Abstract
Selenocysteinyl-tRNASec, cysteinyl-tRNACys, glutaminyl-tRNAGln, and asparaginyl-tRNAAsn in many organisms are formed in an indirect pathway in which a non-cognate amino acid is first attached to the tRNA. This non-cognate amino acid is then converted to the cognate amino acid by a tRNA-dependent modifying enzyme. The in vitro characterization of these modifying enzymes is challenging due to the fact the substrate, aminoacyl-tRNA, is labile and requires a prior enzymatic step to be synthesized. The need to separate product aa-tRNA from unreacted substrate is typically a labor- and time-intensive task; this adds another impediment in the investigation of these enzymes. Here we review four different approaches for studying these tRNA-dependent amino acid modifications. In addition, we describe in detail a [32P]/nuclease P1 assay for glutaminyl-tRNAGln and asparaginyl-tRNAAsn formation which is sensitive, enables monitoring of the aminoacyl state of the tRNA, and is less time consuming than some of the other techniques. This [32P]/nuclease P1 method should be adaptable to studying tRNA-dependent selenocysteine and cysteine synthesis.
Keywords: Gln-tRNA, Asn-tRNA, Cys-tRNA, Sec-tRNA, Sep-tRNA, AdT, GatCAB, GatDE, SepSecS, SepCysS, PSTK, SelA, aminoacyl-tRNA
1. Introduction
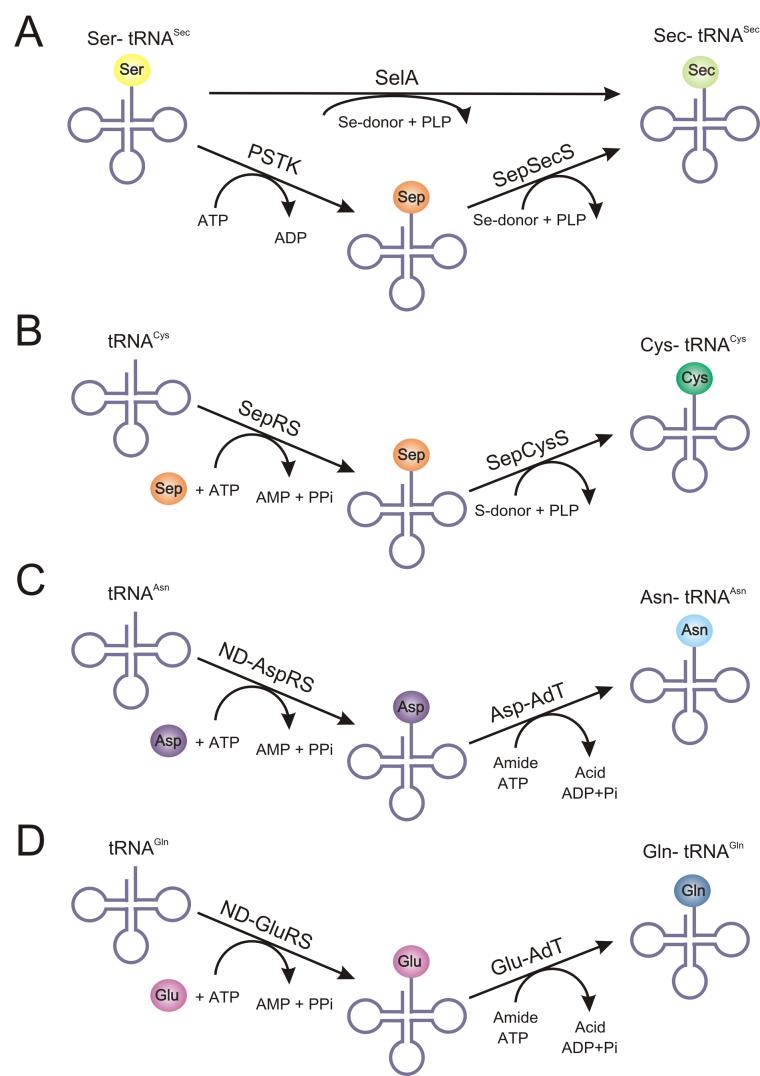
Accurate pairing of an amino acid with its cognate tRNA is an essential step ensuring the fidelity of protein synthesis. This process is usually carried out by the family of aminoacyl-tRNA synthetases (aaRSs), with one enzyme for each of the canonical amino acid [1]. However, many organisms lack one or more members of the aaRS family. In these organisms aminoacyl-tRNA (aa-tRNA) is formed by the pretranslational conversion of a non-cognate amino acid (aa) attached to tRNA by enzymes unrelated to aaRSs. Currently, the indirect formation of four different aa-tRNAs is well known: selenocysteinyl-tRNASec (Sec-tRNASec) [2], cysteinyl-tRNACys (Cys-tRNACys) [3], asparaginyl-tRNAAsn (Asn-tRNAAsn)[4], and glutaminyl-tRNAGln (Gln-tRNAGln) [5]. The four known indirect pathways are presented in Fig. 1.
Fig. 1.
Schematic representation of the known indirect pathways for aa-tRNA formation. The indirect routes for (A) Sec-tRNASec, (B) Cys-tRNACys, (C) Asn-tRNAAsn, and (D) Gln-tRNAGln biosynthesis are shown and described in the text. (A) The first step in Sec-tRNASec formation, serylation of tRNASec by seryl-tRNA synthetase (SerRS), is not shown. Bacteria use the SelA pathway (upper). The PSTK/SepSecS route (lower) is used in eukaryotes and archaea.
Sec-tRNASec is formed only in this manner (Fig. 1A) as no selenocysteinyl-tRNA synthetase (SecRS) has been discovered to date. The first step in the pathway is aminoacylation of tRNASec with Ser by seryl-tRNA synthetase (SerRS) [2]. In bacteria, the Ser attached to the tRNA is then converted to Sec by selenocysteine synthase (SelA) [6]. In archaea and eukaryotes, the Ser moiety is first phosphorylated by O-phosphoseryl-tRNASec kinase (PSTK) [7, 8]. The O-phosphoserine (Sep) formed on the tRNA is then modified to Sec by a Sep-tRNA:Sec-tRNA synthase (SepSecS) [9, 10].
Sep-tRNA is also an intermediate in tRNA-dependent Cys synthesis (Fig. 1B) [3]. Methanogenic archaea which lack CysRS form Cys-tRNACys in an indirect manner in which Sep is first ligated onto tRNACys by O-phosphoseryl-tRNA synthetase (SepRS) [3]. The Sep attached to the tRNA is then converted to Cys by a Sep-tRNA:Cys-tRNA synthase (SepCysS) [3]. In certain archaea, this tRNA-dependent route for Cys formation appears to be the sole means for Cys biosynthesis, including a handful of species that encode CysRS in their genomes [3].
In most prokaryotes Asn-tRNAAsn is also formed via an indirect path (Fig. 1C). In these organisms, a non-discriminating aspartyl-tRNA synthetase (ND-AspRS) aminoacylates tRNAAsn with Asp. The mischarged substrate, Asp-tRNAAsn, is then amidated by an Asp-tRNAAsn-dependent amidotransferase (Asp-AdT) to form Asn-tRNAAsn[11]. Organisms such as Thermus thermophilus and Deinococcus radiodurans have retained this indirect pathway for Asn-tRNAAsn formation even though they also have an AsnRS [12]. Asn in these species is only synthesized via this tRNA-dependent mechanism [12].
The majority of bacteria [13] and all known archaea [14] lack a GlnRS. Gln-tRNAGln is formed in an indirect manner analogous to the Asn-tRNAAsn pathway described above (Fig. 1D). First, tRNAGln is aminoacylated with Glu by a non-discriminating glutamyl-tRNA synthetase (ND-GluRS) [15]. The Glu moiety ligated to the tRNA is then modified to Gln by a Glu-tRNAGln-dependent amidotransferase (Glu-AdT) to form Gln-tRNAGln [5].
Two tRNA-dependent amidotransferases (AdTs) have been discovered in nature, GatCAB [16] and GatDE [14]. The latter is found in all known archaea and solely acts as a Glu-AdT [14]. GatCAB is found in both bacteria [16] and archaea [14]. It can act as both a Glu-AdT and an Asp-AdT [13,17].
The in vitro characterization of the tRNA-dependent modifying enzymes (SelA, PSTK, SepSecS, SepCysS, GatCAB and GatDE) has been hampered by three major factors. First, the substrates for these modifying enzymes require a prior enzymatic step, usually aminoacylation by an aaRS. Second, the aa-tRNA substrates are labile due to the chemical nature of the aminoacyl linkage. Third, given the typically subtle chemical changes in the aa-tRNA introduced by these modifying enzymes, it may be difficult and time consuming to differentiate the product formed from the substrate aa-tRNA.
The ideal assay for these modifying enzymes would monitor the aminoacylation state of the tRNA prior and during the modification reaction while not being labor and time intensive. We will review four different experimental approaches, highlighting the advantages and disadvantages of each methodology accordingly. In addition, we will describe in detail a novel [32P]-tRNA/nuclease P1 based assay for monitoring tRNA-dependent transamidation by the AdTs, which can also be adapted for studies of the other tRNA-dependent amino acid modifying enzymes.
2. Methods for assaying tRNA-dependent amino acid biosynthesis
2.1 [14C]-amino acid/TLC based assay
One of the most commonly used methods for monitoring amino acid modification on the tRNA is a [14C]-amino acid/thin-layer chromatography based assay which has been successfully used to monitor the formation of Gln-tRNAGln [16], Asn-tRNAAsn [11, 18], Cys-tRNACys[3], Sep-tRNASec [9], and Sec-tRNASec [19]. First, the tRNA is aminoacylated with the appropriate 14C-labeled amino acid by a proper aaRS. The aminoacylation reaction can be monitored as previously described [20]. At the end of the reaction, the mixture is phenol (citrate-buffered, pH 4.5)/chloroform extracted and then ethanol precipitated. It is necessary to separate the [14C]-aa-tRNA from the remaining free [14C]-aa; this is usually accomplished by multiple rounds of ethanol precipitation. Passage over a size exclusion column such as a Bio-spin 30 column (Bio-Rad, 732-6231) can also be used. Running the mixtures over such a column has the added benefit of removing excess ATP from the aminoacylation reaction that might precipitate with the tRNA [21, 22].
The [14C]-aa-tRNA substrate is then incubated with the modifying enzyme and appropriate other substrates. The progress of the reaction is followed by removing aliquots of the reaction mixture; they are then quenched usually with 3 M sodium acetate (pH 5) and then ethanol precipitated. The precipitation step separates the [14C]-aa-tRNA from any free [14C]-aa that deacylated from the tRNA over the course of the reaction. Mild alkaline treatment is then applied to deacylate the [14C]-aa-tRNA. The product amino acid generated by the modifying enzyme can then be separated by TLC from the unreacted substrate amino acid based on the chemical difference between the two amino acids. For example, Glu and Gln can be separated from each other on a cellulose plate based on their hydrophobicity; when the plate is developed under basic or acidic conditions due to the differences in the polar nature of their side chains under those conditions. Phosphorimaging is used to visualize the separation, and densitometry software will quantify the results.
The use of [14C]-aa allows for a straightforward method for determining the amount of substrate aa-tRNA formed in the initial aminoacylation reaction [20] as well as a means of monitoring the deacylation of the tRNA over the course of the reaction [23]. The subsequent ethanol precipitation and deacylation steps make it a labor and time consuming assay to carry out for studying a tRNA-dependent modifying enzyme, especially when taking multiple time points. In addition, the lower specific activity of [14C] relative to [32P] necessitates greater quantities of tRNA than the [32P]-tRNA/nuclease P1 based assay described below. Since the amino acids are radiolabeled and are deacylated from the tRNA prior to separation, care must be taken to ensure that the conversion of the amino acid corresponds to that on the tRNA and not to a tRNA-independent modification. In addition, deacylation conditions must be selected that minimize possible side reactions [22]. When separating amino acid by TLC, smearing can be an issue, making quantification challenging.
2.2 [14C]-amino acid/TLE based assay
Recently a new method for transamidation has been developed that is very similar to the [14C]-amino acid/TLC based assay described above. The only difference is that instead of separating the product amino acid from the substrate amino acid by TLC, the two are separated by thin-layer electrophoresis (TLE) [24]. The mobility of the amino acids is based on their charged states allowing for separation of the two amino acids (e.g. Glu from Gln). This system is reported to give rise to improved resolution over the TLC method described above while typically taking less time to separate the two amino acids than TLC does [24]. The assay requires the same care as the TLC one above with regards to ensuring conversion is on the tRNA and that the deacylation conditions do not produce side products. Finally, it is just as cumbersome to perform as the TLC method due to the ethanol precipitation and deacylation steps.
2.3 HPLC-based assay
A high-performance liquid chromatography (HPLC) based system has also been used to monitor the AdT catalyzed transamidation reaction [25]. The system is very similar to the two assays described above. Instead of using [14C]- aa though, unlabeled aa is acylated onto the tRNA [25]. After being released from the tRNA, the amino acids are derivatized with the AccQ-Tag fluorescence reagent (Waters) before being loaded onto an AccQ-Tag HPLC column and separated by reverse-phase elution [25]. This method has low sensitivity, making certain kinetic measurements difficult [25]. The derivatization step makes this assay even more labor and time intensive than the TLE and TLC based systems described above.
2.4 TCA precipitation
In the presence of 10% (w/v) trichloroacetic acid (TCA), tRNA, including aa-tRNA, will precipitate out of solution, while free amino acids, nucleotides and other small molecules do not. This fact has been exploited to successfully monitor SelA conversion of Ser-tRNASec to Sec-tRNASec [26]. [75Se]selenite is the selenium donor in the reaction. The Sec-tRNASec formed by SelA is therefore radiolabeled while the substrate Ser-tRNASec is not. The TCA precipitation separates [75Se]Sec-tRNASec from free [75Se]selenite. The precipitated tRNA is collected on Whatman 3MM filter paper, and the radioactivity is then measured by liquid scintillation counting. Since only Sec-tRNASec is labeled, the radioactivity measured corresponds to Sec formed on the tRNA. The assay is sensitive and relatively easy to perform, avoiding the deacylation and ethanol precipitation steps of the three systems discussed above. The method does not readily lend itself to monitoring whether the aa-tRNA substrate deacylates over the course of the reaction. The assay is limited to situations where a radiolabeled donor is available.
2.5 [32P]tRNA/Nuclease P1 Based Assay
Given the cumbersome nature of the [14C]-aa and HPLC methods and the fact that the TCA precipitation based assay [27] has been ineffective in monitoring the AdT catalyzed transamidation with purified enzyme [22, 25], we developed a new method for monitoring tRNA-dependent transamidation (Fig. 2) [13, 28] by adapting a [32P]tRNA/nuclease P1 assay that has been successfully used to monitor tRNA aminoacylation by aaRSs [21, 29, 30].
Fig. 2.
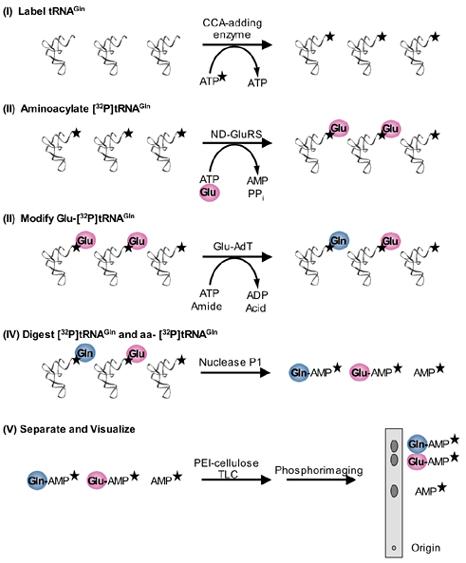
[32P]tRNA/Nuclease P1 amidotransferase assay for Gln-tRNAGln formation. (I) The 3′ terminal AMP of tRNAGln is 32P-labeled by using the E. coli CCA-adding enzyme and [α-32P]ATP. The stars denote the radioactive label. Excess [α-32P]ATP is removed as described in the text. (II) The [32P]tRNAGln is then glutamylated by a ND-GluRS and then purified as detailed in the text. (III) The Glu-[32P]tRNAGln is modified by a Glu-AdT (GatCAB or GatDE) into Gln-[32P]tRNAGln. (IV) The [32P]tRNAGln is digested into mononucleotides by nuclease P1. Only the radiolabeled products (Gln-[32P]AMP, Glu-[32P]AMP and [32P]AMP) are shown. (V) The Gln-[32P]AMP, Glu-[32P]AMP and [32P]AMP, corresponding to Gln-[32P]tRNAGln (product formed), unreacted Glu-[32P]tRNAGln (substrate), and uncharged [32P]tRNA, respectively, can be separated by PEI-cellulose TLC and visualized by phosphorimaging.
In this method, the terminal 3′ AMP of the tRNA is exchanged with [α-32P]ATP by the CCA-adding enzyme (step I) [29]. The tRNA is then aminoacylated with cold amino acid by the appropriate aaRS (step II). After purifying the tRNA following aminoacylation by ethanol precipitation and passage over a size exclusion column to remove free amino acid and ATP, the 32P-labeled aa-tRNA can be incubated with AdT along with ATP and an amide donor, the other two substrates required for transamidation (step III). Aliquots of the reaction mix are quenched under acidic conditions, and the tRNA is digested with nuclease P1 (step IV). This enzyme cleaves the tRNA to mononucleotides release releasing [32P]AMP and aa-[32P]AMP as the reaction products. In the case of the Glu-AdT catalyzed transamidation of Glu-tRNAGln three different [32P]AMP species are left after digestion: Glu-[32P]AMP corresponding to unreacted Glu-[32P]-tRNAGln, Gln-[32P]AMP corresponding to Gln-[32P]-tRNAGln formed, and [32P]AMP corresponding to uncharged [32P]-tRNAGln in the reaction. The three radiolabeled species can be separated from one another based on charge using polyethylenimine (PEI)-cellulose TLC (step V); the digested reaction mixture is spotted on a PEI-cellulose plate, which is developed with acidic solvent (100 mM ammonium acetate, 5 % acetic acid) [21]. Asp-AdT activity can be assayed for similarly except that the three 32P-labeled species (Asp-[32P]AMP, Asn-[32P]AMP, and [32P]AMP) are separated under slightly different solvent conditions (1 mM ammonium acetate, 5 % acetic acid) [13]. The separation is visualized by phosphorimaging, and densitometry software is then used to quantify the amount of product formed.
The [32P]tRNA/nuclease P1 assay is more sensitive than the [14C] and HPLC based assays, thus requiring less aa-tRNA substrate. In addition, it is a less time and labor-intensive method as the ethanol precipitation and deacylation steps of the other assays are avoided. Because the radioactive label is in the tRNA, the aminoacylation state of the tRNA over the course of the reaction can be monitored in parallel to assaying for amino acid conversion on the tRNA. The label on the tRNA has the added benefit of ensuring that any modification of the amino acid is tRNA-dependent. Working with tRNA species that are poorly aminoacylated by aaRSs can be challenging, especially when the assays require high concentrations of aa-tRNA. For poorly charged aa-tRNA preparations higher concentrations of total tRNA must be added to the reaction mixture in order to provide the same amount of aa-tRNA as compared to a well-charged tRNA sample. After digestion with nuclease P1, this corresponds to a greater concentration of [32P]AMP in the mixture. The more total [32P]AMP spotted on the plate, the greater the smearing and streaking of the spot that increases background and makes it difficult to differentiate signal from noise. Decreasing the amount of digested sample spotted will decrease this but with a corresponding decrease in signal. Optimizing aminoacylation conditions is thus important to successfully use this method, especially in kinetic studies.
Given the novelty of the [32P]tRNA/nuclease P1 based assay for measuring tRNA-dependent transamidation and its potential application for studying other modifying enzymes such as PSTK, SepSecS and SepCysS, we will describe in more detail how we carryout the method to study tRNA-dependent transamidation by the AdTs.
3. [32P]tRNA/Nuclease P1 Amidotransferase Assay Protocol
3.1 32P-labeling of tRNA
The 3′ terminal AMP of purified tRNA (tRNAGln or tRNAAsn) is 32P-labeled by using the Escherichia coli CCA-adding enzyme and [α-32P]ATP (GE Healthcare, PB10200) [21]. In 50 mM Tris-HCl (pH 8.0), 20 mM MgCl2, 5 mM DTT and 50 μM NaPPi, 15 μM tRNA is incubated with the CCA-adding enzyme (50 μg/ml) and 1.6 μCi/μL of [α-32P]ATP (GE Healthcare, PB10200) for 35 minutes at room temperature, normally in a reaction volume of 48 μL. We stop the reaction by adding equal volumes of phenol (Tris buffered, pH 7.9, American Bioanalytical, AB01616) and chloroform. The samples are spun for 2 minutes at 13,000 rpm. The aqueous phase is passed over a Bio-Spin 30 (Bio-Rad, 732-6231) column to remove excess [α-32P]ATP [21]. Liquid scintillation counting can be used to determine the efficiency of labeling. If [32P]tRNA is not being used the same day, the sample may be ethanol precipitated and stored at - 20 °C.
3.2 Aminoacylation of [32P]tRNA
As mentioned above, optimization of the tRNA aminoacylation step of the protocol is essential. For glutamylation of the Helicobacter pylori tRNAGln, incubation of 3 μM H. pylori GluRS2, 1 mM L-Glu, 10 μM unlabeled H. pylori tRNAGln and 600 nM 32P-labeled H. pylori tRNAGln carried out in 100 mM Hepes-KOH, pH 7.2, 30 mM KCl, 12 mM MgCl2, 4 mM ATP, 2 mM DTT and 24 μg/μL of pyrophosphatase (Roche) for 45 minutes at 37 °C is optimal to aminoacylate up to 70 % of the tRNA [13]. For aminoacylation of H. pylori tRNAAsn with Asp we incubate 21 μM unlabeled H. pylori tRNAAsn and 1.33 μM 32P-labeled H. pylori tRNAAsn for 14 hours at 37°C in 50 mM Hepes-NaOH, pH 7.2, with 25 mM KCl, 15 mM MgCl2, 4 mM ATP, 5 mM DTT, 3 μM Pseudomonas aeruginosa AspRS, 1 mM L-Asp. By reducing the concentration of unlabeled H. pylori tRNAAsn to 10 μM while increasing the L-Asp added to 3.3 mM, the reaction could be carried out over 90 minutes. We can reliably aminoacylate 75% of the H. pylori tRNAAsn with Asp using this method. By incubating 10 μM unlabeled and 400 nM 32P-labeled Methanothermobacter thermautotrophicus tRNAGln with 50 mM Hepes-KOH buffer (pH 7.2), 25 mM KCl, 10 mM MgCl2, 4 mM ATP, 5 mM DTT, 24 μg/μL of pyrophosphatase (Roche), 3 μM M. thermautotrophicus GluRS, and 1 mM L-Glu for 90 minutes at 37 °C, we reliably aminoacylate 65% of the tRNA [28].
To check levels of aminoacylation, 2 μL aliquots at the start and end of the reactions are quenched with 3 μL 1x nuclease P1 mix and the tRNA is digested for 30 minutes at room temperature. Nuclease P1 from Sigma (N8630-1VL) comes lyophilized. We resuspend it in 100 mM sodium citrate (pH 4.74) to make a 10x stock (6.6 mg/mL of nuclease P1), which can be stored at -20 °C for up to two months. The 10x stock is diluted to a 1x mix (0.66 mg/mL nuclease P1) in 100 mM sodium citrate (pH 4.74) before use. We spot 2 μL of the digested reaction mixture on a PEI-cellulose plate (J.T. Baker, 4474-04 or EMD, 5725-7) and develop the plate under acidic conditions. For the glutamylation reactions, a 100 mM ammonium acetate, 5% acetic acid solvent system is used to develop the plates [21], while for the aspartylation reactions a 1 mM ammonium acetate, 5% acetic acid solvent system is used [13]. The chromatography separates [32P]AMP from aa-[32P]AMP which can be visualized by phosphorimaging. The developed PEI-cellulose plates are exposed to Fuji imaging screens for 16 hours and the screens are then scanned on a Molecular Dynamics Storm 860 phosphoimager. Densitometry software (ImageQuant, Molecular Dynamics) is used to quantify the percentage of tRNA that is aminoacylated. We do this by dividing the intensity of the aa-[32P]AMP spot by the sum of the intensities of the aa-[32P]AMP and [32P]AMP spots after subtracting out background.
At the end of the aminoacylation reaction, the reaction is quenched by adding 1/10 volume 3 M sodium acetate (pH 5.5). Equal volumes of phenol (citrate buffered, pH 4.5, American Bioanalytical, AB01617) and chloroform are added to the reaction mixture. The samples are spun at 13,000 rpm for 2 minutes. To the aqueous phase, 2.5 times the volume of cold ethanol is added and the sample is incubated at -80 °C for a minimum of 20 minutes before centrifuging the sample at 13,000 rpm at 4 °C for 20 minutes to precipitate the tRNA out of solution. The pellets are washed with cold 70 % ethanol and spun again at 13,000 rpm at 4 °C for 5 minutes. If the sample is not going to be used that day, we store the pellet at -20 °C. When ready to use, the pellet is resuspended in deionized water. For a 1 mL aminoacylation reaction, we usually resuspend the pellet in 200 μL of water. The sample is then passed over two Bio-spin 30 columns (Bio-Rad, 732-6231) to remove excess ATP from the aminoacylation reaction.
3.3 32P/nuclease P1 amidotransferase assays
The 32P/nuclease P1 amidotransferase assays are carried out with 4 mM ATP, 2 mM amide donor (Gln or Asn) and 9-11 μM 32P-labeled aa-tRNA (Asp-tRNAAsn or Glu-tRNAGln). For Km determination, we vary one substrate while keeping constant the other two substrates for transamidation. Typically, for such calculations we vary amide donor concentrations from 4 μM - 4 mM. For ATP, the range is between 20 μM - 4 mM. For Km determination of mischarged tRNA substrate, we vary the concentration from 37.5 nM - 10 μM of mischarged tRNA added. The total amount of tRNA added will vary depending on the fraction of tRNA aminoacylated. The enzyme concentration added can vary from 1-20 nM depending on the concentration necessary to obtain a signal while still holding true to steady-state assumptions. For H. pylori GatCAB we carry out the reactions in 50 mM Hepes-KOH, pH 7.2, 15 mM MgCl2, 25 mM KCl and 1 mM DTT. For the M. thermautotrophicus AdTs, reactions are incubated in 100 mM Hepes-KOH, pH 7.2, 30 mM KCl, 12 mM MgCl2, and 5 mM DTT. For time courses, aliquots (1 to 2 μL) of the reaction mixture are taken at time points and quenched in 1x nuclease P1 mix. The quenched reaction mixtures are kept on ice until the last time point is taken. All the quenched reactions are then incubated at room temperature for 30 minutes to digest the tRNA.
PEI-cellulose TLC plates are spotted with 1 to 2 μL of digested sample. The [32P]AMP and the two aa-[32P]AMP species are better resolved and develop approximately 30 minutes faster using the glass-backed plates from EMD (5725-7) as opposed to plastic-backed plates (J.T. Baker, 4474-04). When spotting multiple digested mixtures on a plate, the different samples are spaced 3 cm apart to avert signal bleed from one lane to another, which would prevent accurate quantification of the data. To minimize smearing and streaking, plates should be developed to go with their grain. The glass-backed plates typically take 90 minutes to develop. For separation of Asp-[32P]AMP, Asn-[32P]AMP, and [32P]AMP, we develop the samples in 1 mM ammonium acetate, 5% acetic acid [13], and for separation of Glu-[ 32P]AMP, Gln-[32P]AMP, and [32P]AMP, we develop the plates in 100 mM ammonium acetate, 5% acetic acid [28]. After the plates are run, they are air dried for 15 minutes.
3.4 Quantification
The separation of the three [32P]AMP species can be observed exposing the developed PEI-cellulose plates to a Fuji phosphoimaging screen for 16 hours and then scanning the screen on a Molecular Dynamics Storm 860 phosphoimager. The Rf values for Gln-[32P]AMP, Glu-[32P]AMP, and [32P]AMP are 0.87, 0.76, and 0.49 respectively (Fig. 3) . For Asn-[32P]AMP, Asp-[32P]AMP, and [32P]AMP, the Rf values are 0.91, 0.75, and 0.39 respectively [13]. ImageQuant (Molecular Dynamics) is used to quantify the results. By dividing the intensity of the amide-[32P]AMP by the total intensity after correcting for background, the percentage of amide-[32P]AMP in the mixture can be determined, which corresponds to the percentage of amide-[32P]tRNA formed. The total intensity equals the sum of the intensities of the amide aa-[32P]AMP (Gln- [32P]AMP or Asn-[32P]AMP), acidic aa-[32P]AMP (Glu-[32P]AMP or Asp-[32P]AMP), and [32P]AMP spots after correcting for background. Given that the concentration of total tRNA added to the reaction is known, we can calculate the concentration of amide aa-[32P]tRNA formed in the reaction mixture at each time point. For time courses, we plot our data using Excel (Microsoft, Redmond, WA) or Kaleidagraph (Synergy Software, Reading, PA). Steady-state kinetic parameters (Km and kcat) are determined using Kaleidagraph by nonlinear regression of plots of initial velocity vs. substrate concentration (Fig. 4).
Fig. 3.
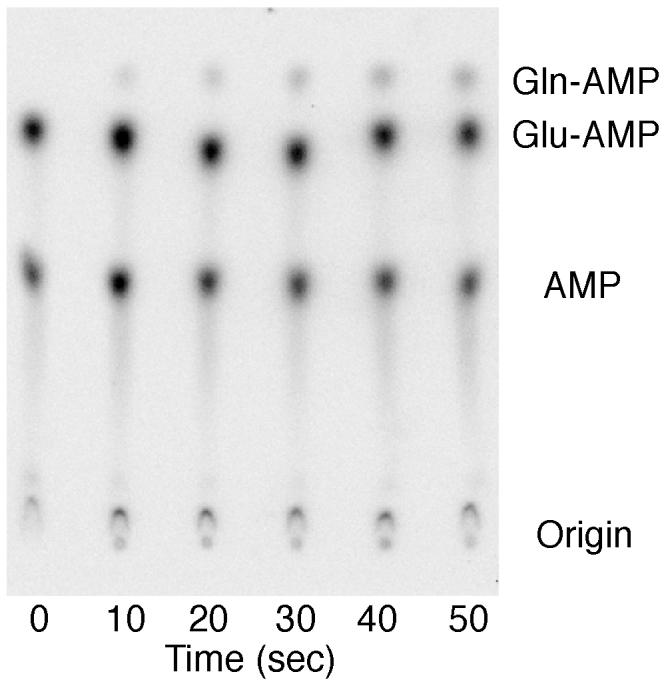
Representative phosphorimage of the separation of Gln-[α-32P]AMP, Glu-[α-32P]AMP and [α-32P]AMP by PEI-cellulose chromatography. Aliquots (2 μL) of the amidotransferase reaction were taken at the time points indicated and quenched on ice with 3 μL of 100 mM sodium citrate, pH 4.74, and 0.66 mg/mL of nuclease P1. Samples were digested at room temperature and then spotted onto a 20 × 20 cm PEI-cellulose glass plate (EMD, 5725-7), which was developed in 100 mM ammonium acetate, 5% acetic acid for 90 minutes. The Rf values are 0.49 (AMP), 0.76 (Glu-AMP) and 0.87 (Gln-AMP).
Fig. 4.
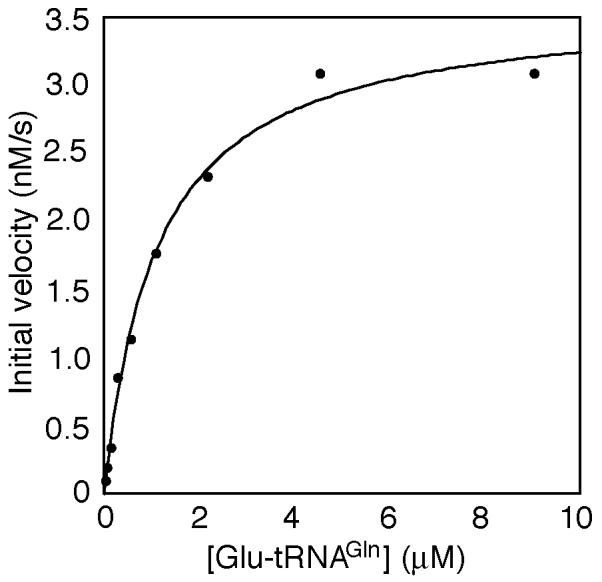
Representative plot of initial velocity (nM/s) vs Glu-tRNAGln concentration (μM) with 1 nM H. pylori GatCAB. The data was fit by nonlinear regression using Kaleidagraph.
We also determine whether the aa-tRNA deacylates over the course of the reaction by calculating the percentage of total aa-tRNA at the beginning and end of the time course. This is done by adding the intensity of the amide aa-[ 32P]AMP spot to the intensity of the acidic aa-[32P]AMP spot and dividing the sum by the total intensity. If the aa-tRNA deacylates during the course of the reaction, conditions may need to be varied; this may include lowering the reaction temperature or shortening the length of the time course.
3.5 Applications
The [32P]tRNA/nuclease P1 amidotransferase assay is more sensitive and easier to carry out than the [14C] and HPLC based methods. We have successfully used it to study the tRNA-dependent transamidations catalyzed by the M. thermautotrophicus GatDE and the H. pylori GatCAB [13, 28]. Given that the separation of aa-[32P]AMP species following digestion by nuclease P1 is based on anion exchange chromatography and the differences in the charged natures of Sep, Ser, Cys and Sec, the method should be amendable for studying the tRNA-dependent amino acid modifications catalyzed by PSTK, SepSecS and SepCysS. Monitoring the aminoacylation state of the aa-tRNA, which this assay permits, is especially important in studies involving SepSecS, since the substrate aa-tRNA for that enzyme, Sep-tRNASec, requires two enzymatic steps for its formation; serylation of the tRNA by SerRS and then phosphorylation of the Ser by PSTK. For this reason, a [32P]tRNA/nuclease P1 based assay for Sec-tRNASec formation by SepSecS might be better than the TCA precipitation assay that was used to monitor tRNA-dependent Sec formation by SelA [26].
Acknowledgements
We thank R. Lynn Sherrer and Sotiria Palioura for useful discussions, and John J. Perona (University of California, Santa Barbara) for experimental suggestions with the [32P]tRNA/nuclease P1 assay. The work in the authors’ laboratory was supported by National Institute of General Medical Sciences grant GM22854, National Science Foundation grant MCB-0645283, and Department of Energy grant DE-FG02-98ER20311.
The abbreviations used are
- aaRS
aminoacyl-tRNA synthetase
- aa-tRNA
aminoacyl-tRNA
- aa
amino acid
- Sec-tRNA
selenocysteinyl-tRNA
- Cys-tRNA
cysteinyl-tRNA
- Asn-tRNA
asparaginyl-tRNA
- Gln-tRNA
glutaminyl-tRNA
- SerRS
seryl-tRNA synthetase
- SelA
selenocysteine synthase
- PSTK
O-phosphoseryl-tRNASec kinase
- Sep
O-phosphoserine
- SepSecS
Sep-tRNA:Sec-tRNA synthase
- CysRS
cysteinyl-tRNA synthetase
- SepRS
O-phosphoseryl-tRNA synthetase
- SepCysS
Sep-tRNA:Cys-tRNA synthase
- GlnRS
glutaminyl-tRNA synthetase
- AsnRS
asparaginyl-tRNA synthetase
- ND
non-discriminating
- AdT
tRNA-dependent amidotransferase
- GluRS
glutamyl-tRNA synthetase
- Glu-AdT
glutamyl-tRNAGln-dependent amidotransferase
- AspRS
aspartyl-tRNA synthetase
- Asp-AdT
aspartyl-tRNAAsn-dependent amidotransferase
- PEI
polyethylenimine
- TLC
thin layer chromatography
The three letter code for amino acids is used througout. Modifying enzyme in this article refers to SelA, PSTK, SepSecS, SepCysS, GatDE, and GatCAB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Ibba M, Söll D. Annu Rev Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- [2].Leinfelder W, Zehelein E, Mandrand-Berthelot MA, Bock A. Nature. 1988;331:723–725. doi: 10.1038/331723a0. [DOI] [PubMed] [Google Scholar]
- [3].Sauerwald A, Zhu W, Major TA, Roy H, Palioura S, Jahn D, Whitman WB, Yates JR, 3rd, Ibba M, Söll D. Science. 2005;307:1969–1972. doi: 10.1126/science.1108329. [DOI] [PubMed] [Google Scholar]
- [4].Curnow AW, Ibba M, Söll D. Nature. 1996;382:589–590. doi: 10.1038/382589b0. [DOI] [PubMed] [Google Scholar]
- [5].Wilcox M, Nirenberg M. Proc. Natl. Acad. Sci. U. S. A. 1968;61:229–236. doi: 10.1073/pnas.61.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hatfield DL, Gladyshev VN. Mol Cell Biol. 2002;22:3565–3576. doi: 10.1128/MCB.22.11.3565-3576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kaiser JT, Gromadski K, Rother M, Engelhardt H, Rodnina MV, Wahl MC. Biochemistry. 2005;44:13315–13327. doi: 10.1021/bi051110r. [DOI] [PubMed] [Google Scholar]
- [8].Carlson BA, Xu XM, Kryukov GV, Rao M, Berry MJ, Gladyshev VN, Hatfield DL. Proc. Natl. Acad. Sci. U. S. A. 2004;101:12848–12853. doi: 10.1073/pnas.0402636101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Yuan J, Palioura S, Salazar JC, Su D, O’Donoghue P, Hohn MJ, Cardoso AM, Whitman WB, Söll D. Proc. Natl. Acad. Sci. U. S. A. 2006;103:18923–18927. doi: 10.1073/pnas.0609703104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Xue-Min X, Carlson BA, Mix H, Zhang Y, Kazima S, Glass RS, Berry MJ, Gladyshev VN, Hatfield DL. PLoS Biology. 2007;5:96–105. doi: 10.1371/journal.pbio.0050004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Curnow AW, Tumbula DL, Pelaschier JT, Min B, Söll D. Proc. Natl. Acad. Sci. U. S. A. 1998;95:12838–41283. doi: 10.1073/pnas.95.22.12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Min B, Pelaschier JT, Graham DE, Tumbula-Hansen D, Söll D. Proc. Natl. Acad. Sci. U. S. A. 2002;99:2678–2683. doi: 10.1073/pnas.012027399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sheppard K, Akochy PM, Salazar JC, Söll D. J. Biol. Chem. 2007;282:11866–11873. doi: 10.1074/jbc.M700398200. [DOI] [PubMed] [Google Scholar]
- [14].Tumbula DL, Becker HD, Chang WZ, Söll D. Nature. 2000;407:106–110. doi: 10.1038/35024120. [DOI] [PubMed] [Google Scholar]
- [15].Lapointe J, Duplain L, Proulx M. J Bacteriol. 1986;165:88–93. doi: 10.1128/jb.165.1.88-93.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Curnow AW, Hong K, Yuan R, Kim S, Martins O, Winkler W, Henkin TM, Söll D. Proc. Natl. Acad. Sci. U. S. A. 1997;94:11819–11826. doi: 10.1073/pnas.94.22.11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Raczniak G, Becker HD, Min B, Söll D. J. Biol. Chem. 2001;276:45862–45867. doi: 10.1074/jbc.M109494200. [DOI] [PubMed] [Google Scholar]
- [18].Bailly M, Giannouli S, Blaise M, Stathopoulos C, Kern D, Becker HD. Nucleic Acids Res. 2006;34:6083–6094. doi: 10.1093/nar/gkl622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Tormay P, Wilting R, Lottspeich F, Mehta PK, Christen P, Bock A. Eur. J. Biochem. 1998;254:655–661. doi: 10.1046/j.1432-1327.1998.2540655.x. [DOI] [PubMed] [Google Scholar]
- [20].Lapointe J, Levasseur S, Kern D. Methods Enzymol. 1985;113:42–49. doi: 10.1016/s0076-6879(85)13009-0. [DOI] [PubMed] [Google Scholar]
- [21].Bullock TL, Uter N, Nissan TA, Perona JJ. J. Mol. Biol. 2003;328:395–408. doi: 10.1016/s0022-2836(03)00305-x. [DOI] [PubMed] [Google Scholar]
- [22].Feng L, Sheppard K, Tumbula-Hansen D, Söll D. J. Biol. Chem. 2005;280:8150–8155. doi: 10.1074/jbc.M411098200. [DOI] [PubMed] [Google Scholar]
- [23].Schmidt E, Schimmel P. Science. 1994;264:265–267. doi: 10.1126/science.8146659. [DOI] [PubMed] [Google Scholar]
- [24].Cathopoulis TJ, Chuawong P, Hendrickson TL. Anal. Biochem. 2007;360:151–153. doi: 10.1016/j.ab.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Horiuchi KY, Harpel MR, Shen L, Luo Y, Rogers KC, Copeland RA. Biochemistry. 2001;40:6450–6457. doi: 10.1021/bi002599l. [DOI] [PubMed] [Google Scholar]
- [26].Thanbichler M, Bock A. Methods Enzymol. 2002;347:3–16. doi: 10.1016/s0076-6879(02)47003-6. [DOI] [PubMed] [Google Scholar]
- [27].Wilcox M. Cold Spring Harb. Symp. Quant. Biol. 1969;34:521–528. doi: 10.1101/sqb.1969.034.01.059. [DOI] [PubMed] [Google Scholar]
- [28].Oshikane H, Sheppard K, Fukai S, Nakamura Y, Ishitani R, Numata T, Sherrer RL, Feng L, Schmitt E, Panvert M, Blanquet S, Mechulam Y, Söll D, Nureki O. Science. 2006;312:1950–1954. doi: 10.1126/science.1128470. [DOI] [PubMed] [Google Scholar]
- [29].Wolfson AD, Pleiss JA, Uhlenbeck OC. Rna. 1998;4:1019–1023. doi: 10.1017/s1355838298980700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wolfson AD, Uhlenbeck OC. Proc. Natl. Acad. Sci. U. S. A. 2002;99:5965–5970. doi: 10.1073/pnas.092152799. [DOI] [PMC free article] [PubMed] [Google Scholar]