiency of at least one additional pituitary hormone was included in the study.
Results: In five patients, three types of heterozygous missense mutations in LHX4 that result in substitution of conserved amino acids were identified. One substitution is between the LIM domains (R84C); the others are in the homeodomain (L190R; A210P). The patients have GH deficiency; some also display reductions in TSH, LH, FSH, or ACTH, and aberrant pituitary morphology. Structural models predict that the aberrant L190R and A210P LHX4 proteins would have impaired DNA binding and gene activation properties. Consistent with these models, EMSAs and transfection experiments using pituitary gene promoters demonstrate that whereas the R84C form has reduced activity, the L190R and A210P proteins are inactive.
Conclusions: LHX4 mutations are a relatively rare cause of combined pituitary hormone deficiency. This report extends the range of phenotypes associated with LHX4 gene mutations and describes three novel exonic mutations in the gene.
Screening pituitary transcription factor genes in patients with combined pituitary hormone deficiency finds three novel mutations in the LIM homeobox protein 4 gene, LHX4. All these patients presented with hypoplastic pituitaries and GH deficiency, with some additionally having reductions in TSH, LH, FSH, or ACTH.
After early inductive events, the development of the specialized hormone-secreting cells of the anterior pituitary gland is dependent on the actions of multiple transcription factors such as LHX3, LHX4, PIT1 (POU1F1 gene), PROP1, PITX1, PITX2, SF1, and TPIT (1). Of these, the structurally related LHX3 and LHX4 proteins are members of the LIM-homeodomain (HD) family of transcription factors (2). LIM-HD proteins feature two amino-terminal LIM domains, required for multiple roles, including protein-protein interactions, and a central DNA-binding HD.
Studies of humans and rodents have observed expression of the LHX4 (or Gsh4) gene in the developing hindbrain, cerebral cortex, pituitary gland, and spinal cord (3,4). In mice, Lhx3 and Lhx4 are expressed at embryonic d 9.5 in Rathke’s pouch, the precursor of the anterior/intermediate lobes of the pituitary (5). By embryonic d 12.5, Lhx4 expression is concentrated in the tissue that will become the anterior lobe of the gland, whereas Lhx3 continues expression throughout the pouch. Later, transcription from the Lhx4 gene is reduced, and transcripts are found at lower levels than Lhx3 in the mature gland (5). Interestingly, Lhx3 and Lhx4 are differentially expressed in subpopulations of adult anterior pituitary cells exhibiting stem/progenitor characteristics (6).
Mice homozygous for a Lhx4 gene disruption die shortly after birth with immature lungs that do not inflate, whereas animals heterozygous for Lhx4 inactivation are apparently normal (4). Lhx4 null mice also exhibit incomplete pituitary gland development. Lhx4 is required in conjunction with Lhx3 to form a definitive Rathke’s pouch: in mice lacking both Lhx3 and Lhx4 genes, pituitary development stops at a rudimentary pouch stage, a more severe phenotype than that of the single gene ablations (5). This observation suggests that these regulatory genes have some functional redundancy in pituitary organogenesis. Subsequently, whereas Lhx3−/− mutant pituitary precursor cells cease to proliferate before differentiation of the characteristic hormone-secreting lineages, in Lhx4−/− mutants these cells differentiate, albeit in reduced numbers (5,7). The lack of cellular proliferation associated with Lhx4 mutation has been attributed to increased cell death resulting from a failure to respond to inductive signals, leading to the misregulation of other transcription factor genes, including Lhx3 (8). Molecular studies have shown that, like LHX3, LHX4 can activate transcription from reporter genes containing the promoters of pituitary hormone component genes, including the α-glycoprotein subunit (αGSU) and FSHβ genes (9,10,11). Studies of mouse models have also revealed that Lhx3 and Lhx4 act with other LIM-HD genes in the assignation of motor neuron subtypes during development (12).
The human LHX4 gene extends over approximately 45 kb on chromosome 1 (3,13,14). Two reports have described patients with LHX4 gene mutations. In the first, analysis of a consanguineous family with members exhibiting combined pituitary hormone deficiency (CPHD), short stature, small sella turcicas, hypoplastic anterior pituitaries, and cerebellar defects revealed a heterozygous mutation in an intron of LHX4, suggesting a possible dominant effect of the mutant allele (14). These patients presented with deficiencies in GH, TSH, and ACTH (LH and FSH not investigated). The mutation is predicted to result in the generation of aberrant LHX4 proteins from the use of alternative cryptic splice sites within exon 5 (14). A recent analysis suggested that the disease is likely a result of deficits in activation of pituitary genes such as PIT1/POU1F1 rather than a dominant negative effect (15). A second type of patient has a heterozygous mutation (P366T) affecting a residue in the carboxyl terminus of LHX4 (16). This patient has deficiencies of GH, prolactin, TSH, LH, FSH, ACTH, a hypoplastic anterior lobe, an ectopic posterior pituitary, a poorly developed sella turcica, Chiari malformation, and respiratory distress syndrome. In this study we report the analysis of patients with hypopituitarism featuring three novel types of mutations in LHX4.
Subjects and Methods
Experimental subjects
We studied 253 patients from 245 families with GH deficiency (GHD) combined with deficiency of at least one additional pituitary hormone. Subjects were recruited from either the Genetics and Neuroendocrinology of Short Stature International Study program or from our own clinics. The distribution of hormone deficiencies in this patient pool has been described (17). After written informed consent was given, blood samples were collected from patients and, whenever possible, from first-degree relatives. The study was approved by the local institutions’ Ethics Review Boards according to the declaration of Helsinki.
Screening for LHX4 mutations
DNA was extracted from blood samples of patients. All six coding exons of the LHX4 gene and the intron-exon boundaries were amplified by PCR in seven fragments using primers recognizing intronic sequences. Screening for the presence of sequence aberrations within amplified LHX4 fragments was performed by temperature-modulated heteroduplex HPLC using the WAVE System (Transgenomics, Elancourt, France). Equal amounts of a patient’s LHX4 PCR product and from an unaffected control subject were mixed and heat denatured. Possible heteroduplex formation was allowed to occur, samples were analyzed on the WAVE System, and DNA fragments were detected by an UV detector. Temperature profiles were optimized using WaveMaker software (WaveMaker Software, Inc., San Francisco, CA). Samples that indicated sequence aberrations were sequenced using an ABI 310 sequencer (PerkinElmer, Waltham, MA). Detailed descriptions of the amplification primers and denaturing high pressure liquid chromatography analysis of each LHX4 gene fragment are provided as supplemental data, which are published as supplemental data on The Endocrine Society’s Journals Online web site at http://jcem.endojournals.org.
Cloning, plasmid construction, and site-directed mutagenesis
A human LHX4 cDNA expression vector was constructed in pcDNA3.1/Myc-His(-)C (Invitrogen, Carlsbad, CA). Site-directed mutagenesis to generate R84C, L190R, and A210P derivatives was performed using the QuikChange kit (Stratagene, La Jolla, CA) using oligonucleotide primers containing the mutations. Plasmid integrity was confirmed by DNA sequencing. The αGSU, TSHβ, and PIT1 pituitary hormone promoter reporter genes have been described (10,18,19,20).
Homology structural modeling
The LHX3 and LHX4 HD sequences were used to screen the protein data bank (PDB) database (Swiss model). A PDB file corresponding to an Engrailed HD DNA-complex at 2.8 Å resolution (the PDB code name is 1hddC) (21) was selected as the best match (BLAST score 1e−8), displaying homology higher than 50%. The LHX3 and LHX4 sequences were structurally aligned to the 1hddC model, subjected to energy minimization, using the Swiss PDB-Viewer 3.7 (www.expasy.org/spdbv/) and evaluated with ANOLEA (Swiss model). Both models have most of the φ and ψ angle pairs in the allowed regions of the Ramachandran plot. The accepted model of each HD was docked into the DNA structure of the 3hdd protein-DNA complex structure (22). The leucine (L190) residue of the LHX4 model was mutated to arginine. Different rotamer predictions for this residue were tested to find putative contacts with the third, DNA-interacting, helix of the HD. A210 (residue 53 of the alignment) was mutated to valine for the LHX3 model or proline for LHX4. A β-structure was assessed to the proline residue. The resultant PDB files were exported to pov3.5 format and rendered using PovRay 3.6 (Persistence of Raytracer Pty. Ltd., Williamstown, Australia).
In vitro transcription/translation
Radiolabeled wild-type and mutant proteins were synthesized in vitro from transcribed cDNAs using Quick Coupled Transcription/Translation System rabbit reticulocyte lysate reagents (Promega, Madison, WI) and 35S-cysteine, and then were analyzed by electrophoresis in 12% acrylamide gels, followed by fluorography as described (19). Dried gels were imaged with a Storm phosphorimager (Amersham Biosciences, Piscataway, NJ).
EMSAs
EMSAs were performed as described using radiolabeled probes representing the pituitary glycoprotein basal element (PGBE) of the αGSU gene (19). Proteins for EMSA were generated by in vitro translation as described above, except that cold cysteine was substituted for 35S-cysteine. Results were visualized by autoradiography or using a phosphorimager.
Cell culture and transfection
Mouse pituitary GHFT1 cells were a gift from Dr. Pamela Mellon (University of California, San Diego, CA). GHFT1 and human embryonic kidney 293T cells were cultured and transfected as described (19). Luciferase assays were performed as reported. Briefly, 500,000 cells were cultured per well of a six-well dish and were transfected with Lipofectamine 2000 (Invitrogen)/DNA mixtures. Five hundred nanograms of reporter plasmid and 250 ng expression vector were added per dish, and all groups received equal final DNA concentrations. Control cultures received empty expression vector. Luciferase activity was measured 48 h after transfection. All assay points were performed in triplicate, and experiments were repeated at least three times. Total cell protein was determined by the Bradford method (Bio-Rad Laboratories, Hercules, CA), and luciferase activity was normalized to the amount of protein present as described (10,19).
Western blot analysis
Western blot analyses of cells transfected with expression vectors encoding myc epitope-tagged LHX4 proteins were performed as described (19). Briefly, whole cell extracts from cells transfected as described above were prepared (23), and 25 μg protein per sample was separated on 12% sodium dodecyl sulfate polyacrylamide gels, then transferred by wet transfer to polyvinylidene difluoride membranes. Mouse anti-myc monoclonal antibody no. 4A6 (Upstate/Millipore, Charlottesville, VA) was used to detect LHX4-myc proteins, and goat anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) polyclonal antibody no. SC-20357 (Santa Cruz Biotechnology, Santa Cruz, CA) was used as a positive control to determine gel loading equivalency. Primary antibodies were used at 0.5 μg/ml. The secondary antibodies were antimouse or antigoat-horseradish peroxidase conjugates (Roche Biochemical, Indianapolis, IN) used at 1:20,000. Results were visualized using SuperSignal West Dura substrate (Pierce Biotechnology, Rockford, IL) and BioMax MR film (Kodak, Rochester, NY).
Protein/protein interaction assays
Recombinant GST-LHX4 proteins were expressed in Escherichia coli BL21 (DE3) pLysS and affinity purified as previously described (19). Expressed proteins were analyzed on 12% sodium dodecyl sulfate-PAGE gels, followed by staining with Coomassie brilliant blue. Radiolabeled substrate PIT1 proteins were synthesized in vitro using rabbit reticulocyte lysate reagents and 35S-methionine as described previously. Protein/protein interaction assays using labeled PIT1 incubated with wild-type and mutant GST-LHX4 proteins were performed as above described (20,24). Briefly, GST-LHX4 proteins bound to glutathione agarose beads were added to 500,000 cpm labeled PIT1 in 20 mm HEPES (pH 7.9), 100 mm NaCl, 1 mm EDTA, 4 mm MgCl2, 1 mm dithiothreitol, 0.02% NP-40, 10% glycerol, 0.5 mm phenylmethylsulfonylfluoride, 1 mm ZnCl2, 50 μg/ml ethidium bromide, and incubated at 37 C. After binding, the bead/protein complexes were washed with the same buffer. Retained proteins were separated on 12% sodium dodecyl sulfate polyacrylamide gels, followed by fixation and treatment with Amplify fluorography reagent (Amersham). Dried gels were imaged with a Storm phosphoimaging screen or BioMax MR film.
Results
Patients with LHX4 mutations
Overall, we identified five individuals (1.98%) with LHX4 mutations from three families in a group of 253 patients with pituitary hormone deficiency.
Family A
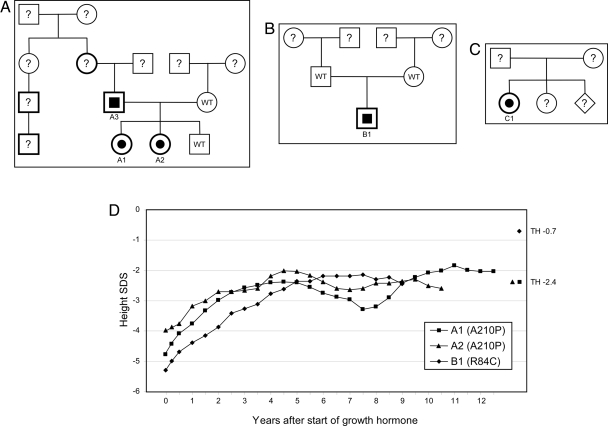
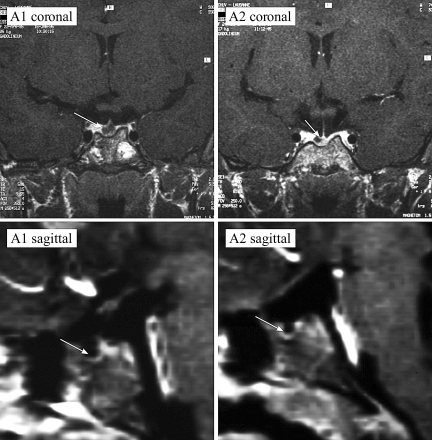
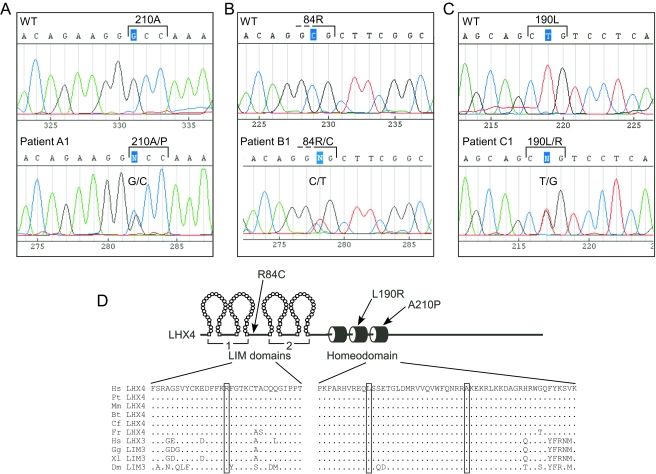
Two female siblings (A1 and A2, Fig. 1A) were investigated for short stature due to GHD. The parents are Swiss, of short stature [target height = 152.0 cm, −2.4 sd score (SDS)] (25). After healthy full-term pregnancies, the girls were delivered vaginally, having normal birth weights. The index case (A1) was referred at the age of 7.0 yr, when her height was already 4.8 sd below the mean for age. Three years later, her sister (A2), at the age of 7.8 yr, was referred for short stature (−4.0 SDS). Both girls were prepubertal (Tanner stage I), had a puppet-like face, mild frontal bossing, and truncal obesity. Both had delayed bone age (according to the Greulich and Pyle atlas) low serum levels of IGF-I (7–47 ng/ml; normal range 99–376), and impaired GH response on arginine-hydrochloride and insulin testing (peak GH ranged from 0.4–4.8 ng/ml in various tests). Magnetic resonance imaging (MRI) showed hypoplastic anterior pituitary glands and cystic lesions within the pituitary for both patients A1 and A2 (Tables 1–3 and Fig. 2). The posterior pituitary “bright spot” was in the normal position in both patients. On recombinant human GH, IGF-I levels increased significantly into the normal range, and both patients presented successful catch-up growth with a first-year height increment of 1.0 and 0.8 SDS for patients A1 and A2, respectively. Both reached a final height SDS of ±0.5 of their target height. After completion of growth, GH therapy was stopped. At least 1 month later, arginine-GHRH testing confirmed persistent complete GHD for patient A1 at the age of 20 yr (peak GH 1.2 ng/ml, normal > 20 ng/ml; IGF-I level of 49.3 ng/ml, normal 115–732) but only partial GHD for patient A2 at the age of 19 yr (peak GH 17 ng/ml, normal > 20; IGF-I levels of 259 ng/ml, normal 115–732) (26). Follow-up MRI showed no changes compared with the initial finding; in particular, the character and size of the pituitary cysts did not change during 12-yr observation. DNA analysis revealed that the father (A3) and his two daughters have a heterozygous G→C transversion within exon 5 of LHX4 resulting in substitution of a conserved alanine in the recognition helix of the HD with a proline (A210P; Fig. 3A). One son and the mother showed no mutation and had no hormonal deficiencies. Therefore, patient A1 is deficient in GH, TSH, ACTH, and gonadotropins, but her sister (A2) has only partial GHD and a partial deficiency for TSH. The father (A3) showed a relatively low peak hormone level of GH (20 ng/ml) on GHRH/arginine stimulation (expected values > 20 ng/ml) when he was tested at the age of 46 yr with an adult height of 162 cm. He also had low serum levels of IGF-I (92 ng/ml; normal 92–267) and IGF binding protein (IGFBP)-3 (2.8 mg/liter; normal 2.9–3.5). Maximal TSH levels were relatively low on TRH stimulation (3.8 mU/liter), but he had a normal free T4 (fT4) level of 16 pmol/liter and did not show any clinical signs of hypothyroidism. There were also normal responses to CRF and GnRH stimulation.
Figure 1.
A–C, Pedigrees of families A–C. Patients with heterozygous LHX4 gene mutations are denoted by half-filled symbols. Bold lines on symbols indicate family members with short stature. D, Growth profiles of patients on GH treatment. TH, Target height; WT, normal LHX4 genotype.
Table 1.
Clinical and genetic features of the patients
| Patient/family member | A-1 | A-2 | A-3 | B-1 | C-1 |
|---|---|---|---|---|---|
| Mutation | A210P | A210P | A210P | R84C | L190R |
| Target height SDS | −2.4 | −2.4 | −1.7 | −0.7 | a |
| Height SDS before GH therapy | −4.8 | −4.0 | −5.3 | a | |
| Additional clinical features | Obesity | ||||
| Therapy | GH, LT4, cortisol, estradiol | GH | None | GH, LT4, testosterone | GH, LT4, cortisol |
Patient put on GH treatment at 2.5 months of age.
Table 2.
MRI findings
| Patient/family member | A-1 | A-2 | A-3 | B-1 | C-1 |
|---|---|---|---|---|---|
| Pituitary size | Small | Small | Normal | Small | Small |
| Location of anterior pituitary | In situ | In situ | In situ | In situ | In situ |
| Location of posterior pituitary | In situ | In situ | In situ | Ectopic | Ectopic |
| Additional morphological anomalies | Pituitary cyst | Pituitary cyst |
Table 3.
Hormone deficiency profiles of the patients
| Patient/family member | A-1 | A-2 | A-3a | B-1 | C-1 | Normal range |
|---|---|---|---|---|---|---|
| Sex | F | F | M | M | F | |
| GH max [on conventional testing (arginine or insulin), ng/ml] | 2.9 | 4.4 | nd | 2.25 | 0.3b | >8.0 |
| GH max [on arginine-GHRH, ng/ml]a | 1.2 | 17 | 20 | nd | nd | <20c |
| TSH (not stimulated, mU/liter) | 0.01 | 0.5 | 0.5 | 1.74 | 6.7b | 0.2–3.1 |
| Free T4 (without thyroxine replacement, ng/dl) | 1.01d | 1.32 | 1.24 | 0.55 | 0.5 | 0.9–1.58 |
| TSH max (stimulated on TRH, mU/liter) | 0.1 | 3.9 | 3.8 | 7.52 | nd | Increase > 2.5 times basal value |
| Cortisol (not stimulated, ng/ml) | 14.5 | 269 | 131 | 63 | 1.2b | 104−278 |
| Cortisol max (stimulated on CRH, ng/ml) | 28.2 | 264 | 181 | 184 | nd | Increase > 50% |
| Estradiol (F) (nmol/liter) | <73 | <71 | nd (age) | F (E): >67 | ||
| Testosterone (M) (nmol/liter) | 24 | 0.4 | M (T): >5.6 | |||
| LH/FSH (not stimulated, U/liter) | <0.1/<0.1 | 1.9/7.1 | 4.0/5.4 | 0.4/0.7 | nd (age) | F: 2–20/2–20 |
| M: 0.8–8/1.2–10.1 | ||||||
| LH/FSH (stimulated, U/liter) (age at testing in yr) | <0.1/<0.1 (19) | 8.7/13 (17) | 24/9.7 (46) | <0.5/0.5 (15) | nd (age) | F: two times basal/>10; M: >12/>4.5 |
| PRL (not stimulated, mU/liter) | 185 | 812 | 146 | 231 | nd | 115–550 |
| PRL max (stimulated on TRH, mU/liter) | 390 | 1365 | 617 | 343 | nd | Increase two times |
E, Estradiol; F, female; M, male; max, maximum; nd, not done; nd (age), not done because still at prepubertal age; PRL, prolactin; T, testosterone.
Patients were tested in adulthood.
Levels obtained on critical sample at the time of hypoglycemia.
Donaubauer et al. (26). Hormone measurement of patients in families A–C was performed in three different pediatric endocrinology centers with different assays. Therefore, absolute values may differ.
l-T4 was stopped just 4 d before testing; total T4 was 5.08 μm/dl.
Figure 2.
Pituitary morphologies of affected patients. T1-weighted section after gadolinium MRI coronal and sagittal scans of the brains of the patients A1 (Left) and A2 (Right). Arrows indicate position of the anterior pituitary gland.
Figure 3.
Three point mutations in the LHX4 gene. A–C, Sequence analyses of patients revealed heterozygous mutations causing R84C, L190R, and A210P amino acid substitutions. D, Diagram of LHX4 protein alterations. The amino acid sequences of the relevant regions of the human (Hs) LHX4 protein are shown in alignment with LHX4, LHX3, and LIM3 proteins from chimp (Pt), mouse (Mm), cow (Bt), dog (Cf), puffer fish [Fugu rubripes (Fr)], chicken (Gg), frog [Xenopus laevis (Xl)], and fruit fly [Drosophila melanogaster (Dm)].
Family B
Patient B1 of a Macedonian family (Fig. 1B) presented at 5.75 yr to the clinic with a standing height 4.8 sd below the mean. He had a small anterior pituitary and an ectopic posterior pituitary. Patient B1 had GHD, and the hormonal workup showed secondary hypothyroidism with decreased fT4 levels in addition to inadequately low serum TSH levels (Tables 1–3). However, there were no clinical or laboratory signs of hypocortisolism or diabetes insipidus. He later developed gonadotropin insufficiency with pubertal failure. Under recombinant human GH and T4 replacement, he showed catch-up growth, but he has yet to reach his final height (Fig. 1D). At 8 yr of age, he experienced a weight gain, and at 10 yr he crossed the 97th weight percentile. At 15 yr of age, he was additionally substituted with depot testosterone. Patient B1 has a heterozygous C→T transition predicted to cause a change in the amino acid sequence between the LIM domains of the protein (R84C, Fig. 3B).
Family C
Patient C1 was born at term by normal vaginal delivery with a birth weight of 2807 g. Her perinatal course was complicated by jaundice requiring phototherapy and hypoglycemia. She was admitted to the hospital at 2.5 months of age for evaluation and management of prolonged jaundice and staring spells indicative of seizure activity. Hypoglycemia was noted on admission. A controlled fast was performed with rapidly ensuing hypoglycemia. GHD was suspected when a critical sample obtained at a venous glucose level of 21 mg/dl showed a GH level of 0.3 ng/ml, T4 serum levels of 2.74 μg /dl (normal 5–12) and fT4 levels of 0.5 ng/dl (normal 0.8–2.3) suggested secondary hypothyroidism in the face of an only inadequately increased TSH level of 6.67 mIU/liter (normal 0.4–4.2). In addition, a serum cortisol level of 1.2 μg/liter showed hypocortisolism, therefore, CPHD involving the GH, TSH, and ACTH axes was diagnosed. A brain MRI revealed the presence of a small anterior pituitary and an ectopic posterior pituitary. No other central nervous system abnormalities were visualized, and normal optic nerves were present. Hormone replacement was begun with T4 and hydrocortisone. GH therapy was added later due to persistent hypoglycemia and blood glucose levels normalized before hospital discharge. The patient developed normally, and her linear growth followed the 50th percentile since the age of 2 yr. She has a clinically healthy younger sister (DNA not tested). In patient C1, a T→G transversion is predicted to cause a missense change (leucine to arginine) in the second helix of the HD (L190R; Fig. 3C).
For all three families (A, B, C), in contrast to the patients reported previously with LHX4 mutations, there were no abnormalities seen in other regions of the brain, i.e. there was no cerebellar hypoplasia reminiscent of an Arnold-Chiari malformation.
Structural consequences of LHX4 mutations
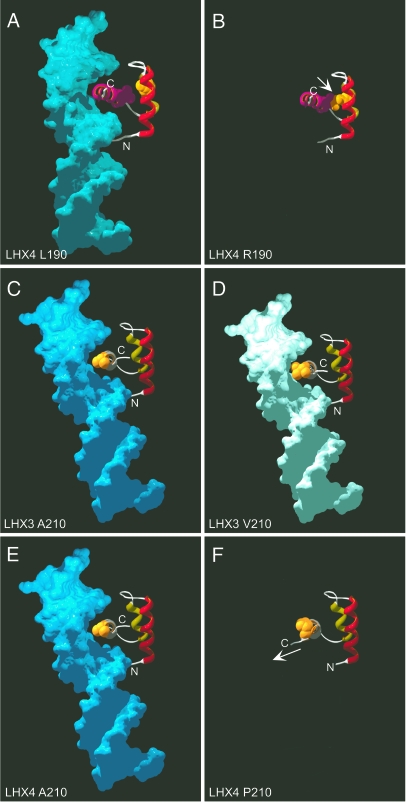
All of the changed LHX4 amino acid residues are conserved in known LHX3 or LHX4 sequences (Fig. 3D), including mammalian, fish, bird, amphibian, and insect proteins. Because two of the three identified mutations cause alterations in the HD, a structure critical for DNA binding and, therefore, gene regulation, we made predictions of the structures of the altered HDs. Models of possible LHX4 interactions with target DNA were performed using the Engrailed HD/DNA complex structure (21) as a template. In the prediction for the wild-type protein, the leucine at position 190 can be seen as part of the HD helix-turn-helix structure (Fig. 4A). Substitution of this amino acid with an arginine can result in several possible structures, including many that result in inappropriate contact with the valine residue (V201) within the third DNA-recognizing α-helix of the HD, a condition that would likely destabilize the structure and negatively affect DNA binding (Fig. 4B).
Figure 4.
Structural predictions of LHX3 and LHX4 HD/DNA interactions. Models of LHX3 and LHX4 interaction with target DNAs were performed using the Engrailed HD/DNA complex as a template (21). A, Ribbon model of wild-type LHX4 HD/DNA with leucine 190 shown in space filling format. The predicted positions of the amino (N) and carboxyl (C) termini of the full protein are shown. B, Predicted LHX4 HD with an arginine at position 190. Different rotamers for this residue are possible. In this prediction there is contact with the V201 residue within the third α-helix of the HD (arrow), destabilizing the structure and inhibiting DNA binding. C, Ribbon model of wild-type LHX3 HD/DNA with alanine 210 highlighted. D, Predicted LHX3 HD/DNA with a valine at position 210. E, Ribbon model of wild-type LHX4 HD/DNA with alanine 210 highlighted. F, Predicted LHX4 HD with a proline at position 210. Note the altered configuration of the carboxyl terminus (C) (arrow).
In a previous study, we have described a patient with CPHD with a mutation in the LHX3 gene that results in a valine substitution for a conserved alanine at position 210 (17). This change is relatively conservative, and the more bulky valine is predicted to reduce but not abolish DNA binding (Fig. 4, C and D). Indeed, reduced DNA binding is observed for this protein (17). The LHX3 and LHX4 proteins have identical sequence in the third helix of the HD, and the A210P mutation described here alters the equivalent alanine of LHX4 (A210) to a proline. In comparison to the subtle effects observed for the LHX3 A210V substitution, the LHX4 A210P change is predicted to result in a protein conformation in which the carboxyl terminus is placed in a position inconsistent with DNA binding (Fig. 4, E and F).
Biochemical, DNA binding, and transcriptional properties of aberrant LHX4 proteins
Expression plasmids encoding each of the aberrant LHX4 proteins were constructed and used to produce radiolabeled proteins by in vitro translation. The resulting proteins were analyzed by denaturing gel electrophoresis, and they migrated similarly to wild-type LHX4 (Fig. 5A), consistent with the predicted point mutations. When expressed in either pituitary GHFT1 cells or heterologous 293T cells, wild-type and “mutant” LHX4 proteins of similar sizes were detected by western blotting (Fig. 5B). In both cell types, the detected level of LHX4 R84C was lower than the other variants, suggesting that this aberrant protein may have a different stability from the wild type.
Figure 5.
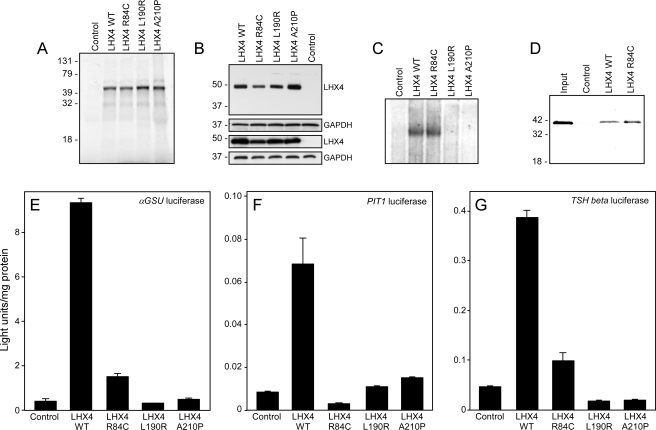
Biochemical and functional properties of proteins encoded by mutated LHX4 genes. A, Radiolabeled wild-type (WT) and mutant LHX4 proteins were generated from cDNA expression vectors by in vitro transcription/translation using rabbit reticulocyte lysates in the presence of 35S-cysteine. Labeled products were separated by sodium dodecyl sulfate electrophoresis, and dried gels were visualized by fluorography. The migration positions of protein standards (in kilodaltons) are shown. Control is a reaction from a lysate programmed with empty vector (negative control). B, Western blot analysis of wild-type and mutant LHX4 proteins from transfected pituitary GHFT1 (two upper panels) or heterologous human embryonic kidney 293T cells (two lower panels). Whole cell extracts were separated on polyacrylamide gels, then transferred to membranes. LHX4-myc proteins were detected with an anti-myc monoclonal antibody (LHX4), and equal protein loading was conformed by detecting GAPDH using an anti-GAPDH polyclonal antibody (GAPDH). The migration positions of protein standards (in kilodaltons) are shown. Control is transfection with empty vector. C, EMSA experiments were performed using nonradiolabeled LHX4 proteins translated in vitro in rabbit reticulocyte lysates and radiolabeled probes representing the PGBE of the αGSU gene promoter. Unprogrammed lysates were used as negative controls (control). D, Interaction between the PIT1 and LHX4 proteins is unaffected by the R84C substitution. Affinity resins containing either recombinant wild-type or R84C LHX4 proteins (as GST fusions) were incubated with radiolabeled PIT1 proteins. After washing, the bound PIT1 protein was detected by electrophoresis and fluorography. The migration positions of protein standards (in kilodaltons) are shown. Control is GST resin (negative control). E, Proteins encoded by mutated LHX4 genes display impaired transactivation properties. Expression vectors for wild-type and mutant LHX4 proteins were transiently cotransfected into pituitary GHFT1 cells with a luciferase reporter gene under the control of the αGSU promoter. Promoter activity was assayed by measuring luciferase activity 48 h after transfection. Negative controls (control) received equivalent amounts of empty expression vector plasmid. Activities are mean (light units/10 sec/μg total protein) of triplicate assays ± sem. A representative experiment of at least three experiments is depicted. F, Gene activation assay using a PIT1 promoter/enhancer reporter gene. G, Gene activation assay using the TSHβ promoter.
To test the DNA binding properties predicted by the structural modeling for the altered LHX4 proteins, we performed EMSA using probes representing the αGSU gene PGBE. The R84C protein, which has an intact HD, bound with similar efficiency to the wild-type protein (Fig. 5C). By contrast, consistent with the structural predictions, L190R and A210P did not bind to the PGBE site.
The LIM domains of the related LHX3 protein interact with the PIT1 pituitary transcription factor protein (19,20). Therefore, we tested whether the LHX4 protein displayed similar interactions and whether the R84C substitution affected such an interaction. Using in vitro pull-down assays, we found that the R84C protein retained interaction with PIT1 (Fig. 5D).
We also tested the gene activation capacities of the altered LHX4 proteins using pituitary gene regulatory region reporter genes. LHX4 has previously been demonstrated to activate the promoter of the αGSU gene (10,11). We also tested LHX4 activation of the PIT1 transcription factor and TSHβ hormone subunit promoters. LHX4 expression vectors were cotransfected with reporter genes into pituitary GHFT1 cells, and gene activity was recorded. Wild-type LHX4 activates the αGSU reporter (∼23- fold), whereas R84C has reduced capacity (∼4-fold) compared with a negative control (Fig. 5E). Neither L190R nor A210P is able to activate the αGSU promoter. Similar activities were seen in experiments using PIT1 and TSHβ reporters except that the R84C was inactive on the PIT1 reporter gene (Fig. 5, F and G). To our knowledge, this is the first demonstration of LHX4 activation of the TSHβ promoter. The GHFT1 pituitary cells used in these experiments express PIT1 protein, so these observations likely reflect the combined actions of LHX4 and PIT1 on the PIT1 and TSHβ promoters, as we have reported for LHX3 (27). Indeed, the data trends are the same in assays in which LHX4 is cotransfected with a PIT1 expression vector (data not shown).
Discussion
A mutation screen within pituitary transcription factor genes of patients with CPHD identified patients harboring three types of heterozygous mutations in LHX4. Whereas two of the resulting amino acid changes, A210P and L190R, are located in the HD of the molecule, a third mutation, R84C, alters a conserved residue between the LIM domains of LHX4. All of the patients with LHX4 aberrations presented with hypoplastic pituitaries and hormone deficiencies. Morphological variations included ectopically located posterior pituitaries in two patients and pituitary cysts in another two individuals. Deficiencies of anterior pituitary hormones were variable across patients with different genotypes as well as within the same family.
The molecular assays presented here demonstrate that the L190R and A210P proteins are inactive in DNA binding and pituitary gene activation assays. LHX4 R84C has impaired activity on the tested αGSU, PIT1, and TSHβ promoters, and exhibits normal DNA binding to the αGSU PGBE and interaction with the PIT1 protein. Because the patients all have GHD, we also tested the ability of LHX4 to activate a GH promoter reporter gene. As has been observed for the related LHX3 factor (20), we found that wild-type LHX4 is a very poor activator of GH regulatory regions (≤2-fold), and the mutant proteins are relatively inactive (C.S.H., R.D.M., and S.J.R., unpublished data). The data presented here showing the loss of DNA binding and gene activation function by the affected LHX4 proteins likely reflect a deficiency of LHX4 activity at both early and intermediate stages of pituitary development. Indeed, the inactivity of the aberrant proteins on the PIT1 promoter is consistent with an inability to activate transcriptional cascades important for the establishment and differentiation of the hormone-secreting cell types.
The LIM domains of LIM-HD proteins, including the related LHX3 protein, are multifunctional, mediating interactions that modulate target gene transactivation, DNA binding affinity, protein stability, and other parameters (28,29). Although we have observed no effects of the R84C substitution on LHX4 interaction with tested proteins and DNA sites, we do observe impaired transactivation function. The nonconservative substitution of an arginine with a cysteine in LHX4 R84C might also cause inappropriate disulfide bonds or incorrect coordination of the zinc ions that are part of the LIM structure, resulting in protein conformations that affect interactions with factors regulating transcription properties.
The heterozygous condition of the LHX4 mutations described here could be associated with several mechanisms, including dominant negative action of aberrant proteins or by a reduction in the activity of LHX4 to a level below critical thresholds for developmental steps. To date, in experiments using pituitary reporter genes, we have not observed any specific dominant-negative effect of the aberrant proteins (J.J.S., C.S.H., and S.J.R., unpublished data). Alternatively, it is possible that the phenotypes result from the lack of production of LHX4 protein or inaction of the protein. Classically, haploinsufficiency is defined as an abnormal condition resulting from 50% reduction in the protein produced from a gene (30). It is possible that the point mutations result in the production of unstable proteins, and for the R84C protein, our data are consistent with a reduced stability profile. A broader definition for haploinsufficiency is that the mutation results in a reduction in gene function. We have shown that the LHX4 mutations are associated with disabled or partially active proteins. Many clinical disorders are caused by heterozygous mutations in transcription factor genes (30). Gene dosage of transcription factor genes is a critical parameter in the regulation of organogenesis of the mouse pituitary gland. For example, studies of mice carrying varying numbers of Lhx3, Lhx4, or Pitx gene family alleles show a gradient of effects on anterior pituitary development (5,31,32,33). However, the hypothesis that LHX4 haploinsufficiency explains the disease phenotypes described here is tempered by the observation that, to date, pituitary disease has not been reported for Lhx4± mice (4,5,8). It may be that gene dosage has differential effects in mice and humans. It is possible that mutations in LHX4 of the kind described here are actually semidominant traits and that patients with homozygous mutations of LHX4 are nonviable, as is true for mice (4). It is also possible that the patients are compound heterozygotes with additional mutations affecting LHX4 gene function or that they are doubly heterozygous with mutations in other genes that are important in pituitary development (“synergistic heterozygosity”). We have not observed any mutations in coding regions of the LHX3, PROP1, or PIT1/POU1F1 genes in these patients (data not shown). We also cannot exclude gene inactivation or epigenetic effects.
The phenotypical variation documented in this study for patients with CPHD with mutations in LHX4, including dissimilarity within probands from the same pedigree as in family A, is likely partly due to the impact of other genes in these patients. Genetic background has affected the phenotypes associated with the loss of pituitary transcription factor gene function, including Lhx4 in mice (8). Similarly, the phenotype of homozygous mice lacking Prop1 is influenced by the genetic background (34), and the loss of hormones in mouse models is often less severe than that of human patients (35). Genetic background may also determine whether patients with LHX4 gene defects also display nervous system abnormalities, such as cerebellar defects. Such features have been described in other patients with LHX4 mutations (14,16) but were not observed in the patients described here. Alternately, the three types of mutation found in this study may only affect the pituitary functions of LHX4, and sufficient function remains for nervous system development.
Although there is some redundancy of function in the actions of the Lhx3 and Lhx4 genes during specific stages of pituitary development (5), studies in mice demonstrate that Lhx4 is required for the correct temporal expression of regulatory genes such as Lhx3 (8), which suggests that there may be a partial loss of LHX3 function in patients carrying LHX4 gene mutations. To understand better the mechanisms underlying LHX4-associated diseases, it will be important to more completely identify LHX4 (-specific) target genes that are involved in the different stages of pituitary development.
In recent years, characterization of mutations in pituitary transcription factor genes have significantly advanced our knowledge of the mechanisms involved in pituitary development, have improved our understanding of pituitary diseases, and have allowed the development of diagnostic and genetic counseling tools. This study describes three new types of exonic mutations in the LHX4 gene that are associated with CPHD and broadens the described phenotypes associated with such mutations.
Supplementary Material
Acknowledgments
We thank the patients and their families for participation in the study. We also thank Drs. Lisa Cushman, Kyle Sloop, and Emily Walvoord for advice, and Dr. Laurent Chapuis, Clinique de la Source, Lausanne, Switzerland, for magnetic resonance imaging analyses.
Footnotes
This work was supported by a grant from the National Institutes of Health (HD42024) (to S.J.R.). J.J.S. was an Elizabeth Steele Creveling Memorial Scholar during this work. The Genetics and Neuroendocrinology of Short Stature International Study DNA Analysis Sub-study is part of an observational research program for long-term follow up of pediatric patients sponsored by Eli Lilly & Company (to R.W.P.).
Disclosure Statement: C.S.H., J.J.S., M.D.-P., R.D.M., Z.P.N., U.E., V.H., N.G.H., H.M.S., J.F.W.W., and S.J.R have nothing to declare. R.W.P. is on an Eli Lilly advisory board and received lecture fees from Pfizer, Lilly, Ferring, and Novo Nordisk. W.F.B. is employed by Eli Lilly & Company and has equity interests in that company.
First Published Online December 11, 2007
Abbreviations: αGSU, α-Glycoprotein subunit; CPHD, combined pituitary hormone deficiency; fT4, free T4; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GHD, GH deficiency; HD, homeodomain; IGFBP, IGF binding protein; MRI, magnetic resonance imaging; PDB, protein data bank; PGBE, pituitary glycoprotein basal element; SDS, sd score.
References
- Zhu X, Lin CR, Prefontaine GG, Tollkuhn J, Rosenfeld MG 2005 Genetic control of pituitary development and hypopituitarism. Curr Opin Genet Dev 15:332–340 [DOI] [PubMed] [Google Scholar]
- Mullen RD, Colvin SC, Hunter CS, Savage JJ, Walvoord EC, Bhangoo AP, Ten S, Weigel J, Pfaffle RW, Rhodes SJ 2007 Roles of the LHX3 and LHX4 LIM-homeodomain factors in pituitary development. Mol Cell Endocrinol 265- 266:190–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Fan M, Yu S, Zhou Y, Wang J, Yuan J, Qiang B 2002 cDNA cloning, chromosomal localization and expression pattern analysis of human LIM-homeobox gene LHX4. Brain Res 928:147–155 [DOI] [PubMed] [Google Scholar]
- Li H, Witte DP, Branford WW, Aronow BJ, Weinstein M, Kaur S, Wert S, Singh G, Schreiner CM, Whitsett JA 1994 Gsh-4 encodes a LIM-type homeodomain, is expressed in the developing central nervous system and is required for early postnatal survival. EMBO J 13:2876–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng HZ, Moriyama K, Yamashita T, Li H, Potter SS, Mahon KA, Westphal H 1997 Multistep control of pituitary organogenesis. Science 278:1809–1812 [DOI] [PubMed] [Google Scholar]
- Chen J, Hersmus N, Van Duppen V, Caesens P, Denef C, Vankelecom H 2005 The adult pituitary contains a cell population displaying stem/progenitor cell and early embryonic characteristics. Endocrinology 146:3985–3998 [DOI] [PubMed] [Google Scholar]
- Sheng HZ, Zhadanov AB, Mosinger Jr B, Fujii T, Bertuzzi S, Grinberg A, Lee EJ, Huang SP, Mahon KA, Westphal H 1996 Specification of pituitary cell lineages by the LIM homeobox gene Lhx3. Science 272:1004–1007 [DOI] [PubMed] [Google Scholar]
- Raetzman LT, Ward R, Camper SA 2002 Lhx4 and Prop1 are required for cell survival and expansion of the pituitary primordia. Development 129:4229–4239 [DOI] [PubMed] [Google Scholar]
- West BE, Parker GE, Savage JJ, Kiratipranon P, Toomey KS, Beach LR, Colvin SC, Sloop KW, Rhodes SJ 2004 Regulation of the follicle-stimulating hormone β gene by the LHX3 LIM-homeodomain transcription factor. Endocrinology 145:4866–4879 [DOI] [PubMed] [Google Scholar]
- Sloop KW, Dwyer CJ, Rhodes SJ 2001 An isoform-specific inhibitory domain regulates the LHX3 LIM homeodomain factor holoprotein and the production of a functional alternate translation form. J Biol Chem 276:36311–36319 [DOI] [PubMed] [Google Scholar]
- Kawamata N, Sakajiri S, Sugimoto KJ, Isobe Y, Kobayashi H, Oshimi K 2002 A novel chromosomal translocation t(1;14)(q25;q32) in pre-B acute lymphoblastic leukemia involves the LIM homeodomain protein gene, Lhx4. Oncogene 21:4983–4991 [DOI] [PubMed] [Google Scholar]
- Sharma K, Sheng HZ, Lettieri K, Li H, Karavanov A, Potter S, Westphal H, Pfaff SL 1998 LIM homeodomain factors Lhx3 and Lhx4 assign subtype identities for motor neurons. Cell 95:817–828 [DOI] [PubMed] [Google Scholar]
- Sloop KW, Parker GE, Rhodes SJ 2001 Transcriptional regulation in mammalian pituitary development and disease. Curr Genomics 2:379–398 [Google Scholar]
- Machinis K, Pantel J, Netchine I, Leger J, Camand OJ, Sobrier ML, Dastot-Le Moal F, Duquesnoy P, Abitbol M, Czernichow P, Amselem S 2001 Syndromic short stature in patients with a germline mutation in the LIM homeobox LHX4. Am J Hum Genet 69:961–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machinis K, Amselem S 2005 Functional relationship between LHX4 and POU1F1 in light of the LHX4 mutation identified in patients with pituitary defects. J Clin Endocrinol Metab 90:5456–5462 [DOI] [PubMed] [Google Scholar]
- Tajima T, Hattori T, Nakajima T, Okuhara K, Tsubaki J, Fujieda K 2007 A novel missense mutation (P366T) of the LHX4 gene causes severe combined pituitary hormone deficiency with pituitary hypoplasia, ectopic posterior lobe and a poorly developed sella turcica. Endocr J 54:637–641 [DOI] [PubMed] [Google Scholar]
- Pfaeffle RW, Savage JJ, Hunter CS, Palme C, Ahlmann M, Kumar P, Bellone J, Schoenau E, Korsch E, Bramswig JH, Stobbe HM, Blum WF, Rhodes SJ 2007 Four novel mutations of the LHX3 gene cause combined pituitary hormone deficiencies with or without limited neck rotation. J Clin Endocrinol Metab 92:1909–1919 [DOI] [PubMed] [Google Scholar]
- Savage JJ, Hunter CS, Clark-Sturm SL, Jacob TM, Pfaeffle RW, Rhodes SJ 2007 Mutations in the LHX3 gene cause dysregulation of pituitary and neural target genes that reflect patient phenotypes. Gene 400:44–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloop KW, Parker GE, Hanna KR, Wright HA, Rhodes SJ 2001 LHX3 transcription factor mutations associated with combined pituitary hormone deficiency impair the activation of pituitary target genes. Gene 265:61–69 [DOI] [PubMed] [Google Scholar]
- Bach I, Rhodes SJ, Pearse 2nd RV, Heinzel T, Gloss B, Scully KM, Sawchenko PE, Rosenfeld MG 1995 P-Lim, a LIM homeodomain factor, is expressed during pituitary organ and cell commitment and synergizes with Pit-1. Proc Natl Acad Sci USA 92:2720–2724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissinger CR, Liu BS, Martin-Blanco E, Kornberg TB, Pabo CO 1990 Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: a framework for understanding homeodomain-DNA interactions. Cell 63:579–590 [DOI] [PubMed] [Google Scholar]
- Fraenkel E, Rould MA, Chambers KA, Pabo CO 1998 Engrailed homeodomain-DNA complex at 2.2 A resolution: a detailed view of the interface and comparison with other engrailed structures. J Mol Biol 284:351–361 [DOI] [PubMed] [Google Scholar]
- Parker GE, Sandoval RM, Feister HA, Bidwell JP, Rhodes SJ 2000 The homeodomain coordinates nuclear entry of the Lhx3 neuroendocrine transcription factor and association with the nuclear matrix. J Biol Chem 275:23891–23898 [DOI] [PubMed] [Google Scholar]
- Meier BC, Price JR, Parker GE, Bridwell JL, Rhodes SJ 1999 Characterization of the porcine Lhx3/LIM-3/P-Lim LIM homeodomain transcription factor. Mol Cell Endocrinol 147:65–74 [DOI] [PubMed] [Google Scholar]
- Prader A, Largo RH, Molinari L, Issler C 1989 Physical growth of Swiss children from birth to 20 years of age. First Zurich longitudinal study of growth and development. Helv Paediatr Acta Suppl 52:1–125 [PubMed] [Google Scholar]
- Donaubauer J, Kiess W, Kratzsch J, Nowak T, Steinkamp H, Willgerodt H, Keller E 2003 Re-assessment of growth hormone secretion in young adult patients with childhood-onset growth hormone deficiency. Clin Endocrinol (Oxf) 58:456–463 [DOI] [PubMed] [Google Scholar]
- Sloop KW, Meier BC, Bridwell JL, Parker GE, Schiller AM, Rhodes SJ 1999 Differential activation of pituitary hormone genes by human Lhx3 isoforms with distinct DNA binding properties. Mol Endocrinol 13:2212–2225 [DOI] [PubMed] [Google Scholar]
- Bach I 2000 The LIM domain: regulation by association. Mech Dev 91:5–17 [DOI] [PubMed] [Google Scholar]
- Bridwell JA, Price JR, Parker GE, McCutchan Schiller A, Sloop KW, Rhodes SJ 2001 Role of the LIM domains in DNA recognition by the Lhx3 neuroendocrine transcription factor. Gene 277:239–250 [DOI] [PubMed] [Google Scholar]
- Seidman JG, Seidman C 2002 Transcription factor haploinsufficiency: when half a loaf is not enough. J Clin Invest 109:451–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles MA, Suh H, Hjalt TA, Drouin J, Camper SA, Gage PJ 2005 PITX genes are required for cell survival and Lhx3 activation. Mol Endocrinol 19:1893–1903 [DOI] [PubMed] [Google Scholar]
- Suh H, Gage PJ, Drouin J, Camper SA 2002 Pitx2 is required at multiple stages of pituitary organogenesis: pituitary primordium formation and cell specification. Development 129:329–337 [DOI] [PubMed] [Google Scholar]
- Savage JJ, Mullen RD, Sloop KW, Colvin SC, Camper SA, Franklin CL, Rhodes SJ 2007 Transgenic mice expressing LHX3 transcription factor isoforms in the pituitary: effects on the gonadotrope axis and sex-specific reproductive disease. J Cell Physiol 212:105–117 [DOI] [PubMed] [Google Scholar]
- Nasonkin IO, Ward RD, Raetzman LT, Seasholtz AF, Saunders TL, Gillespie PJ, Camper SA 2004 Pituitary hypoplasia and respiratory distress syndrome in Prop1 knockout mice. Hum Mol Genet 13:2727–2735 [DOI] [PubMed] [Google Scholar]
- Ward RD, Raetzman LT, Suh H, Stone BM, Nasonkin IO, Camper SA 2005 Role of PROP1 in pituitary gland growth. Mol Endocrinol 19:698–710 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.