Abstract
The piggyBac (IFP2) short inverted terminal repeat transposable element from the cabbage looper Trichoplusia ni was tested for gene transfer vector function as part of a bipartite vector–helper system in the Mediterranean fruit fly Ceratitis capitata. A piggyBac vector marked with the medfly white gene was tested with a normally regulated piggyBac transposase helper at two different concentrations in a white eye host strain. Both experiments yielded transformants at an approximate frequency of 3–5%, with a total of six lines isolated having pigmented eyes with various levels of coloration. G1 transformant siblings from each line shared at least one common integration, with several sublines having an additional second integration. For the first transformant line isolated, two integrations were determined to be stable for 15 generations. For five of the lines, a piggyBac-mediated transposition was verified by sequencing the insertion site junctions isolated by inverse PCR that identified a characteristic piggyBac TTAA target site duplication. The efficient and stable transformation of the medfly with a lepidopteran vector represents transposon function over a relatively large evolutionary distance and suggests that the piggyBac system will be functional in a broad range of insects.
After more than a decade of concerted efforts to achieve transposon-mediated germ-line transformation in insect species other than Drosophila melanogaster, the first successes have only recently been reported. The medfly Ceratitis capitata has been transformed with the Drosophila hydei Minos element (1), Drosophila virilis has been transformed with the hobo (2, 3) and mariner elements (4), and very recently, transformation of Aedes aegypti with mariner (5) and Hermes (6) vectors has been reported. It is particularly interesting that after many attempts with several mosquito and tephritid fruit fly species, neither the P nor hobo vectors from D. melanogaster have yielded unambiguous germ-line transformants (7). The phylogenetic restriction for these vectors, however, was predicted by transient embryonic excision and transposition assays that indicated their lack of or limited mobility in several nondrosophilid species (8–12). Similar mobility assays with the Trichoplusia ni piggyBac element (originally referred to as IFP2) in cell lines (13, 14) and embryos (M.J.F., unpublished data) suggested that piggyBac may be less restricted as a gene-transfer vector, which encouraged the testing of this potential.
The piggyBac element is a short inverted terminal repeat (ITR) transposable element, 2.5 kb long, with 13-bp ITR sequences and a 2.1-kb ORF (15, 16). It is part of a subclass of ITR elements that are thus far found only in lepidopterans and that insert exclusively into TTAA target sites (17–19). On insertion, the target site is duplicated with excision occurring only in a precise fashion, restoring the insertion site. Beyond this functional similarity, the TTAA elements share no apparent structural identities.
piggyBac was originally discovered as the causative agent of FP (few polyhedra) mutations in baculoviruses passed through the T. ni TN-368 cell line (20). The element is dispersed and repeated in the genome of this cell line, but it has not been found in any others (16, 20). Its presence in vivo is discontinuous among T. ni strains, and thus far, it has not been detected in any other lepidopteran or other species. A piggyBac element isolated from the FP mutation 3E1 (15) was found to have autonomous function based on transposition assays in cell lines (18). Preliminary evidence from transient excision assays in embryos also support piggyBac function in several insect species, with similar rates of movement in lepidopteran and dipteran species (M.J.F., unpublished data). These results suggested that piggyBac vectors might also mediate germ-line transformation in these and, perhaps, other species.
The testing of vector function in most nondrosophilid insects has been limited by the lack of suitable transformant marking systems. Several chemical selection systems have been developed, but none have proven generally reliable, and most insect transformations have depended on less ambiguous visible markers (7). A previous medfly transformation with the Minos vector (1) used the medfly white (w) gene marking system (21), suggesting that piggyBac vector function could be similarly tested. Thus, a piggyBac transposon having a medfly w gene insertion was used with a nonautonomous piggyBac transposase-encoding helper plasmid to test its ability to mediate germ-line transformation as part of a bipartite vector–helper system. Herein we report the relatively efficient creation of piggyBac-mediated transformant lines in medfly, one of which has remained stable for more than 15 generations. The ability of this lepidopteran vector system to efficiently transform a dipteran species is encouraging for its use in other dipteran species and, perhaps, in a broad spectrum of other economically and medically important insects.
MATERIALS AND METHODS
Insect Strains and Rearing.
The C. capitata white-eye (we; ref. 22), wild-type, and transformant strains were maintained in a quarantine facility at the Department of Entomology, University of Hawaii. Standard larval and adult rearing methods were used (23).
Plasmids.
The pB[Ccw] vector was created by insertion of the medfly w cDNA gene into the 3E1 piggyBac element within the 6.0-kb p3E1.2 plasmid (15). The w gene, under D. melanogaster hsp70 promoter regulation within pDM30/hspCcWhite (21), was isolated as a 3.6-kb NotI fragment and ligated into the p3E1.2 NotI-linked HpaI site. The inserted w gene interrupts the piggyBac ORF, but otherwise leaves the piggyBac element intact, with the respective promoters in opposite orientation. The medfly w cDNA clone has been shown (1) to complement the we host strain mutation after transformation. The piggyBac transposase helper plasmid (pBΔSac) was created by digestion of p3E1.2 with SacI and religation, which deletes the 5′ piggyBac terminal sequences but maintains the putative piggyBac promoter region.
Injections.
Oviposited eggs were collected and dechorionated in 1.6% hypochlorite solution followed by several washes in 0.02% Triton X-100. Eggs were placed on double-stick tape, desiccated in room-air for times determined empirically (usually 10–15 min) and submerged in Halocarbon 700 oil. Injections followed standard Drosophila microinjection procedures (24) using vector and helper concentrations of 500 μg/ml and 150 μg/ml or 500 μg/ml and 300 μg/ml, respectively, in injection buffer (5 mM KCl/0.1 mM sodium phosphate, pH 6.8). Injected eggs were placed in an oxygenated tissue culture chamber at 22–23°C, and hatched larvae were collected 3–4 days later and placed on larval diet. Starting from early third instar, larvae and pupae were heat-shocked every day at 37°C for 1 h (for some transformants, heat shock was found to be unnecessary in subsequent generations for full eye pigmentation). Eclosed G0 male adults were mated individually to three we host-strain virgin females, and G0 virgin female adults were mated either individually to three we host-strain males or in groups of three females to six males. G1 eggs were collected for 2 weeks and reared under standard conditions.
Southern Blot Hybridization.
Approximately 10 μg of genomic DNA was digested with indicated restriction enzymes and separated on 0.8% agarose gels. DNA was stained with ethidium bromide, blotted to nylon filters, and immobilized by UV irradiation. Hybridization probes were labeled with [32P]dCTP by random priming (GIBCO/BRL) according to the manufacturer’s specifications. Hybridizations were performed in 0.25 M sodium phosphate buffer, pH 7.5/1% BSA/7% SDS at 65°C with an initial wash in 2× SSC/0.2% SDS at room temperature and two washes in 1× SSC/0.1% SDS at 55°C for 30 min. Autoradiography was performed by exposure on Kodak X-Omat film at −90°C.
PCR and Sequence Analysis.
Inverse PCR (25) was performed by initial digestion of 1–3 μg of transformant genomic DNA with HaeIII for 5′ and 3′ junctions, TaqI for 5′ junctions, or MspI for 3′ junctions. After 4-h digestions, restriction fragments were circularized by ligation at 16°C for 16 h. PCR was performed on the circularized fragments by using primer sequences in opposite orientation within the piggyBac restriction site and terminus for each junction. For the 5′ junction, the forward primer (574F) 5′-TCTTGACCTTGCCACAGAGG-3′ and reverse primer (157R) 5′-TGACACTTACCGCATTGACA-3′ were used. For the 3′ junction the forward primer (2388F) 5′-GTCAGTCCAGAAACAACTTTGGC-3′ and reverse primer (2123R) 5′-CCTCGATATACAGACCGATAAAAACACATG-3′ were used. PCR products were separated in low-melting-temperature agarose, and fragments were selected that were longer than the respective restriction site–terminus distances and different from those expected from the p3E1.2 based vector and helper plasmids. These products were directly subcloned into ddT vectors (Invitrogen), which were sequenced by using primers to vector sequence proximal to the respective termini. Sequence analysis was performed by using geneworks 2.5 software (Oxford Molecular Group, Oxford, UK) to align the junction sequences to piggyBac. Chromosomal insertion site sequences were subjected to blastn analysis (26).
RESULTS
Transformation Experiments.
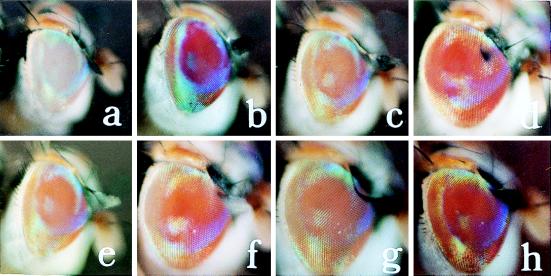
Germ-line transformation of a medfly we host strain mediated by the piggyBac vector was tested in two experiments using a vector marked with the medfly w gene and a nonautonomous helper that had a normally regulated transposase. In experiment I, 816 eggs from the C. capitata we host strain were injected with a vector–helper mixture of 500 μg/ml and 150 μg/ml, respectively, in injection buffer (Table 1). Approximately 190 G0 larvae hatched, with 73 emerging as adults. All of the adults were individually backcrossed to we host-strain adults, which were allowed to oviposit for 2 weeks. G1 progeny resulted from 19 of the G0 lines, yielding a fertility rate of 26%. All G0 and G1 adults were inspected for any eye coloration, with no pigmentation observed in the G0 individuals. From the G1 adults in line 41, two females (Cc[pBw]41F1 and 41F2) and one male (Cc[pBw]41M1), were observed to have the same dark-orange eye color (Table 2 and Fig. 1). These G1 adults were individually backcrossed to we adults, as were pigmented flies from each succeeding generation to maintain heterozygous lines. Homozygous G1 sublines were established by inbreeding 10 individual pairs of pigmented flies from G2 and G3 and continuing those lines yielding only pigmented flies.
Table 1.
Transformation experiments
| Exp. | Vector–helper, μg/ml | No. eggs injected | No. G0 adults mated | % fertility | No. G1 progeny | No. transformant G0 lines | Transformant frequency | No. pB[Ccw] integrations | Integration frequency |
|---|---|---|---|---|---|---|---|---|---|
| I | 500/150 | 816 | 73 | 26 | 3,752 | 1 | 0.053 | 2 | 0.11 |
| II | 500/300 | 1,211 | 253 | 62* | 30,715 | 5 | 0.03 | 9 | 0.06 |
*Calculated from individual G0 male matings.
Table 2.
Cc[pBw] G1 transformant lines
| Line | No. transformants
|
No. G1we | % transformed | Eye phenotype | No. total integrations | |
|---|---|---|---|---|---|---|
| Male | Female | |||||
| 41 | 1 | 2 | 30 | 9.1 | Dark-orange | 2 |
| 5 | 2 | 1 | 156 | 1.9 | Pale-orange | 1 |
| 53 | 1 | 6 | 223 | 3.0 | Orange | 3 |
| 69 | 6 | 3 | 287 | 3.0 | Red-orange | 2 |
| 134 | 1 | — | 236 | 0.8 | Yellow-orange* | 1 |
| — | 1 | Peach | ||||
| 220 | 8 | 7 | 79 | 16.0 | Yellow-orange | 2 |
*Phenotype is a yellow to red-orange gradient.
Figure 1.
Eye color phenotypes of the medfly strains we (a), wild-type (b), and the Cc[pBw] transformant sublines 220M1 (c), 69M1 (d), 53F4 (e), 5M2 (f), 134M1 (g), and 41F1 (h). Actual visual descriptions are given in Table 2.
In experiment II, 1,211 we host-strain eggs were injected with a vector–helper mixture of 500 μg/ml and 300 μg/ml, respectively. Approximately 700 larvae hatched, with 287 G0 adults emerging, none of which exhibited eye pigmentation (Table 1). All of the adults were initially backcrossed individually to we adults, including 139 G0 males and 148 G0 females. Of the male lines, 87 gave offspring yielding a fertility rate of 62%. A significant number of the G0 females failed to oviposit, and 114 surviving females were subsequently put into 38 groups of three, all of which yielded progeny. Because at least one female per group had to be fertile, females had a minimum fertility rate of 33%, but we presume it to be actually closer to the male fertility rate, which was used to conservatively estimate the frequency of transformation. In experiment I, many individually mated females also failed to oviposit, but they were not group mated, which may explain their lower overall fertility.
Four of the G0 female groups and one G0 male line gave rise to G1 offspring with pigmented eyes, with the number of putative transformant siblings from each line ranging from 2 to 15 (Table 2). Each of the sibling G1 flies was individually backcrossed to we and maintained as a separate G1 subline. For line 5, of the three sibling G1 flies, only 5M2 survived. Eye coloration, which ranged from a light-peach to dark-orange among the lines (Table 2 and Fig. 1), was consistent among the sublines within each line except for line 134, where 134F1 was peach and 134M1 exhibited a coloration gradient from yellow dorsally to red-orange ventrally. Pigmentation was enhanced visibly by heat shock only in the 5M2 and 134F1 lines.
DNA Hybridization.
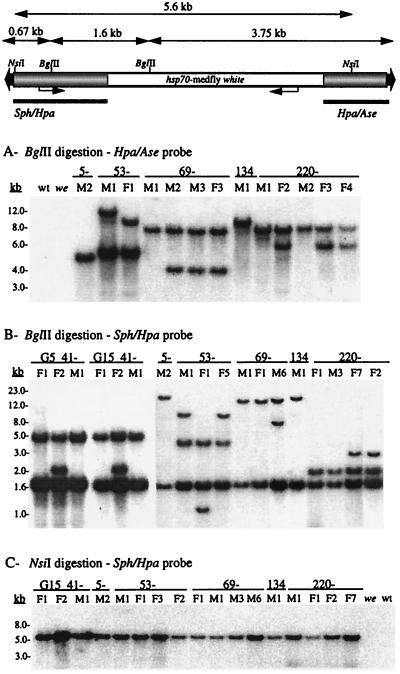
Verification of transformation was initially determined by Southern DNA blot hybridizations to BglII-digested genomic DNA with probe to the 5′ and 3′ vector arms surrounding the w marker gene (Fig. 2). Hybridization to a radiolabeled 0.95-kb piggyBac HpaI–AseI probe detects a single fragment for each integration that includes 3.75 kb of the 3′ vector arm with additional adjacent genomic DNA. Hybridization to a 1.4-kb piggyBac SphI–HpaI probe detects an internal 1.6-kb fragment on the 5′ vector arm, with an additional fragment for each integration that includes 0.67 kb of the vector arm terminus and adjacent genomic DNA. Because of the large number of sublines, most of the putative G1 or G2 transformants were initially screened to make an initial assessment of transformation and to determine common sublines. Presence of the piggyBac vector was verified in all of the sublines, with no hybridization detected in the wild-type or we control lines (also after extended autoradiographic exposure; data not shown). The data presented herein represent sublines exhibiting different molecular phenotypes from each G0 line.
Figure 2.
Southern DNA blot hybridization analysis of Cc[pBw] transformant sublines, and we host strain and wild-type (wt) control samples. At the top is a schematic diagram (not to scale) of the pB[Ccw] vector showing the BglII and NsiI restriction sites used to digest the genomic DNA and the 1.4-kb SphI–HpaI and 0.95-kb HpaI–AseI piggyBac vector fragments used as hybridization probes (bars). Above the schematic are distances used to calculate internal restriction fragment sizes and minimum sizes for junction fragments. Vector sequences are shaded, and the hsp70-white marker sequences are open. DNA size markers are shown to the left of the autoradiograms.
Hybridizations with either probe detected one or two piggyBac integrations in each subline (Fig. 2 A and B). For line 53, each of the sublines tested has two integrations, one of which is shared, with two independent secondary integrations. In the Sph–Hpa hybridization, sublines 53M1 and F5 share the same second integration (11-kb fragment), and 53F1 has a different second integration (1.1-kb fragment). The Hpa–Ase hybridization of 53M1 and F1 yields a similar result. For line 69, the first emerging G1 flies, 69M1 and 69F1, share a single common integration, with an additional integration appearing in the 13 subsequently emerging G1 flies (not all data shown). Similarly, in line 220, all the G1 males and the first emerging G1 female (220F1) have the same single integration. Subsequently emerging G1 females exhibited a second integration. Of the six G0 lines, only lines 5 and 134 exhibited unique single integrations; but both gave rise to the smallest number of G1 sublines, of which two of the three in line 5 died before reproducing. From this analysis, it appears that piggyBac transposition is strongly penetrant, with a high frequency of secondary integrations when the initial integration occurs early in gametogenesis. The apparent existence of a common integration in all the sublines suggests that the four transformants that arose from the group-mated females each came from only one of the females.
The data from the two hybridizations are generally consistent, and the minimum sizes of bands including the vector termini and chromosomal insertion site DNA are consistent with intact vector integrations. This was further verified by an NsiI digestion with hybridization to the Sph–Hpa probe. NsiI releases a 5.6-kb internal fragment, including more than 90% of the 6.0-kb vector, and Fig. 2C shows that in all sublines only a 5.6-kb fragment is detected regardless of the number of integrations, which is consistent with an intact vector for each integration.
The original transformant line 41 from the first experiment has been maintained for more than 15 generations. The visible phenotype has remained consistent in all three sublines and is the same even though 41F2 exhibits two integrations, with a single integration in 41F1 and 41M1. Stability of the vector integrations in line 41 homozygotes was assessed by Southern blot analysis at different generations. BglII digestion and Sph–Hpa probe hybridization resulted in a consistent number and pattern of integrations in all three sublines at generations 5 (G5) and 15 (G15), indicating piggyBac vector stability in this line (Fig. 2B).
Insertion-Site Sequencing.
Confirmation of a chromosomal vector integration or an intact vector does not ensure that a transposon-mediated event occurred. Indeed, P vector integrations were observed in several mosquito species (27–29), but all appeared to occur by random insertion independent of the transposon integration process. To verify that piggyBac-mediated transpositions had occurred, insertion sites were isolated by inverse PCR and sequenced from sublines 41F1, 5M2, 69M1, and 220M1 that had single integrations and subline 53M1 that had two integrations. PCRs usually resulted in only one amplified product consistent with single integrations. Subcloned PCR products were sequenced and compared with piggyBac terminal sequences by DNA alignment and blastn analysis (26) to identify genomic insertion site sequences and distinguish them from those in the injected plasmids. As shown in Fig. 3, for all integrations both the 5′ and 3′ junctions yielded the piggyBac inverted terminal repeat sequences immediately adjacent to a TTAA sequence and proximal insertion site DNA. The TTAA duplicated target site is characteristic of all piggyBac integrations and typically indicates a vector-mediated transposition.
Figure 3.
Inverse PCR strategy to isolate and sequence the pB[Ccw] vector insertion site in transformant sublines. At the top is a schematic diagram (not to scale) of the vector insertion in the host plasmid showing the approximate location of the restriction sites and primers used for PCR. Forward (F) and reverse (R) primers are numbered according to their nucleotide position in piggyBac. The piggyBac sequence is shown shaded, the medfly white marker gene is open, and chromosomal sequence is hatched. Below are shown the piggyBac insertion site sequence in p3E1.2 and the proximal insertion site sequences from several of the transformant sublines.
For line 220M1, a HaeIII site, used for inverse PCR, was adjacent to the 5′ TTAA insertion site thereby revealing only very proximal sequence. Although 53M1 yielded both 5′ and 3′ sequences, hybridizations indicated two integrations in this line (Fig. 2), and therefore these junctions may be from independent insertions. Fig. 3 shows the most proximal sequences, though for most insertion sites inverse PCR yielded several hundred base pairs. Comparisons to p3E1.2 sequence by direct alignment indicated that none of these insertion sites contained plasmid or vector DNA, which argues against the integrations resulting from fortuitous recombination events or being in tandem arrays; furthermore, they are not from contaminating or perduring plasmid. blastn comparisons reaffirmed this observation and, additionally, did not show strong identity to DNA sequences from any known genes within the database, and the insertion sites, thus, are concluded to be medfly chromosomal DNA. Beyond the TTAA insertion site, no obvious consensus was detected by inspection (or blast analyses) in the proximal sequences. Taken with the transformant white marker expression and Southern blot analyses, the vector insertion site sequences provide convincing evidence for an intact piggyBac vector mediating germ-line transformation in the medfly.
DISCUSSION
Results of two experiments show the ability of the piggyBac transposable element from T. ni to mediate germ-line transformation in the Mediterranean fruit fly C. capitata. Although the first experiment yielded a single transformant line, with the second experiment yielding five transformant lines, a similar transformation frequency of 3–5% per fertile G0 was obtained. A total of 11 integrations occurred, with at least two integrations detected in four of the six lines, yielding approximately a 2-fold higher total frequency of transformation events. This frequency is similar to that achieved with the Minos transposon vector (1) and is within the typical frequency range for Drosophila transformation. Notably, these systems normally take advantage of a heat-shock-regulated helper, whereas the piggyBac experiments used a self-regulated helper, reserving the possibility that the efficiency of the piggyBac system can be improved. Use of its own promoter, however, shows that the piggyBac transposon retains autonomous function in a very distantly related insect species, which is supportive of its function in other insects.
For many years there have been sporadic reports of insect transformation (30), but most of these were never characterized or were found to be somatic transient expression or random integrations. The Minos medfly transformation was the first unambiguous nondrosophilid transformation that clearly resulted from chromosomal integrations that were most likely transposon-mediated. For piggyBac, integrations into the medfly genome have also been demonstrated, and sequencing of five independent insertion sites, revealing characteristic piggyBac TTAA target site duplications, conclusively demonstrates that the transformations were due to chromosomal transpositions mediated by the piggyBac vector, as opposed to random recombination events. Stability for piggyBac vector integrations was shown for the first transformant line (Cc[pBw]41), in which two integrations have been stably maintained for at least 15 generations. Thus, these data show that the piggyBac transposable element from T. ni can function as an effective and stable vector for gene transfer in a dipteran species.
All of the sublines for each transformant line shared at least one integration, with many of the sublines containing an additional integration. Multiple integrations in D. melanogaster w transformants often result in darker eye color phenotypes (31, 32), although in this study, except for line 134, we could not visibly discern differing phenotypes among the sublines. Although this requires more quantitative analysis by pigmentation assays, some of the phenotypes are very pale or required heat shock to be detected at all. This study, as well as the Minos transformants, indicate that w gene position effect suppression in medfly is quite prevalent, as it is in Drosophila (31, 32). Indeed, none of the transformants from the two studies showed full w gene expression, even with heat-shock stimulation. Thus, it is not unlikely that some secondary integrations may be silent. Single integrations that are silent would not be detected, and if they are common, the actual transformation frequency might be somewhat higher than that reflected by visible reversion of the mutant phenotype. One of the piggyBac transformant sublines expressed an unusual phenotype also observed in Drosophila that appears as a dorsal–ventral pigmentation gradient. Pirrotta et al. (32) provide an interesting interpretation of this phenomena that suggests a positional–temporal gene expression gradient. Comparison of w position effects between medfly and Drosophila species is only the first of numerous biological phenomena that can now be compared by the study of transgenic insects. The limited number of previous insect transformations mediated by transposon-based vectors all have been accomplished in dipteran insects using vectors discovered in the same or other dipteran species. Although previous evidence suggested that piggyBac could function transiently in dipteran embryos, this report conclusively demonstrates that a vector system from one insect order can mediate stable gene transfer in a different order. This has important implications for the more widespread use of the piggyBac system and provides a functional assay for the activity of this somewhat unique transposon.
Studies to date have only detected piggyBac in T. ni, and other TTAA elements have been found solely in a limited number of lepidopteran species (17–19). Thus, the apparently restricted existence of piggyBac, in comparison to other short inverted terminal repeat elements such as P, hobo, and mariner, is intriguing given the present demonstration of its normal mobility in a distantly related species. In comparison, the P element exists in several Drosophila and nondrosophilid dipteran species (33, 34), but its mobility appears completely restricted to drosophilids (8, 9). For hobo, limited excision and transposition activity has been observed in several dipteran and one lepidopteran species (10–12), and although an effective transformation vector in D. melanogaster, it functions at a relatively low frequency in D. virilis (2, 3), with no reported transformations in nondrosophilids. In an experiment performed in parallel with our first piggyBac transformation, a hobo vector marked with the w gene failed to yield visible medfly transformants after screening nearly 100 fertile G0 lines (A.M.H. and S.D.M., unpublished data), compared with one piggyBac transformant from 19 fertile lines. Despite the limited mobility of hobo in nondrosophilids, closely related elements have been discovered in several dipteran insects (12, 35, 36), some being very distantly related, which suggests that horizontal transmission may have occurred (12). The Minos element was also originally discovered in a drosophilid (37), but similar to piggyBac, it retains function in a distantly related tephritid. However, Minos belongs to the mariner/Tc1 family, which is one of the most widely distributed transposable element families known and for which horizontal transmission has been clearly implicated (38). Thus its broad functional range is not surprising. It is therefore somewhat enigmatic that the piggyBac element is not, similarly, more widespread among the Insecta, if not other organisms, and this demands further investigation.
The relatively efficient and stable transposition of piggyBac in a dipteran genome suggests that it will be equally active in other dipterans and, perhaps, insects in general. For the medfly in particular, which is one of the greatest agricultural pests worldwide, effective gene transfer systems should allow the creation of strains useful for the sterile insect technique (39), such as those allowing genetic sexing and male sterilization (40). Although the Minos vector should facilitate the creation of such strains as well, the most novel transgenic strains, which result in direct biological control, may require multiple germ-line integrations, which will require different noninteracting vectors. It is also critical that vectors and endogenous transposons do not cross-mobilize one another, for the stability of the vector integration and integrity of the host genome. Additional vector systems also widen the possibilities for genetic manipulation including transposon mutagenesis and targeted transposition.
Germ-line transformation of nondrosophilid insects has been a priority of molecular entomologists since Drosophila transformation was first reported. The recent transformations of the medfly make it apparent that progress toward this goal has been limited by the vector and marker systems available. Efforts with P and hobo have not been successful, yet for both Minos and piggyBac, determination of transposon function in nonhost species was quickly followed by successful germ-line transformation. It is equally apparent that these successes and hobo transformation of D. virilis depended on the use of unambiguous visible eye-color marking systems. Visible markers not only are important to the initial identification of transgenic insects but also are critical to the establishment of transgenic lines and determination of stability. Selections that rely on chemical resistance may effectively select G1 transformants, but non-vector-related resistant organisms (possibly caused by resistance in symbionts) may be selected in later generations and only identified by molecular methods. Mixed strains of this sort would be of limited basic or applied use. Further advances in insect gene transfer will require additional visible marker systems for the many species not amenable to eye color mutant rescue and for secondary integrations in species such as the medfly. Additional vectors, especially those less dependent on host-specific factors, will be required for the most efficient and widespread use of this technology.
Acknowledgments
Grateful appreciation is extended to Drs. O. P. Perera, P. D. Shirk, and L. Zwiebel for sharing plasmids. This work was supported by the National Research Initiative Competitive Grants Program/U.S. Department of Agriculture Grant 9702823 (A.M.H.), and California Department of Food and Agriculture Grant 94-0615 (S.D.M. and S.H.S.).
References
- 1.Loukeris T G, Livadaras I, Arca B, Zabalou S, Savakis C. Science. 1995;270:2002–2005. doi: 10.1126/science.270.5244.2002. [DOI] [PubMed] [Google Scholar]
- 2.Lozovskaya E R, Nurminsky D I, Hartl D L, Sullivan D T. Genetics. 1995;142:173–177. doi: 10.1093/genetics/142.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gomez S P, Handler A M. Insect Mol Biol. 1997;6:1–8. doi: 10.1111/j.1365-2583.1997.tb00084.x. [DOI] [PubMed] [Google Scholar]
- 4.Lohe A R, Hartl D L. Genetics. 1996;143:365–374. doi: 10.1093/genetics/143.1.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coates C J, Jasinskiene N, Miyashiro L, James A A. Proc Natl Acad Sci USA. 1998;95:3748–3751. doi: 10.1073/pnas.95.7.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jasinskiene N, Coates C J, Benedict M Q, Cornel A J, Rafferty C S, James A A, Collins F H. Proc Natl Acad Sci USA. 1998;95:3743–3747. doi: 10.1073/pnas.95.7.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Brochta D A, Atkinson P W. Insect Biochem Mol Biol. 1996;26:739–753. doi: 10.1016/s0965-1748(96)00022-7. [DOI] [PubMed] [Google Scholar]
- 8.O’Brochta D A, Handler A M. Proc Natl Acad Sci USA. 1988;85:6052–6056. doi: 10.1073/pnas.85.16.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Handler A M, Gomez S P, O’Brochta D A. Arch Insect Biochem Physiol. 1993;22:373–384. doi: 10.1002/arch.940220306. [DOI] [PubMed] [Google Scholar]
- 10.Atkinson P W, Warren W D, O’Brochta D A. Proc Natl Acad Sci USA. 1993;90:9693–9697. doi: 10.1073/pnas.90.20.9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Brochta D A, Warren W D, Saville K J, Atkinson P W. Mol Gen Genet. 1994;244:9–14. doi: 10.1007/BF00280181. [DOI] [PubMed] [Google Scholar]
- 12.Handler A M, Gomez S P. Genetics. 1996;143:1339–1347. doi: 10.1093/genetics/143.3.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fraser M J, Ciszczon T, Elick T, Bauser C. Insect Mol Biol. 1996;5:141–151. doi: 10.1111/j.1365-2583.1996.tb00048.x. [DOI] [PubMed] [Google Scholar]
- 14.Elick T A, Bauser C A, Fraser M J. Genetica. 1996;98:33–41. doi: 10.1007/BF00120216. [DOI] [PubMed] [Google Scholar]
- 15.Cary L C, Goebel M, Corsaro H H, Wang H H, Rosen E, Fraser M J. Virology. 1989;161:8–17. doi: 10.1016/0042-6822(89)90117-7. [DOI] [PubMed] [Google Scholar]
- 16.Elick T A, Bauser C A, Principe N M, Fraser M J. Genetica. 1995;97:127–139. doi: 10.1007/BF00054620. [DOI] [PubMed] [Google Scholar]
- 17.Beames B, Summers M D. Virology. 1990;174:354–363. doi: 10.1016/0042-6822(90)90089-a. [DOI] [PubMed] [Google Scholar]
- 18.Fraser M J, Cary L, Boonvisudhi K, Wang H H. Virology. 1995;211:397–407. doi: 10.1006/viro.1995.1422. [DOI] [PubMed] [Google Scholar]
- 19.Wang H H, Fraser M J. Insect Mol Biol. 1993;1:109–116. doi: 10.1111/j.1365-2583.1993.tb00111.x. [DOI] [PubMed] [Google Scholar]
- 20.Fraser M J, Smith G E, Summers M D. J Virol. 1983;47:287–300. doi: 10.1128/jvi.47.2.287-300.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zwiebel L J, Saccone G, Zacharapoulou A, Besansky N J, Favia G, Collins F H, Louis C, Kafatos F C. Science. 1995;270:2005–2008. doi: 10.1126/science.270.5244.2005. [DOI] [PubMed] [Google Scholar]
- 22.Rössler Y, Rosenthal H. Ann Entomol Soc Am. 1992;85:525–531. [Google Scholar]
- 23.Saul S H. Ann Entomol Soc Am. 1982;75:480–483. [Google Scholar]
- 24.Rubin G M, Spradling A C. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 25.Ochman H, Ayala F J, Hartl D L. Methods Enzymol. 1993;218:309–321. doi: 10.1016/0076-6879(93)18023-6. [DOI] [PubMed] [Google Scholar]
- 26.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 27.Miller L H, Sakai R K, Romans P, Gwadz R W, Kantoff P, Coon H G. Science. 1987;237:779–781. doi: 10.1126/science.3039658. [DOI] [PubMed] [Google Scholar]
- 28.McGrane V, Carlson J O, Miller B R, Beaty B J. Am J Trop Med Hyg. 1988;39:502–510. doi: 10.4269/ajtmh.1988.39.502. [DOI] [PubMed] [Google Scholar]
- 29.Morris A C, Eggelston P, Crampton J M. Med Vet Entomol. 1989;3:1–7. doi: 10.1111/j.1365-2915.1989.tb00467.x. [DOI] [PubMed] [Google Scholar]
- 30.Handler A M, O’Brochta D A. Annu Rev Entomol. 1990;36:159–183. doi: 10.1146/annurev.en.36.010191.001111. [DOI] [PubMed] [Google Scholar]
- 31.Hazelrigg T, Levis R, Rubin G M. Cell. 1984;64:1083–1092. doi: 10.1016/0092-8674(84)90240-x. [DOI] [PubMed] [Google Scholar]
- 32.Pirrotta V, Steller H, Bozzetti M P. EMBO J. 1985;4:3501–3508. doi: 10.1002/j.1460-2075.1985.tb04109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lansman R A, Shade R O, Grigliatti T A, Brock H W. Proc Natl Acad Sci USA. 1987;84:6491–6495. doi: 10.1073/pnas.84.18.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perkins H D, Howells A J. Proc Natl Acad Sci USA. 1992;89:10753–10757. doi: 10.1073/pnas.89.22.10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Warren W D, Atkinson P W, O’Brochta D A. Genet Res Camb. 1994;64:87–97. doi: 10.1017/s0016672300032699. [DOI] [PubMed] [Google Scholar]
- 36.Handler A M, Gomez S P. Gene. 1997;185:133–135. doi: 10.1016/s0378-1119(96)00658-0. [DOI] [PubMed] [Google Scholar]
- 37.Franz G, Loukeris G T, Dialektaki G, Thomson R L C, Savakis C. Proc Natl Acad Sci USA. 1994;91:4746–4750. doi: 10.1073/pnas.91.11.4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robertson H M. J Insect Physiol. 1995;41:99–105. [Google Scholar]
- 39.Knipling E F. J Econ Entomol. 1955;48:459–462. [Google Scholar]
- 40.Handler A M. In: Advances in Insect Rearing for Research and Pest Management. Anderson T E, Leppla N C, editors. Boulder, CO: Westview; 1992. pp. 11–32. [Google Scholar]