Abstract
A molecular clone of Japanese encephalitis (JE) virus Nakayama strain was used to create intertypic viruses containing either the 5’-C-prM-E or the prM-E region of the attenuated JE SA14-14-2 virus in the JE-Nakayama background. These two intertypic JE viruses, JE-X/5’CprME(S) and JE-X/prME(S) respectively, generally resembled the parental JE virus in cell culture properties. Similar to virus derived from the JE Nakayama molecular clone (JE-XJN), JE-X/prME(S) was highly neuroinvasive and neurovirulent for young adult mice, whereas JE-X/5’CprME(S) was attenuated for neuroinvasiveness and only partially attenuated for neurovirulence. Immunization of young mice with JE-X/5’CprME(S) virus elicited neutralizing antibodies against JE Nakayama virus and conferred protection against encephalitis following challenge with JE Nakayama virus. The sequence of the JE-X/5’CprME(S) virus differed from that of JE-X/prME(S) virus at two nucleotides in the 5’ UTR, 3 amino acid positions in the capsid protein, 4 positions in the prM protein and 1 in the envelope protein. For JE-X/prME(S) virus, the 4 differences in prM and the single substitution in the envelope represented reversions to the sequence of JE-Nakayama virus. Overall, this study reveals that molecular determinants associated with the prM-E region of the attenuated JE SA14-14-2 virus are insufficient by themselves to confer an attenuation phenotype upon JE Nakayama virus. This suggests a role for determinants in the 5’ UTR and/or the capsid protein of the JE SA 14-14-2 virus genome in influencing the virulence properties of the JE Nakayama virus in the mouse model.
INTRODUCTION
Japanese encephalitis (JE) virus is the principal member of the JE serogroup, which includes several agents of acute neurologic disease in humans (Monath and Heinz, 1996). JE is the most important cause of arthropod-transmitted acute viral encephalitis on a worldwide basis (Tsai, 1994). The virus causes seasonal epidemic outbreaks and sporadic endemic disease in many countries of Western and Southeast Asia (Burke and Leake, 1988; Halstead and Jacobson, 2003). It is estimated that as many as 30,000 cases of acute encephalitis due to JE virus occur annually within the Peoples Republic of China alone. Moreover, the geographic distribution of the virus has changed in recent years, with extension of its westward range into provinces of India and southward into Indonesian islands adjacent to Australia (Mackenzie et al., 2004). Neurologic disease caused by JE is often serious, with a mortality rate as high as 30%, and permanent neurological sequellae are frequently observed among survivors (Solomon et al., 2000). Traditionally, vaccine products for prevention of JE have included inactivated virus prepared from mouse-brain, and the live-attenuated JE-SA14-14-2 strain, which is not licensed for use outside of China (Tsai, 1994). Second generation vaccines for worldwide use are needed, primarily because of adverse reactions associated with mouse-brain-derived vaccines (Marfin et al., 2005; Takahashi et al., 2005).
Various approaches have been taken to investigate the molecular basis of JE virus virulence, including comparisons of nucleotide sequences of virus strains differing in virulence properties, and of engineered viruses as well as mutants selected for neutralization resistance or receptor escape. Mutations in the envelope protein have generally been regarded as critical in governing the attenuation of JE virus in mouse model systems (Cecelia and Gould, 1991; Hasegawa et al., 1992; Ni et al. 1994; 1995; Sumiyoshi et al., 1995; Ni and Barrett, 1996; 1998). Molecular characterization of the JE-SA14-14-2 vaccine and related attenuated strains suggests that there are multiple attenuating determinants within the E proteins relative to their parental JE SA14 viruses (Nitayaphan et al., 1990; Aihara et al., 1991; Ni et al., 1994; 1995). To further evaluate this hypothesis, we tested whether the structural proteins of the JE-SA14-14-2 strain would attenuate neuroinvasiveness and neurovirulence of the virulent JE Nakayama virus in the mouse model. This was done by constructing and testing intertypic structural region JE viruses. One such virus, containing the 5’ UTR, C, prM and E regions from JE SA14-14-2 virus, exhibited an attenuated phenotype in this model and was shown to have efficacy as an experimental vaccine.
RESULTS
Recovery of JE-X/5’CprME(S) and JE-X/prME(S) viruses
Figure 1 indicates the genomic organization of the viruses used in this study. JE-XJN represents the parental JE Nakayama virus infectious clone. For JE-X/prME(S), the 5’ terminus through the capsid protein region was derived from the JE-XJN clone, whereas the prM and E regions were derived from the JE SA14-14-2 virus. For JE-X/5’CprME(S), the 5’ terminus through the E region was derived from the JE SA14-14-2 virus. For both intertypic viruses, the region from NS1 through the 3’ UTR was derived from the JE-XJN clone. Thus, the two intertypic viruses were designed to differ only with respect to the region encoded from the 5’ terminus through the capsid protein.
Figure 1.
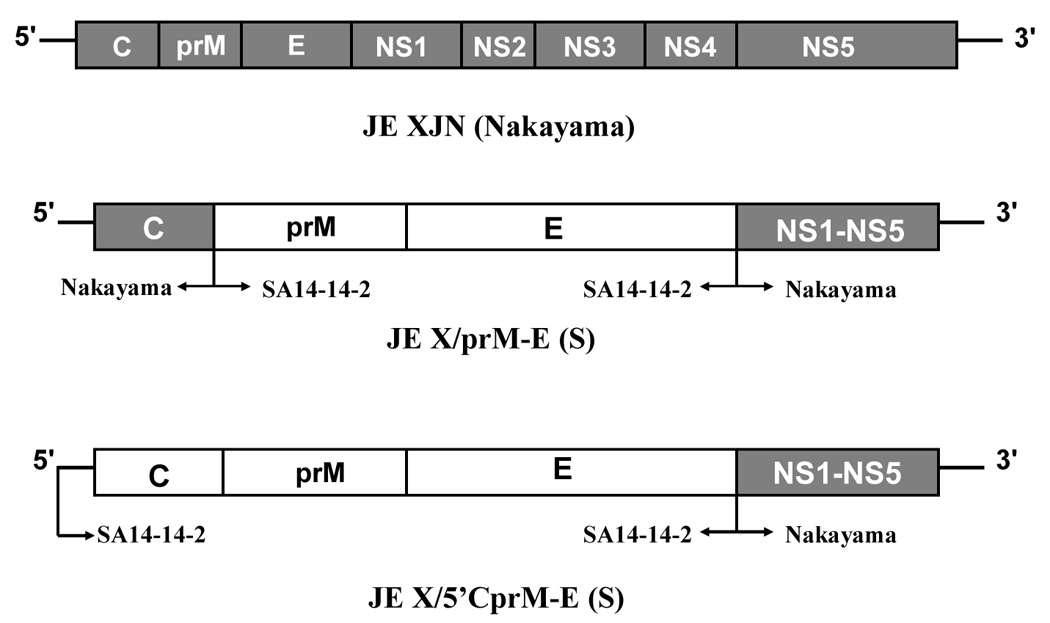
Structure of the JE XJN and intertypic viruses. At top is shown the parental JE XJN virus molecular clone. Below are engineered viruses containing JE SA14-14-2 structural proteins. Shaded regions depict the regions of the JE SA14-14-2 virus incorporated into the JE-XJN molecular clone, as also indicated by arrows below the diagrams. For clarity, the nonstructural regions of the chimeras are not drawn to scale.
Yields of infectious JE-XJN virus after transfection were in the range of 3 × 107 to 1 × 108 PFU/ml. The plaque size was identical to that of parental JE-Nakayama virus (approximately 3 to 4 mm after 4 to 5 days of incubation). Transfection harvests of the two intertypic viruses yielded concentrations on the order of 5.5 × 106 to 7.5 × 106 PFU/ml. The JE-X/prME(S) virus formed plaques on Vero cell monolayers that resembled those of the parental JE-XJN clone. The JE-X/5'CprME(S) virus formed plaques of 2–3 mm that were less distinct and required 1–2 days longer to form than those of JE-XJN virus.
Nucleotide sequence analysis
The nucleotide sequence of the JE-XJN virus was described previously, and is homologous to other virulent JE virus strains (Chambers et al., 2006). For the JE-X/5’CprME(S) and JE-X/prME(S) viruses, the nucleotide sequences from the 5’ terminus through E regions were determined for the plasmid templates used for generation of infectious RNA transcripts, and also from cDNA derived by RT/PCR of virus-infected Vero cell monolayers, using the same virus stocks employed for the cell culture and mouse experiments. Table 1 indicates the results. Comparison is made with the virulent JE-XJN virus, and the attenuated JE-SA14-14-2 virus (Nitayaphan et al., 1990). In the case of both intertypic viruses, a few unexpected differences were observed between the plasmid templates and the recovered viruses, resulting in some differences in their prM and E proteins, as described below.
Table 1.
Comparison of JE virus sequencesa
| Position | JE SA14-14-2b | JE-X/5’CprME(S) | JE-X/prME(S) | JE-XJN |
|---|---|---|---|---|
| 5’UTR-26 | g | u(g) | - | - |
| 5’UTR-39 | a | - | u | u |
| C40 | G | S(G) | - | - |
| C66 | S | - | L | L |
| C100 | R | - | G | G |
| prM14 | I | - | V(I) | V |
| prM16 | N | - | K(N) | K |
| prM129 | I | - | I, Vc(V) | V |
| prM140 | V | - | I, Vd (I) | I |
| E91 | S | - | - | V |
| E107 | F | - | - | L |
| E138 | K | - | - | E |
| E176 | V | - | - | I |
| E177 | A | - | - | T |
| E227 | S | - | - | P |
| E242 | F | - | - | E |
| E244 | G | - | - | E |
| E264 | H | - | - | G |
| E279 | M | - | - | K |
| E315 | V | - | A(V) | A |
| E345 | L | - | - | P |
| E439 | R | - | - | K |
Dashed lines indicate the same nucleotide or amino acid as in JE SA14-14-2.
Nitayaphan et al., 1990 substitutions in parentheses indicate nucleotides and amino acids observed in plasmid templates.
sequence data indicate mixed residues at these positions
JE-X/5’CprME(S) differed unexpectedly from JE-SA14-14-2 virus at 1 nucleotide in the 5’ UTR (u instead of g at position 26), and 1 amino acid in the capsid region (serine instead of glycine at position 40), both of which emerged after recovery of JE-X/5’CprME(S) virus from its plasmid template. JE-X/5’CprME(S) differed from JE-XJN virus at two nucleotide substitutions in the 5’ UTR (u instead of g at position 26 and a instead of u at position 39), as well as 3 amino acid substitutions in the capsid, 4 in the prM region, and 13 in the E protein. JE-X/5’CprME(S) differed from JE-X/prME(S) virus at the two nucleotide positions in the 5’ UTR mentioned above, at 3 amino acids in the capsid protein (residues 40, 66, and 100), at 2 amino acids in the prM protein (residues 14 and 16, where reversion of JE-X/prME(S) sequence to that of JE-XJN occurred), and at 1 amino acid in the E protein (residue 315). At two positions in the prM protein, the JE-X/prME(S) virus contained a mixture of residues found in both JE-X/5’CprME(S) and JE-XJN viruses.
JE-X/prME(S) differed from JE-XJN virus at the two 2 amino acid positions in the prM region where mixed residues were found, and at 12 amino acids in the E region.
The lack of complete similarity in the prM-E regions of the intertypic viruses was therefore due to clonal substitutions in the 5’ UTR and capsid protein of JE-X/5’CprME(S) virus and reversions in the prM and E proteins of JE-X/prME(S) virus (see Discussion).
Additional nucleotide sequencing of a 2500 base pair region from the carboxy-terminus through the 3' UTR was conducted to determine if the intertypic viruses harbored a high level of substitutions as a result of possible genetic incompatibilities between JE-SA14-14-2 and JE-XJN virus. Results are included in the Supplementary Material. Each virus contained only a few silent nucleotide substitutions and a single unique amino acid substitution.
Growth properties of the JE viruses
The growth kinetics of the JE-XJN virus and the parental JE Nakayama virus from which it was derived are compared in Figure 2A. The peak titer of JE Nakayama virus in LLC-MK2 cells occurred at 48 hours and was 7.25 log PFU/ml. The peak titer of JE-XJN virus in LLC-MK2 cells occurred at 60 hours and was 8.5 log PFU/ml. In C6/36 cells, JE Nakayama virus was slightly more rapid in initial virus production over the initial 36 hours, but reached a peak titer at 60 hours which was similar to that of JE XJN, approximately 7.5 logs.
Figure 2.
Growth curve analysis of the JE XJN, and intertypic JE viruses. Infections were done as described in the Methods. Panels A and B shows results for parental JE Nakayama (open symbols) and JE XJN (closed symbols) viruses on LLC-MK2 and C6/36 cells, respectively. Panels C and D shows results for JE Nakayama (open squares), JE-X/prME(S) (closed circles) and JE-X/5’CprME(S) (open diamonds) viruses on LLC-MK2 and C6/36 cells, respectively. Values represent the mean +/− standard deviation (SD) for three independent samples harvested and tested for each time point. In most cases, SDs were small and error bars are not visible.
Virus production for the two intertypic viruses was compared to that of JE Nakayama virus in LLC-MK2 and C6/36 cells (Figures 2C and D). JE-Nakayama was used in these experiments because no apparent differences in growth properties were found between it and JE-XJN in these cell lines. In LLC-MK2 cells, JE Nakayama virus reached a peak titer of approximately 7.0 logs. The JE-X/prME(S) virus exhibited very similar kinetics of virus production with the same peak titer. In contrast, the JE-X/5’CprME(S) virus exhibited reduced virus production at early time points, but eventually reached a peak titer similar to those of the other viruses. In C6/36 cells, JE Nakayama virus reached a peak titer of approximately 9.0 logs. The JE-X/prME(S) virus initially displayed a similar rate of virus production, but reached a titer of only 8.0 logs at the conclusion of the experiment. JE-X/5’CprME(S) virus was reduced in initial rate of virus production, and eventually reached a peak titer of only 6.75 logs.
Mouse virulence testing
The virulence properties of the parental and the intertypic JE viruses were evaluated by intracerebral (i.c.) and intraperitoneal (i.p.) inoculation of weaned (3–4 week old) ICR mice. In an initial pilot experiment to evaluate the neurovirulence of the JE Nakayama and JE-XJN viruses, 4 week old mice were inoculated with a dose of 4.0 logs PFU of either virus by the i.c. route. 100% mortality was observed for these two viruses and average survival times were similar (9 days for each virus). In a separate preliminary experiment to evaluate the neurovirulence of the intertypic viruses, i.c. inoculation of 4 week old ICR mice with fixed doses was also tested. At a dose of 6.1 logs, 12 of 12 mice inoculated with the JE-X/prME(S) virus succumbed to infection. In contrast, at a dose of 5.6 logs only 7/12 mice inoculated with the JE-X/5’CprME(S) succumbed to infection. Average survival times for these groups of mice were 5.6 and 7.6 days, respectively (data not shown).
Further evaluation of the neurovirulence properties of the JE-XJN and intertypic viruses was done by dose titration experiments using the same mouse model as for the pilot experiments. Table 2 shows the results of these experiments. The JE-XJN virus exhibited less than 100% mortality when the dose was lowered below 1.0 log, and the LD50 was 0.50 PFU. JE-X/prME(S) virus was tested in three independent experiments (shown in Table 2). In one experiment, the calculated LD50 was 0.20 PFU. In 2 of the 3 experiments, less than 100% mortality at the highest doses tested did not allow calculation of the LD50. In contrast to both JE-XJN and JE X/prME(S) viruses, the JE-X/5’CprME(S) virus exhibited less neurovirulence, based on less than 100% mortality at or below a dose of 4 log. The LD50 was 1.40 PFU. At two of four comparable doses where JE-X/5'CprME(S) and JE-X/prME(S) were tested the same LD50 experiment, the mortality percentages were significantly for JE-X/prME(S) (p=.023 at 1.8 vs. 2.3 log PFU, [JE-X/5'CprME(S) vs. JE-X/prME(S), respectively]; p=.023 at 5 vs. 4 log PFU, [JE-X/5'CprME(S) vs. JE-X/prME(S), respectively]). Average survival times in these experiments ranged from 5 to 12 days among the different doses tested, with longer survival times observed at the lower doses.
Table 2.
Neurovirulence Testinga
| VIRUS | DOSE RANGEb | LD50 | Average Survival Times (days)c |
|---|---|---|---|
| JE-XJN | 0.3–1.9 | 0.50 | 5.4–8.6 |
| JE-X/prME(S)d | 2.3–5.4 | NA | 5.9–6.4 |
| JE-X/prME(S)d | 0.3–3.5 | < 0.3 | 5.2–6.0 |
| JE-X/prME(S)d | 0.1–3.6 | 0.20 | 6.0–9.5 |
| JE-X/5’CprME(S) | 0.8–5 | 1.40 | 7.0–12 |
intracerebral inoculation; groups of 5–7 mice per dose
log PFU/dose
Range of average survival times among different dose groups
Three independent experiments with JE-X/prME(S) virus were performed
In initial experiments to evaluate the neuroinvasiveness of the JE Nakayama and JE-XJN viruses, 3 week old mice were inoculated with a dose of 4.7 logs PFU by the intraperitoneal route (Table 3). Mortality percentages were similar for the two viruses (60% for JE-XJN and 50% for JE Nakayama virus, respectively). Average survival times were 5 days in both cases. Neuroinvasiveness of the two intertypic viruses was initially evaluated by inoculation of 3 week old mice with doses of approximately 6 logs PFU by the i.p. route. The mortality for mice inoculated with JE-X/prME(S) was 70%, with an average survival time of 6.3 days. In contrast, the mortality rate for mice inoculated with the JE-X/5’CprME(S) was 0%, and no mice exhibited signs of illness.
Table 3.
Neuroinvasiveness of JE virusesa
| VIRUS | DOSEb | MORTALITY | % |
|---|---|---|---|
| Experiment 1 | |||
| JE-XJN | 4.7 | 6/10 | 60 |
| JE-Nakayama | 4.7 | 5/10 | 50 |
| Experiment 2c | |||
| JE-X/prME(S) | 6.0 | 7/10 | 70 |
| JE-X/5’CprME(S) | 5.6 | 0/10 | 0 |
| Experiment 3 | |||
| JE-XJN | 4.4 | 2/7 | 28.6 |
| 4.3 | 3/6 | 50 | |
| 3.2 | 2/5 | 40 | |
| 2.5 | 2/7 | 28.6 | |
| JE-Nakayama | 4.5 | 2/5 | 40 |
| 3.7 | 4/6 | 66 | |
| 2.7 | 4/8 | 50 | |
| JE-X/prME(S) | 5.5 | 2/7 | 28.6 |
| 4.6 | 3/7 | 42.8 |
intraperitoneal inoculation of 4 week old ICR mice
log PFU/dose
p = .003 for difference in mortality percentage
Since the JE-X/prME(S) virus exhibited neuroinvasiveness, further studies were done to characterize this property. Dose titration studies were conducted using the parental JE-XJN virus and JE Nakayama viruses as controls (Table 3). At doses in the range of 2.5 to 4.4 log PFU, all mice inoculated with JE-XJN had similar levels of mortality ranging from 28 to 50%. The JE Nakayama virus exhibited generally similar mortality percentages over the same dose range (40–66%). In the same experiment the JE-X/prME(S) virus also exhibited partial neuroinvasiveness at doses of 4.6 and 5.5 log PFU, with mortality rates of 28 to 42.8%, respectively. LD50 values could not be calculated for any group due to incomplete mortality even at the highest doses tested.
Immunogenicity of intertypic viruses
To determine what levels of neutralizing antibodies against JE-Nakayama virus were produced after immunization with the intertypic viruses, sera obtained from mice which had been inoculated by the i.p. route with either JE-X/prM-E(S) or JE-X/5’CprME(S) from Experiment #2 shown in Table 3. (Experiment 2). Table 4 shows the results of plaque-reduction titers using 50% endpoints. Both viruses elicited neutralizing antibodies, with titers ranging from 320–2560 (GMT of 910) for JE-X/5’CprME(S), and a titer of 1280 (GMT of 1280) for JE-X/prME(S), respectively. Only the three mice which survived inoculation with JE-X/prME(S) virus were tested for neutralizing antibodies in this experiment. There was no significant difference in the GMTs of the samples. A true comparison of the maximal antibody responses generated by these two viruses is not available, as only a single time point was measured in these experiments.
Table 4.
Plaque-reduction neutralization titersa
| SAMPLE | DAYb | N | GMT | RANGE |
|---|---|---|---|---|
| Immunization | ||||
| JE-X/5'CprME(S) | 38 | 6 | 910c | 320–2560 |
| JE-X/prME(S) | 38 | 3 | 1280c | 1280 |
| Immunization and Challenge | ||||
| Mock-immunized/Challenged | 5 | 4 | 80d | 40–160 |
| Immunized/Challenged | 5 | 3 | 3175d, e | 2500–5120 |
| Immunized/Challenged-4 | 21 | 5 | 6756e | 2560–10240 |
50% plaque reduction titers, as reciprocal of serum dilution
day serum obtained post-immunization or post-challenge
p = .45
p < .001
p = .11
Protective immunity induced by JE-X/5’CprME(S) virus
Because the JE-X/5’CprME(S) virus was attenuated for neuroinvasiveness and induced neutralizing antibodies against JE-Nakayama virus, it was evaluated for the capacity to generate protective immunity against challenge with this virus. C57BL/6 mice were mock-immunized or immunized at 5 weeks of age with 6.1 log PFU of JE-X/5’CprME(S) virus, and then challenged at 14 weeks of age with 20,000 PFU of JE-XJN virus by the i.c route. Figure 3 shows that all mock-immunized mice succumbed to infection with typical signs of acute encephalitis, and the average survival time was 5.9 days (range 5 to 8 days). In contrast, only 2/12 mice immunized with JE-X/5’CprME(S) virus developed a lethal infection and survived for 8 and 10 days, respectively. The difference in the mortality percentages and average survival times were significant between the mock-immunized and immunized groups (p < .001 and p = .02, respectively).
Figure 3.
Protection of C57BL/6 mice against JE virus encephalitis after immunization with the JE-X/5’CprME(S) virus. Mice were mock-immunized or immunized with JE-X/5’CprME(S) virus and challenged with JE Nakayama virus by the i.c. route and observed until moribund or sacrificed as described in the text. Statistical comparisons for mortality rates and average survival time were significant, as described in the text.
Brains of some of the mock-immunized and immunized mice were recovered on post-challenge day 5 to determine if there were differences in the amounts of brain-associated virus. Brains of 4 mock-immunized mice contained a mean of 10.44 (+/− 1.2 logs (PFU/gram) of brain tissue at a time when these mice appeared moribund. Brains of 3 mice immunized with JE-X/5’CprME(S) virus contained a mean of 7.15 +/− 1.2 logs (PFU/gram), and these mice exhibited no signs of illness at this time. These latter mice were not used for calculations of the mortality percentages.
To evaluate the magnitude of change in circulating neutralizing antibody titers associated with protection from encephalitis after the immunization and challenge, postchallenge titers were measured in both surviving immunized mice and moribund mock-immunized mice harvested on day 5 of the experiment (Table 4). Mock-immunized mice which succumbed by day 5 post-challenge exhibited titers in the range of 40–160 (GMT of 80). Among the three immunized mice sacrificed at day 5, the titer at this time was already 37-fold higher (GMT of 3175). Among mice which survived challenge, the level had increased to a GMT of 6756 by day 21 post-challenge. Although a control of immunized and nonchallenged mice was not tested, the large increase in titers of immunized mice indicates a substantial anamnestic response occurs in those immunized mice which survived challenge.
DISCUSSION
In this study, a JE-Nakayama molecular clone, referred to as JE-XJN, was used as the basis for constructing two intertypic JE viruses, JE-X/5’CprME(S) and JE-X/prME(S). These expressed either the 5’ UTR, capsid, prM and E regions, or only the prM and E regions respectively, of the attenuated JE SA14-14-2 virus, in the JE Nakayama background. The intertypic viruses exhibited cell culture properties that were generally similar to those of the parental JE viruses, including virus production in cell lines of mammalian and mosquito origin, and plaque formation on Vero cells. Overall, the results indicate no severe defect in virus replication resulting from replacement of the prM and E proteins of JE Nakayama virus with those of JE SA14-14-2. However, inclusion of the 5’ UTR and capsid region from the JE SA-14-14-2 virus did cause some deleterious effects on virus production in cell culture.
JE Nakayama virus is a virulent strain of JE virus, characterized by high neuroinvasiveness and high neurovirulence for mice (Cao et al., 1995; Ni and Barrett, 1996; Lee and Lobigs, 2002). As demonstrated in weaned mice, the JE-XJN virus and its parental JE Nakayama virus exhibited neuroinvasive and neurovirulent phenotypes. However, uniform neuroinvasiveness was not observed even at the highest doses tested (Table 2). This likely results from these viruses being maintained as cell culture-passaged virus stocks rather than as suckling mouse brain suspension, which retains higher neuroinvasiveness for mice of this age (Lee and Lobigs, 2002).
JE SA-14-14-2 is a highly attenuated strain of JE virus, characterized by lack of both neuroinvasiveness and intracerebral neurovirulence in weanling mice (Eckels et al., 1988). Introduction of the prM and E proteins of the JE SA14-14-2 virus into the JE Nakayama background was predicted to result in attenuation of the virulence properties of the latter virus. This expectation is based on the fact that the E protein harbors critical determinants of virulence for JE virus, as shown for several different strains of JE virus (Sumiyoshi et al., 1995; Cecelia and Gould, 1991; Hasegawa et al., 1992, Ni and Barrett, 1998). Virulence testing of the JE-X/prME(S) virus in 3–4 week old mice revealed it to be neurovirulent and neuroinvasive, with overall properties generally similar to those of the JE-XJN virus. In contrast, the JE-X/5’CprME(S) virus was less virulent than JE-XJN and JE-X/prME(S), with no evidence of neuroinvasiveness or signs of illness among mice inoculated intraperitoneally with doses as high as 5.6 logs PFU, and incomplete mortality at doses as high as 4 log PFU given intracerebrally. Although we did not test the JE SA14-14-2 virus in these experiments, comparison of the JE-X/prME(S) and JE-X/5’CprME(S) viruses for mortality percentages revealed differences in both neurovirulence (Table 2) and neuroinvasiveness (Table 3), indicating a relative attenuation phenotype of the JE-X/5’CprME(S) virus. However, based on available data for JE SA14-14-2 (Eckels et al., 1988), JE-X/5’CprME(S) virus is only partially attenuated with respect to neurovirulence. We therefore conclude that the prM and E proteins of the JE-SA14-14-2 virus are not capable of fully attenuating the virulence properties of the JE Nakayama virus. Rather, the 5’ UTR and capsid protein regions of JE-SA14-14-2 virus are required to attenuate neuroinvasiveness, but only partially attenuate neurovirulence. These observations do not mean that the E protein of the JE SA14-14-2 virus lacks attenuating determinants for the virulent JE-SA14 virus from which it was derived. In this regard, JE SA14 virus is quantitatively less virulent in the mouse model than JE Nakayama virus (Cao et al., 1995; Ni and Barrett, 1996), so attenuating determinants within the JE-SA14-14-2 virus E protein alone may be sufficient to eliminate neuroinvasiveness and reduce neurovirulence of the JE SA14 strain.
In other experimental systems where chimeric flaviviruses have been used to test effects of structural proteins from attenuated viruses, the prM and E proteins have often been observed to confer strong attenuating effects. For instance, the prM and E proteins of the highly attenuated dengue-2 strain were sufficient to abolish the neuroinvasiveness and lower the neurovirulence of JE Nakayama virus (Chambers et al., 2006). Similar results were observed with a corresponding JE/dengue-4 chimeric virus (Mathenge et al., 2004). In the context of a chimeric yellow fever/JE virus, the prM and E proteins of JE SA14-14-2 profoundly attenuated the neurovirulence of the yellow fever 17D strain, even at doses as high as 106 PFU delivered intracerebrally (Chambers et al., 1999). In the case of the JE-X/prME(S) virus, the prM and E proteins of JE-SA14-14-2 virus may not be able to overcome an intrinsically high level of virulence contributed by the remainder of JE Nakayama virus genome.
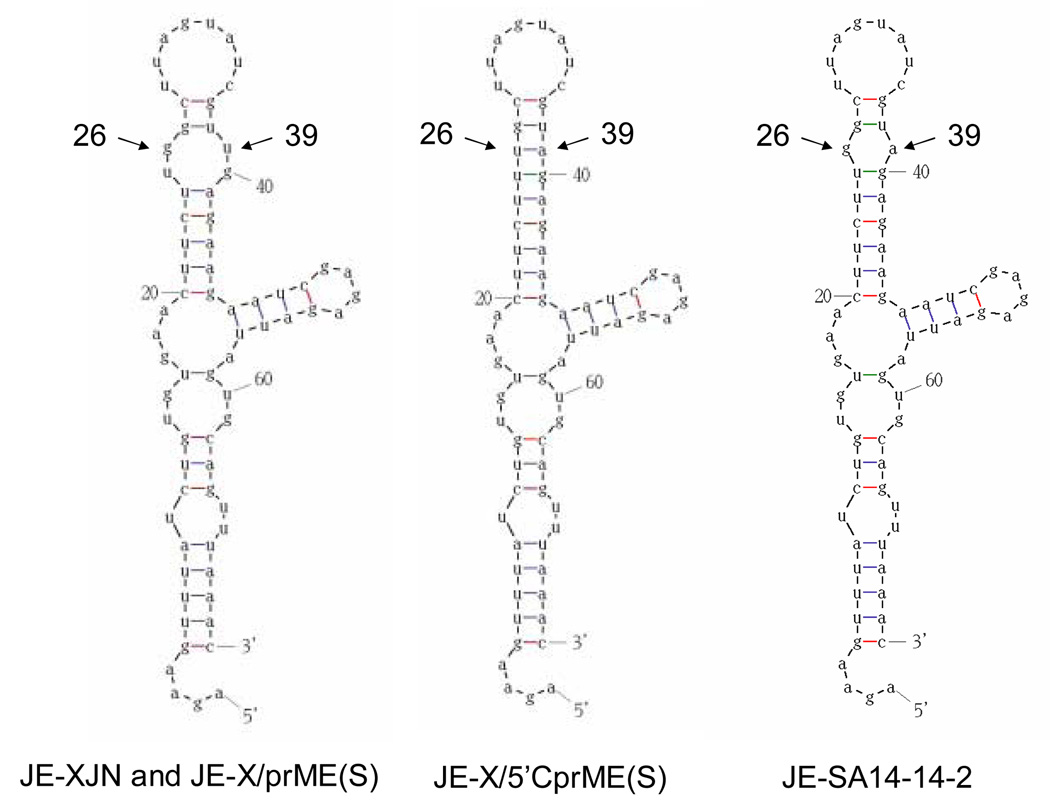
Genetic differences between the JE-X/5’CprME(S) and JE-X/prME(S) viruses in the region from the 5’ UTR through the E protein included multiple nucleotide and amino acid substitutions that presumably contribute to differences in their virulence properties. For the 5’ UTR, insight into the possible significance of the differences was sought by generating secondary structure models for the two viruses (Figure 4). The structures formed for the parental JE-XJN and the two intertypic viruses resemble the most stable predictions for the 5’ UTR of flaviviruses, consistent with those modelled for JE, dengue and other viruses (Brinton and Dispoto 1988; Cauhour et al., 1995; Thurner et al, 2004). For JE-XJN and JE-X/prME(S), nucleotides 26 and 39 are contained within a small bulge at the end of the long stem formed by nucleotides 20 to 45. In contrast, the 5’ UTR of JE-X/5’CprME(S) lacks this bulge because the uridine substitution at position 26 compensates for the adenine at position 39, to create a longer stem structure. The JE-SA14-14-2 virus contains an adenine nucleotide at position 39, but a guanosine at position 26, resulting in a bulge slightly smaller than that observed for the JE-XJN virus, indicating that this feature is not specific to the virulent JE strains. The lack of any major effect of the substitutions in the 5’ UTRs on predicted RNA structure, and the presence of similar structures for JE-SA14-14-2 and JE-XJN viruses suggests that these substitutions may not affect the functional properties of the 5’ UTR. Ni et al have analyzed the 5’ UTR sequences of multiple JE viruses in the lineage of the JE SA14-14-2 vaccine strain from its JE SA14 precursor (Ni et al., 1994), and concluded that the adenine substitution at position 39 is not a marker for JE virus virulence. Furthermore, positions 26 and 39 lie within a region of the 5’ UTR structure that has been found non-essential for viral replication or mosquito competence in the context of dengue 4 virus (Cauhour et al., 1995).
Figure 4.
Modelled RNA structures for the 5’ UTR of the parental JE-XJN and JE-X/prME(S) viruses (left) and the JE-X/5’CprME(S) virus (middle), and JE-SA14-14-2 virus (right). Positions of the nucleotide substitutions 29 and 36 are indicated. Modelling procedures were as described in the Methods.
Amino acid substitutions were observed between JE-X/prME(S) and JE-X/5’CprME(S) viruses in the capsid, prM and E protein regions. One or more of these substitutions could affect steps in polyprotein processing, RNA synthesis, and/or virus assembly, and contribute to the moderate defect in virus production in LLC-MK2 cells observed with the JE-X/5’CprME(S) virus. A corresponding or even worse degree impairment of replication efficiency of this virus in mouse tissues could explain its lack of neuroinvasiveness, based on a low virus burden being generated in peripheral tissues prior to induction of an immune response capable of controlling the infection. The lower neurovirulence of JE-X/5’CprME(S) virus, as reflected by an LD50 7.5 fold higher than that of the JE-X/prME(S) virus after intracerebral inoculation, is consistent with this hypothesis, as rate of virus production in the central nervous system is a marker for virulence of flaviviruses (Schlesinger et al., 1996).
The three amino acid substitutions in the capsid protein where JE-X/5’CprME(S) differed from JE-X/prME(S) virus do not occur in regions of conserved RNA sequences (Markhoff, 2003), and therefore effects on RNA structure were not modelled. To gain insight into possible effects of the substitutions at positions 40 and 66, a homology model for the JE virus capsid was derived based on the structure for the dengue 2 virus (Ma et al., 2004). Relative positions of the substitutions found in the JE-X/5’CprME(S) capsid are shown in Figure 5. Modelling of the predicted local effects of the substitutions at positions 40 and 66 of the JE virus capsid is discussed in Supplementary Information. Position 40 lies within the loop connecting the alpha 1 and alpha 2 segments, whereas position 66 lies within the helical alpha 3 segment. Based on the solution structure for dengue-2 virus (Ma et al., 2004), both of these substitutions lie within regions that are involved in stabilization of the capsid monomer core, and in formation of the hydrophobic cleft that serves to associate the capsid with intracellular membranes. In the case of the JE-X/5’CprME(S), the two serine substitutions may result in deleterious effects on the function of the capsid protein dimer due to loss of hydrophobic character in this region of the protein. In this regard, studies with TBE virus have shown that deletions encompassing this region of the capsid protein reduce the virulence of TBEV for mice, probably through destabilization of the capsid dimers and their interaction with membrane surfaces (Kofler et al., 2002; 2003). The possible effects of the substitution at position 100 were not analyzed, since this position does not lie within the model.
Figure 5.
Homology model for the JE Nakayama capsid protein, based on the dengue-2 structure. Modelling procedures were as described in the Methods. Capsid monomers are shown in green and magenta, respectively. Positions of residues 40 and 66 are indicated.
Two amino acid substitutions in the pr region (positions 14, 16) differentiated the JE-X/5’CprME(S) and JE-X/prME(S) viruses. These were unexpected reversions to residues found in the parental JE-XJN virus. Selective pressure for reversion also occurred at positions 129 and 140 in the prM region of JE-X/prME(S) virus, where mixtures of valine and isoleucine were observed. These partial and complete reversions were presumably selected during transfection and amplification of JE-X/prME(S) virus on Vero cells, and may reflect a requirement for replicative fitness of the virus in the context of the intertypic capsid/prM-E region. None of these positions in the prM region have been classified as virulence determinants for JE viruses (Nitayaphan et al., 1991; Aihara et al., 1991; Ni et al., 1995).
With regard to attenuating determinants in the JE virus E protein, JE SA14-14-2 contains multiple substitutions compared to its parental JE SA14 strain (Nitayaphan et al., 1991). However, comparison of various attenuated derivatives of JE SA14 viruses, including JE SA14-14-2, JE SA14-5-2 and JE SA14-2-8, has revealed a consensus of four common amino acid substitutions (E-138, E-176, E-315, and E-439) that are associated with attenuation (Nitayaphan et al., 1991; Aihara et al., 1991; Ni et al., 1995). All four substitutions were found in the E protein of JE-X/5’CprME(S) and all but position 315 were found in the E protein of JE-X/prME(S) virus. Eight additional substitutions specific to the JE SA14-14-2 virus (positions E-91, E-107, E-177, E-227, E-244, E-264, E-279, and E-345) were also found in both intertypic viruses. Of particular note, substitution of lysine for glutamate at position 138, which was reported as a strongly attenuating determinant for the JE JaOArS982 strain (Sumiyoshi et al., 1995; Zhao et al., 2005), was not sufficient to attenuate the JE-XJN virus in our experiments. Potential virulence determinants in the nonstructural region and/or the 3’ UTR of JE-XJN virus might negate or counteract effects of position 138 or other attenuating sites in the E protein and account for the lack of attenuation of JE-X/prME(S) virus.
One amino acid substitution in the E protein (position 315) differentiated the JE-X/5’CprME(S) and JE-X/prME(S) viruses, where reversion to the residue found in JE-XJN virus was unexpectedly found in the latter virus. Although the alanine substitution is found among virulent strains of JE virus (Ni et al., 1995), there is some experimental evidence that it does not exert any dominant effect on virulence properties associated with the JE virus E protein (Arroyo et al., 2001).
Because of the attenuated properties of JE-X/5’CprME(S) virus, experiments were done to evaluate its potential as an experimental vaccine. Neutralizing antibody responses elicited against JE-Nakayama virus after peripheral inoculation of young mice were comparable to those induced by the neuroinvasive JE-X/prME(S) virus. Weanling mice are highly susceptible to intracerebral inoculation with the JE Nakayama virus, and challenge with this strain serves as a severe test of a protective immune responses. Using this model, 100% mortality was found in mock-immunized mice, whereas immunization with JE-X/5’CprME(S) virus was able to protect 83% percent of mice, indicating a high degree of efficacy as a prophylactic vaccine. The post-challenge antibody responses in immunized mice were 6 to 20 fold higher than those of mock-immunized mice by day 5 post-challenge and continued to rise another 2 to 3 fold by 3 weeks post-challenge. This anamnestic response was associated with a reduction of brain-associated JE Nakayama challenge virus as early as 5 days post challenge, compared to levels found in mock-immunized mice. Lack of complete protection of immunized mice against challenge with JE Nakayama virus is presumably related to variability in the levels of neutralizing antibody produced in response to immunization with JE-X/5’CprME(S) virus (Table 4). For instance, the magnitude of the anamnestic response has been identified as a critical factor in the control of JE virus encephalitis in the mouse model (Konishi et al 1999).
JE virus causes an acute encephalitis in humans that is preventable using vaccination with live-attenuated JE virus (JE-SA14-14-2, [Chinese vaccine strain, (Eckels et al., 1988), or with inactivated vaccine (JE-VAX), (Tsai, 1994). The attenuation phenotype of the live-attenuated JE-SA14-14-2 vaccine is presumed to involve numerous mutations which occurred during passage in PDK (primary dog kidney cells), and which differentiate it from its JE SA-14 parent and other virulent JE virus strains (Nitayaphan et al., 1990; Ni et al., 1994, 1995). However, the molecular basis of attenuation of JE SA-14 virus has not been fully defined. The results of our study suggest that the 5’ UTR and capsid regions of the JE SA14-14-2 virus may contain attenuating mutations, but their importance may depend on the genetic background of the JE virus strain. Mutations in the prM and E proteins may be sufficient for attenuation of JE SA14 virus, although further studies are required to confirm this hypothesis. Such studies may yield information that can be used for engineering of novel live-attenuated viral vaccines for JE virus.
MATERIALS AND METHODS
Cells and viruses
Vero and C6/36 cells were originally obtained from the American Type Culture Collection (ATCC) and maintained in alpha minimal essential media (alpha-MEM) plus 10% fetal bovine serum. LLC-MK2 cells (ATCC) were maintained in M-199 media plus 20% fetal bovine serum (FBS). The JE SA-14-14-2 virus was previously described (Chambers et al., 1999). The JE Nakayama virus clone (JE-XJN) was described previously (Chambers et al., 2006). Plaque assays for JE viruses were done in confluent Vero cells, using an overlay of 1% ME agarose (Biowhittaker), in alpha MEM plus 5% FBS and incubated at 37°C for 4 to 5 days. Plaques were visualized by staining with .05% neutral red in phosphate-buffered saline, followed by fixation in 7% formalin and staining with 1% crystal violet.
Construction of intertypic JE viruses
The JE-XJN molecular clone utilizes two plasmids (pJE5'NB25 and pCR-XL3'BK2), encoding the 5’ 3.4 kb and the 3’ 7.5 kb of the JE Nakayama virus genome, to generate infectious JE virus (Chambers et al., 2006). The plasmid pJE5'NB25 was therefore used for construction of intertypic JE viruses containing the 5’ UTR, and/or the C, prM and E proteins of JE SA14-14-2 virus in the JE-XJN background.
JE-SA14-14-2 virus RNA was obtained from virus-infected LLC-MK2 cells using Trizol LE reagent (GIBCO/BRL). The final RNA pellet was dissolved in RNAse-free water. The primers for the reverse transcription and long polymerase chain reaction amplification were as follows (5’ to 3’). Sense primers are indicated by (+), and antisense by (−).
JE5'(+): 5’-AGAAGTTTATCTGTGTGAACTTCTT-3’
JE(E/NS1)(−): 5’-CACATCCAGTGTCAGCATGCACATTG-3’
These primers were based on the JE-SA14 sequence (Nitayaphan et al., 1990). cDNA synthesis was done using Superscript II RNAseH(−) reverse transcriptase (Gibco/BRL). 1–2 micrograms of JE virus RNA was mixed with 10 picomoles of JE(E/NS1) primer, heated for 10 min at 70°C, cooled on ice, and then incubated with Superscript II in the appropriate buffer for 2–4 hours at 42°C. Reactions were then treated with DNAse-free ribonuclease H at 37°C for 20 min., followed by heating to 70°C for 10 min, and then extracted with phenol/chloroform. The cDNA products were recovered by ethanol precipitation and dissolved in water for PCR amplification.
PCR amplification was done from JE SA14-14-2 cDNA using the Expand High Fidelity PCR System (Roche). Reaction mixtures contained 10 picomoles each of sense and antisense primers and were run for 30 cycles with the following program after an intitial step of 94°C for 2 minutes: 94°C for 15 seconds, 60°C for 30 seconds, 68°C for 4 minutes. The final elongation step was at 72°C for 10 minutes. The PCR product was isolated by agarose gels electrophoresis, visualized by crystal violet staining, excised and recovered by addition of 3 volumes of 6M sodium iodide and incubation at 70°C and purified with Wizard PCR Preps DNA Purification System (Promega).
The plasmid pJE5’NK/5’CprME(S) was obtained by replacing the sequence between the Not I site immediately upstream of the SP6 promoter sequence and the Mun I site (nt 2413 of the JEV genome) of pJE5’NB25 with a corresponding fragment generated from the JE SA14-14-2 PCR product described above. The PCR fragment was digested with Not I and Mun I and ligated into pJE5'NB25 to produce pJE5’NK/5’CprME(S). The sequence of the resulting plasmid was analyzed from the 5’ terminus through the E-NS1 junction.
The plasmid pJE5’NK/prME(S) was generated using a PCR ligation strategy to exchange the fragment between the 5’ terminus and the C/prM junction in pJE5’NK/5’CprME(S) with sequence from JE Nakayama virus. Based on comparison of the nucleotide sequences of the JE SA14-14-2 and JE Nakayama viruses, there are no amino acid differences immediately upstream of the C/prM junction. An overlapping DNA sequence of 10 base pairs was used for the ligation of two PCR fragments generated from JE Nakayama and JE SA14-14-2 viruses. The JE Nakayama fragment from the 5’ terminus through the 3’ terminus of the capsid region was amplified from pJE5’NB25 using the primer JE5’-Sp6 and a second primer JE (C-): 5’-GAAATTTGACAACTTCATGGCTCCTGCGCAG-3’. The second PCR fragment from the 3’ terminus of the JE SA14-14-2 capsid region through the Nhe I site (nt 1124) near the 5’ terminus of the E protein was amplified from the JE SA14-14-2 PCR product described above using the primers JE prM(+): 5’-CGTCCTCGGTACTTCAACA-3’ and the primer JE 1148-1125(−): 5’-TTCTGACCTCAGCAAGTTGGCTAG-3’. The PCR products were purified and quantitated. 100 nanograms of each PCR product were mixed and diluted 1:500 and 1.0 microliter was used in a 50 microliter PCR reaction in the absence of primers. The program was run as follows: 94°C for 1 minute, 60°C for 1 minute, and 72°C for 5 minutes, followed by addition of 10 picomoles each of primers JE5’Sp6 and JE 1125(−), and the reaction was continued for 25 cycles. The amplified PCR product was purified and the sequence confirmed. The product was digested with Not I and Nhe I and used to replace the corresponding fragment in pJE5’NK/5’CprME(S) to yield pJE5’NK/prME(S). The final clone was analyzed by nucleotide sequencing.
RNA transcription and transfection
Full-length cDNA templates for recovery of infectious intertypic JE-XJN/SA14-14-2 viruses were assembled using a two-plasmid system analogous to that used for the production of JE-Nakayama virus from the JE-XJN clone (Chambers et al., 2006). Full-length transcription templates were made by ligating the 3.4 kb 5’ fragments from either pJE5’NK/5’CprME(S) or pJE5’NK/prME(S) after digestion with NotI and BspEI, together with the 7.4 kb 3’ fragment from pCR-XL3'BK2 after digestion with KpnI and BspEI. Digested fragments were isolated on crystal violet agarose gels, purified and quantitated by comparison to DNA molecular weight standards by ethidium bromide staining. Approximately 3.0 micrograms of the 3.4 kb NotI/BspEI fragment and 12.0 micrograms of the 7.6 kb BspEI/KpnI fragment were added to ligation reactions containing 400 units of T4 DNA ligase (NEB) for 12 hours at 16°C. The ligation reactions were then heat inactivated, extracted with phenol/chloroform and precipitated with ethanol. The final pellet was dissolved in TE (10 mM Tris-HCl pH 7.5, 1 mm EDTA), and the presence of full-length ligation product was verified by agarose gel electrophoresis. In vitro-ligated templates were transcribed using SP6 polymerase (NEB), essentially as described previously (Rice et al., 1989), Transfection of confluent BHK cells was done using a mixture of ¼ of the in vitro-transcribed RNA, and 20 micrograms of Lipofectin Gibco/BRL) and phosphate-buffered saline in a volume of 300 microliters for 15 minutes at 37°C, followed by washing the cells with alpha-MEM plus 3% FBS. Cells were then incubated at 37°C in the same media. Media were harvested at onset of cytopathic effects, between 4 and 5 days posttransfection, and infectious virus was titrated by plaque assay on Vero cells. Viruses were amplified once in Vero cells to produce working virus stocks.
Virus growth curves
Analysis of virus production in cell culture was done in confluent monolayers of LLC-MK2 or C6/36 cells, using low multiplicity infections for each virus tested (0.05 PFU/cell). After initial infection for 1 hour at 37°C (LLC-MK2) or 28°C (C6/36 cells), media were replaced and cells were incubated at 37°C (LLC-MK2 cells) or 28°C (C6/36 cells). Media were harvested at approximately 12 or 24 hour intervals followed by replacement with fresh media (alpha MEM plus 5% FBS). Virus yields were quantitated by plaque titration on Vero cells as described above.
Nucleotide sequence analyses
Plasmids encoding the intertypic JE Nakayama/JE SA14-14-2 viruses were sequenced using Applied Biosystems Big Dye sequence reactions and analyzed on an ABI sequencer. Viruses were also sequenced by recovering RNA from virus-infected cells and sequencing PCR products generated by RT/PCR of the region from the 5’ terminus through the E/NS1 junction.
Molecular modelling
RNA structures were generated with RNA mfold, version 3.2 (Zuker, 2003; Mathews et al., 1999). Folding was determined for nucleotides 1–71 at 37°C over a range of free energies resulting from setting a suboptimality value of 30%, to define the extent of variability of predicted structures for the 5’ UTRs of the JE-XJN, JE-X/5’CprME(S) and JE SA14-14-2 viruses.
A homology model for the JEV capsid protein based on the NMR structure of the dimeric capsid protein of dengue-2 (PDB ID code 1R6R) (Ma et al., 2004), was generated by the automated comparative protein modeling server Swiss-Model using the first approach mode (Peitsch, 1995; Guex and Peitsch, 1997; Schwede et al., 2003). Sequence alignment was determined by Swiss-Model, which aligned amino acids 26 to 100 of the dengue-2, JE Nakayama and JE SA14-14-2 capsid proteins. Rotamers for amino acids at positions 40 and 66 were analyzed for the most favorable energy configuration using calculation provided by the modelling tools. Hydrogen bonds were determined by the Swiss-Pdv viewer.
Plaque reduction neutralization testing
Plaque reduction neutralization assay for JE virus was done as described previously (Chambers et al., 2006). JE hyperimmune asctic fluid and control nonimmune ascitic fluids were obtained from the ATCC. Approximately 100 PFU of input JE Nakayama virus were mixed with serial 2-fold dilutions of immune ascitic fluid, nonimmune control ascitic fluid, or test sera in a standard experiment. The reaction mixture was kept at 4°C for 6 hours, followed by plaque assay on Vero cells using conditions described above.
Mouse experiments
ICR mice were purchased from Harlan Sprague Dawley (Indianapolis, IN), and handled in accordance with a protocol approved by the institutional committee on the care and use of laboratory animals. Mice were used for immunization at 3–5 weeks of age, and for neurovirulence testing at 4 weeks of age. Immunized mice were challenged at 8 weeks post-immunization, by inoculation with JE Nakayama virus. Mice were observed for four weeks, or until moribund, and then euthanized, at which time sera were collected for measurement of neutralizing antibody titers. LD50 values were determined by the method of Reed and Muench (Reed and Muench, 1938).
Statistical analyses
Differences in mortality endpoints were analyzed using Fishers exact test. Differences in average survival times were measured using log rank testing. Differences in neutralizing antibody titers were determined with independent means t-tests of log transformed data. Data calculations were performed with the SPSS package.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants from the NIAID (AI-43512) and CDC (CI-000094).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aihara S, Rao CM, Yu YX, Lee T, Watanabe K, Komiya T, Sumiyoshi H, Hashimoto H, Nomoto A. Identification of mutations that occurred on the genome of Japanese encephalitis virus during the attenuation process. Virus Genes. 1991;5:95–109. doi: 10.1007/BF00571925. [DOI] [PubMed] [Google Scholar]
- Arroyo J, Guirakhoo F, Fenner S, Zhang Z-X, Monath TP, Chambers TJ. Molecular Basis for Attenuation of Neurovirulence of a Yellow Fever Virus/Japanese Encephalitis Virus Chimera Vaccine (ChimeriVax-JE) J. Virol. 2001;75:934–942. doi: 10.1128/JVI.75.2.934-942.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinton MA, Dispoto JH. Sequence and secondary structure analysis of the 5'-terminal region of flavivirus genome RNA. Virology. 1988;162:290–299. doi: 10.1016/0042-6822(88)90468-0. [DOI] [PubMed] [Google Scholar]
- Burke DS, Leake CJ. Japanese encephalitis. In: Monath TP, editor. The Arboviruses: Epidemiology and Ecology. Boca Raton, Fla.: CRC Press, Inc.; 1988. pp. 63–92. [Google Scholar]
- Cahour A, Pletnev A, Vazielle-Falcoz M, Rosen L, Lai CJ. Growth-restricted dengue virus mutants containing deletions in the 5' noncoding region of the RNA genome. Virology. 1995;207:68–76. doi: 10.1006/viro.1995.1052. [DOI] [PubMed] [Google Scholar]
- Cao JX, Ni H, Wills MR, Campbell GA, Sil BK, Ryman KD, Kitchen I, Barrett AD. Passage of Japanese encephalitis virus in HeLa cells results in attenuation of virulence in mice. J. Gen. Virol. 1995;76:2757–2764. doi: 10.1099/0022-1317-76-11-2757. [DOI] [PubMed] [Google Scholar]
- Cecilia D, Gould EA. Nucleotide changes responsible for loss of neuroinvasiveness in Japanese encephalitis virus neutralization-resistant mutants. Virology. 1991;181:70–77. doi: 10.1016/0042-6822(91)90471-m. [DOI] [PubMed] [Google Scholar]
- Chambers TJ, Nestorowicz A, Mason PW, Rice CM. Yellow fever/Japanese encephalitis chimeric viruses: Construction and biological properties. J. Virol. 1999;73:3095–3101. doi: 10.1128/jvi.73.4.3095-3101.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers TJ, Jiang X, Droll DA, Liang Y, Wold WSM, Nickells J. Chimeric Japanese encephalitis virus/dengue-2 virus infectious clone : biological properties, immunogenicity and protection against dengue encephalitis in mice. J. Gen. Virol. 2006;87:3131–3140. doi: 10.1099/vir.0.81909-0. [DOI] [PubMed] [Google Scholar]
- Eckels KH, Yu YX, Dubois DR, Marchette NJ, Trent DW, Johnson AJ. Japanese encephalitis virus live attenuated vaccine, Chinese strain SA14-14-2: Adaptation to primary canine kidney cell cultures and preparation of a vaccine for human use. Vaccine. 1988;6:513–518. doi: 10.1016/0264-410x(88)90103-x. [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-Pdb Viewer: An ebvironment for comparative protein modellimng. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Halstead SB, Jacobson J. Japanese encephalitis. Adv. Virus Res. 2003;61:103–138. doi: 10.1016/s0065-3527(03)61003-1. [DOI] [PubMed] [Google Scholar]
- Hasegawa H, Yoshida M, Shiosake T, Fujita S, Kobayashi Y. Mutations in the envelope protein of Japanese encephalitis virus affect entry into cultured cells and virulence in mice. Virology. 1992;191:158–165. doi: 10.1016/0042-6822(92)90177-q. [DOI] [PubMed] [Google Scholar]
- Kofler RM, Heinz FX, Mandl CW. Capsid protein C of tick-Bbrne encephalitis virus tolerates large internal deletions and is a favorable target for attenuation of virulence. J. Virol. 2002;76:3534–3543. doi: 10.1128/JVI.76.7.3534-3543.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler RM, Leitner A, O’Riordain G, Heinz FX, Mandl CW. Spontaneous mutations restore the viability of tick-borne encephalitis virus mutants with large deletions in protein C. J. Virol. 2003;77:443–451. doi: 10.1128/JVI.77.1.443-451.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi E, Yamaoka M, Win K-S, Kurane I, Takada K, Mason PW. Anamnestic neutralizing antibody response is critical for protection of mice from challenge following vaccination with a plasmid encoding Japanese encephalitis virus premembrane and envelope genes. J. Virol. 1999;73:5527–5534. doi: 10.1128/jvi.73.7.5527-5534.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Lobigs M. Mechanism of virulence attenuation of glycosaminoglycan-binding variants of Japanese encephalitis virus and Murray Valley encephalitis virus. J. Virol. 2002;76:4901–4911. doi: 10.1128/JVI.76.10.4901-4911.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Jones CT, Groesdch TD, Kuhn RK, Post CB. Solution structure of dengue virus capsid protein reveals another fold. Proc. Natl. Acad. Sci. (USA) 2004;101:3414–3419. doi: 10.1073/pnas.0305892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie JS, Gubler DJ, Petersen LR. Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat. Med. 2004;10(12 Suppl):S98–S109. doi: 10.1038/nm1144. [DOI] [PubMed] [Google Scholar]
- Marfin AA, Eidex RS, Kozarsky PE, Cetron MS. Yellow fever and Japanese encephalitis vaccines: indications and complications. Infect. Dis. Clin. North Am. 2005;19:151–168. doi: 10.1016/j.idc.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Markoff L. 5'- and 3'-noncoding regions in flavivirus RNA. Adv. Virus Res. 2003;59:177–228. doi: 10.1016/S0065-3527(03)59006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathenge EG, Parquet Mdel C, Funakoshi Y, Houhara S, Wong PF, Ichinose A, Hasebe F, Inoue S, Morita K. Fusion PCR generated Japanese encephalitis virus/dengue 4 virus chimera exhibits lack of neuroinvasiveness, attenuated neurovirulence, and a dual-flavi immune response in mice. J. Gen. Virol. 2004;85:2503–2513. doi: 10.1099/vir.0.80120-0. [DOI] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- Monath TP, Heinz FX. Flaviviruses. In: Fields BN, Knipe DM, Chanock RM, Melnick JL, Monath TP, Roizman B, editors. Fields Virology. 3rd edition. Philadelphia: Lippincott-Raven; 1996. pp. 961–1034. [Google Scholar]
- Ni H, Burns NJ, Chang G-J, Zhang M-J, Wills MR, Trent DW, Sanders PG, Barrett ADT. Comparison nucleotide and deduced amino acid sequence of the 5' non-coding region and structural protein genes of the wild-type Japanese encephalitis virus strain SA14 and its attenuated vaccine derivatives. J. Gen. Virol. 1994;75:1505–1510. doi: 10.1099/0022-1317-75-6-1505. [DOI] [PubMed] [Google Scholar]
- Ni H, Chang GJ, Xie H, Trent DW, Barrett AD. Molecular basis of attenuation of neurovirulence of wild-type Japanese encephalitis virus strain SA14. J. Gen. Virol. 1995;76:409–413. doi: 10.1099/0022-1317-76-2-409. [DOI] [PubMed] [Google Scholar]
- Ni H, Barrett AD. Molecular differences between wild-type Japanese encephalitis virus strains of high and low mouse neuroinvasiveness. J. Gen. Virol. 1996;77:1449–1455. doi: 10.1099/0022-1317-77-7-1449. [DOI] [PubMed] [Google Scholar]
- Ni H, Barrett AD. Attenuation of Japanese encephalitis virus by selection of its mouse brain membrane receptor preparation escape variants. Virology. 1998;241:30–36. doi: 10.1006/viro.1997.8956. [DOI] [PubMed] [Google Scholar]
- Nitayaphan S, Grant JA, Chang G-J, J, Trent DW. Nucleotide sequence of the virulent SA-14 strain of japanese encephalitis virus and its attenuated vaccine derivative, SA14-14-2. Virology. 1990;177:541–552. doi: 10.1016/0042-6822(90)90519-w. [DOI] [PubMed] [Google Scholar]
- Peitsch MC. Protein modeling by E-mail. Bio/Technology. 1995;13:658–660. [Google Scholar]
- Reed LJ, Muench H. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 1938;27:493–497. WHO Expert Committee on Biological Standardization. [Google Scholar]
- Rice CM, Grakoui A, Galler R, Chambers TJ. Transcription of infectious yellow fever virus RNA from full-length cDNA templates produced by in vitro ligation. New Biol. 1989;1:285–296. [PubMed] [Google Scholar]
- Schlesinger JJ, Chapman S, Nestorowicz A, Rice CM, Ginocchio TE, Chambers TJ. Replication of yellow fever virus in the mouse central nervous system; comparison of neuroadapted and nonneuroadapted virus and partial sequence analysis of the neuroadapted strain. J. Gen. Virol. 1996;77:1277–1285. doi: 10.1099/0022-1317-77-6-1277. [DOI] [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL an automated protein homology-modeling server. Nuc. Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon T, Dung NM, Kneen R, Gainsborough M, Vaughn DW, Khanh VT. Japanese encephalitis. J. Neurol. Neurosurg. Psych. 2000;68:405–415. doi: 10.1136/jnnp.68.4.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumiyoshi H, Mori C, Fuke I, Morita K, Kuhara S, Kondou J, Kikuchi Y, Nagamatu H, Igarashi A. Complete nucleotide sequence of the Japanese encephalitis virus genome RNA. Virology. 1987;161:497–510. doi: 10.1016/0042-6822(87)90144-9. [DOI] [PubMed] [Google Scholar]
- Sumiyoshi H, Tignor GH, Shope RE. Characterization of a highly attenuated Japanese encephalitis virus generated from molecularly cloned cDNA. J. Infect. Dis. 1995;171:1144–1151. doi: 10.1093/infdis/171.5.1144. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Pool V, Tsai TF, Chen RT. Adverse events after Japanese encephalitis vaccination: review of post-marketing surveillance data from Japan and the United States. The VAERS Working Group. Vaccine. 2000;18:2963–2969. doi: 10.1016/s0264-410x(00)00111-0. [DOI] [PubMed] [Google Scholar]
- Thurner C, Witwer C, Hofacker IL, Stadler PF. Conserved RNA secondary structures in Flaviviridae genomes. J. Gen. Virol. 2004;85:1113–1124. doi: 10.1099/vir.0.19462-0. [DOI] [PubMed] [Google Scholar]
- Tsai TF. Japanese encephalitis vaccines. In: Plotkin SA, Mortimer EA, editors. Vaccines. 2nd edition. Philadelphia: WB Saunders; 1994. pp. 671–713. [Google Scholar]
- Zhao Z, Date T, Li Y, Kato T, Miyamoto M, Yasui K, Wakita T. Characterization of the E-138 (Glu/Lys) mutation in Japanese encephalitis virus by using a stable, full-length, infectious cDNA clone. J. Gen. Virol. 2005;86:2209–2220. doi: 10.1099/vir.0.80638-0. [DOI] [PubMed] [Google Scholar]
- Zuker M, Mathews DH, Turner DH. Algorithms and thermodynamics for RNA secondary structure predictions: a practical guide. In: Barciszewski J, Clark BFC, editors. RNA biochemistry. Boston, Mass.: NATO ASI Series, Kluwer Academic Publishers; 1999. pp. 11–43. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.