Abstract
Sedimentation velocity experiments show that only monomers coexist with amyloid fibrils of human islet amyloid-polypeptide. No oligomers containing <100 monomers could be detected, suggesting that the putative toxic oligomers are much larger than those found for the Alzheimer's peptide, Aβ(1-42).
Human Islet amyloid polypeptide (hIAPP or amylin) is a 37-residue peptide that is cosecreted with insulin in the β-cells of the pancreas. It is the main component found in amyloid deposits of type II diabetes and forms amyloid fibrils in vitro (1,2). As is the case with other amyloid peptides, the link between fibril formation and disease is not yet understood (3–8). Early sedimentation equilibrium studies of the amyloid β-peptide, Aβ(1-40), showed that fibril formation in this system exhibits a critical concentration, or solubility, below which no assembly occurs, and that the principal species in equilibrium with fibril is the Aβ monomer (9,10). No such studies have been carried out for hIAPP, presumably because its high polymerization propensity makes detection of monomers at this concentration impossible using conventional probes. In addition, atomic force and electron microscope studies of fibril formation on many amyloid peptides have shown the presence of smaller aggregates, frequently coexisting with mature fibrils and sometimes appearing only as transient species at early stages of aggregation (11–15). The role of these species in fibril assembly and in the disease pathogenesis is still a matter of debate (8,16–19).
In this communication, we report sedimentation studies on metastable solutions of hIAPP for which polymerization is sufficiently slow that fibril formation does not occur in solution during the time required to carry out the sedimentation velocity experiments. The solutions were either cleared of any aggregated hIAPP by sedimentation in the analytical centrifuge or by prior preparative centrifugation at 60 krpm (g = 485,000). The peptide was dissolved directly in aqueous buffer, without the use of organic solvents or denaturants. At room temperature and neutral pH, the peptide aggregates rapidly, reducing the monomer concentration to below the limits of detection. Solutions were therefore prepared at 4.0°C, initially at low pH where solubility appears to be higher and the rate of fibril formation is much slower.
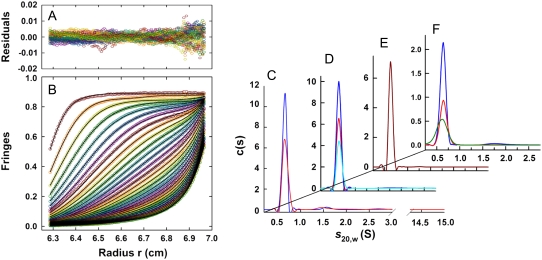
Fig. 1 B shows sedimentation velocity profiles obtained for the soluble component of the sample using interference optics (identical results were obtained with absorbance; see Supplementary Material) on a precleared sample of hIAPP. Also shown are the best-fit curves resulting from an analysis of the distribution of sedimentation coefficients, c(s). The fits obtained in this analysis are excellent as judged by the pooled residuals (Fig. 1 A) and their root mean-square deviation. The c(s) distribution shown on the right (blue curve, Fig. 1 C) shows clearly that the sedimentation profile results from a single species with an estimated molecular mass of 4.0 ± 0.1 kDa (s20,w of 0.65 S). The molecular mass of the amidated and oxidized form of hIAPP is 3.904 kDa (confirmed by electrospray ionization mass spectrometry performed on the same samples). These experiments therefore demonstrate that the soluble species is monomeric hIAPP.
FIGURE 1.
Summary of sedimentation velocity runs for the soluble component of hIAPP (data collected at 60 krpm). (Left) Typical sedimentation velocity profiles (obtained on a precleared sample at pH = 4.9, T = 4°C, and c = 55 μM) shown in terms of interference fringes versus radius (B), and pooled residuals from the fit (A). For clarity, scans are shown at 50-min intervals (every 10th scan with every eighth interference data point shown). Lines are best-fit curves obtained from a continuous c(s) analysis. (Right) The c(s) distributions obtained for: (C) precleared samples at pH = 4.9, T = 4°C, c = 68 μM (blue), and c = 55 μM (red), using interference optics; (D) samples filtered but not prespun in the preparative centrifuge before loading, at pH = 4.9, T = 4.0°C, c = 59 μM, λ = 236 nm (cyan), c = 150 μM, λ = 236 nm (blue), c = 230 μM, and λ = 280 nm data multiplied by 1.5 for clarity (red); (E) pH = 4.9, T = 20°C, and c = 55 μM using interference optics; and (F) pH dependence at 4.0°C, pH 6.2 (red, c = 82 μM, λ = 280 nm), pH 7.4 (green, c = 76 μM, λ = 280 nm), and pH 7.9 (blue, c = 59 μM, λ = 236 nm).
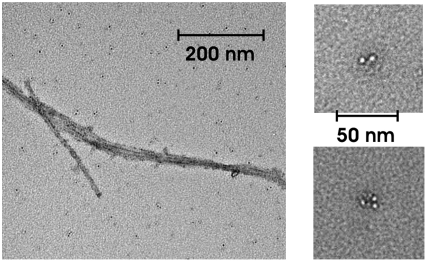
After the sedimentation velocity runs, the hIAPP was resuspended and the absorbance of the solution was measured while spinning at low speed (3 krpm). The absorbance decreased relative to that measured on the initially loaded solution indicating that additional high-molecular-weight species had formed during the velocity run and accumulated at the cell bottom. Transmission electron microscopy (TEM) imaging was performed on samples recovered at the end of each ultracentrifugation run. A typical result is shown in Fig. 2. It shows the presence of fibrils as well as other small aggregates, which include species that have curvy, short, spherical, or sometimes annular shapes. Similar species have been previously described in the literature (12,16,20–22).
FIGURE 2.
Transmission electron micrograph of the resuspended cell contents after completion of the sedimentation velocity runs shows the presence of both fibrillar (left panel) and nonfibrillar (right panels) aggregates.
Equilibrium sedimentation experiments were attempted on the same (or analogous) samples at 4.0°C and 50 krpm, using both absorbance and interference optics. In all cases, the concentration profiles did not indicate the presence of any species larger than monomer; the samples, however, never truly reached equilibrium (even after 86 h) due to slow and steady loss of monomer. This indicates that the soluble monomers of Fig. 1 are only metastable relative to larger aggregates on the timescales of the duration of velocity experiments (24 h), and eventually precipitate out of solution.
Identical results to those in Fig. 1, A and B, were found for the soluble supernatant when sedimentation velocity experiments were repeated for different loading concentrations (Fig. 1 C), without preclearing the samples (Fig. 1 D), at higher temperature (Fig. 1 E) and different pH (Fig. 1 F). In all cases, the soluble material consisted of a single species corresponding to an hIAPP monomer. When samples were not precleared, significant material losses were observed within the first few minutes at 60 krpm. Correspondingly, TEM images showed the presence of more fibrils. When experiments were attempted at pH = 7.4 and higher temperatures (20°C or 30°C) all of the hIAPP precipitated during or before the sedimentation runs, indicating that aggregation kinetics is too fast (and the solubility too low) for metastable monomers to be observed on timescales required for these experiments.
Importantly, the sedimentation velocity experiments did not detect appreciable amounts of species with S-values <15, corresponding to oligomers containing <100 monomers. We note that in most cases equally good fits were obtained when data were analyzed in terms of a single species; that is, the root mean-square deviations for the single species fits and the fits to a c(s) distribution were indistinguishable. The absence of small aggregates shows that there is a significant nucleation barrier to fibril formation. For example, the maximum concentration of small oligomers (dimers, trimers) can be estimated to be <∼2% of the monomer concentration, so even these small aggregates must be unstable by >2.5 kcal/mol relative to monomers.
There is a significant literature to document the presence of oligomers in other amyloid-forming systems. Oligomers varying in size between dimers and 40–50-mers were observed by sedimentation velocity in a recent study of β2 microglobulin, the peptide involved in dialysis related amyloidosis (23). In the case of Aβ(1−42), dimers, trimers, and higher oligomers have been observed and isolated by size exclusion chromatography, in samples prepared by dilution from organic solvents (24–26). These oligomers react with antibodies that do not react with fibrils, indicating significant structural differences. The more striking result is that the same antibodies react with putative oligomers in solutions of hIAPP, but not with fibrils. From these results, as well as those on several other amyloid-forming peptides, Glabe and co-workers have suggested that soluble amyloid oligomers have a common structure and share a common toxicity mechanism (19,26,27).
In their study, Glabe and co-workers did not determine the size of the soluble oligomers for hIAPP, as they did with Aβ(1-42), where they found that antibody binding required a minimum of eight monomers. Although our experiments are not carried out under the identical conditions used in the antibody assay, our results suggest that the minimum number of monomers in these putative toxic oligomers is >100.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Acknowledgments
We thank Robert Tycko and Sorin Luca for discussion, advice, and help in carrying out the TEM measurements.
This work was supported by the Intramural Research Program of The National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health.
Sara M. Vaiana and Rodolfo Ghirlando made equal contributions to this work.
Editor: Kathleen B. Hall.
References
- 1.Westermark, P., C. Wernstedt, E. Wilander, D. W. Hayden, T. D. O'Brien, and K. H. Johnson. 1987. Amyloid fibrils in human insulinoma and islets of Langerhans of the diabetic cat are derived from a neuropeptide-like protein also present in normal islet cells. Proc. Natl. Acad. Sci. USA. 84:3881–3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cooper, G. J. S., A. C. Willis, A. Clark, R. C. Turner, R. B. Sim, and K. B. M. Reid. 1987. Purification and characterization of a peptide from amyloid-rich pancreases of type 2 diabetic patients. Proc. Natl. Acad. Sci. USA. 84:8628–8632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoppener, J. W. M., and C. J. M. Lips. 2006. Role of islet amyloid in type 2 diabetes mellitus. Int. J. Biochem. Cell Biol. 38:726–736. [DOI] [PubMed] [Google Scholar]
- 4.Lansbury, P. T., and H. A. Lashuel. 2006. A century-old debate on protein aggregation and neurodegeneration enters the clinic. Nature. 443:774–779. [DOI] [PubMed] [Google Scholar]
- 5.Lorenzo, A., B. Razzaboni, G. C. Weir, and B. A. Yankner. 1994. Pancreatic-islet cell toxicity of amylin associated with type 2 diabetes mellitus. Nature. 368:756–760. [DOI] [PubMed] [Google Scholar]
- 6.Kapurniotu, A. 2001. Amyloidogenicity and cytotoxicity of islet amyloid polypeptide. Biopolymers. 60:438–459. [DOI] [PubMed] [Google Scholar]
- 7.Jaikaran, E. T. A. S., and A. Clark. 2001. Islet amyloid and type 2 diabetes: from molecular misfolding to islet pathophysiology. Biochim. Biophys. Acta Mol. Basis Dis. 1537:179–203. [DOI] [PubMed] [Google Scholar]
- 8.Janson, J., R. H. Ashley, D. Harrison, S. McIntyre, and P. C. Butler. 1999. The mechanism of islet amyloid polypeptide toxicity is membrane disruption by intermediate-sized toxic amyloid particles. Diabetes. 48:491–498. [DOI] [PubMed] [Google Scholar]
- 9.Harper, J. D., and P. T. Lansbury, Jr. 1997. Models of amyloid seeding in Alzheimer's disease and scrapie: mechanistic truths and physiological consequences of the time-dependent solubility of amyloid proteins. Annu. Rev. Biochem. 66:385–407. [DOI] [PubMed] [Google Scholar]
- 10.Terzi, E., G. Holzemann, and J. Seelig. 1995. Self-association of β-amyloid peptide (1-40) in solution and binding to lipid membranes. J. Mol. Biol. 252:633–642. [DOI] [PubMed] [Google Scholar]
- 11.Huang, T. H. J., D. S. Yang, N. P. Plaskos, S. Gō, C. M. Yip, P. E. Fraser, and A. Chakrabartty. 2000. Structural studies of soluble oligomers of the Alzheimer β-amyloid peptide. J. Mol. Biol. 297:73–87. [DOI] [PubMed] [Google Scholar]
- 12.Green, J. D., C. Goldsbury, J. Kistler, G. J. S. Cooper, and U. Aebi. 2004. Human amylin oligomer growth and fibril elongation define two distinct phases in amyloid formation. J. Biol. Chem. 279:12206–12212. [DOI] [PubMed] [Google Scholar]
- 13.Goldsbury, C., J. Kistler, U. Aebi, T. Arvinte, and G. J. S. Cooper. 1999. Watching amyloid fibrils grow by time-lapse atomic force microscopy. J. Mol. Biol. 285:33–39. [DOI] [PubMed] [Google Scholar]
- 14.Konarkowska, B., J. F. Aitken, J. Kistler, S. P. Zhang, and G. J. S. Cooper. 2006. The aggregation potential of human amylin determines its cytotoxicity towards islet β-cells. FEBS J. 273:3614–3624. [DOI] [PubMed] [Google Scholar]
- 15.El-Agnaf, O. M. A., S. Nagala, B. P. Patel, and B. M. Austen. 2001. Non-fibrillar oligomeric species of the amyloid Aβ peptide, implicated in familial British dementia, are more potent at inducing apoptotic cell death than protofibrils or mature fibrils. J. Mol. Biol. 310:157–168. [DOI] [PubMed] [Google Scholar]
- 16.Lashuel, H. A., and P. T. Lansbury. 2006. Are amyloid diseases caused by protein aggregates that mimic bacterial pore-forming toxins? Q. Rev. Biophys. 39:167–201. [DOI] [PubMed] [Google Scholar]
- 17.Caughey, B., and P. T. Lansbury. 2003. Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu. Rev. Neurol. 26:267–298. [DOI] [PubMed] [Google Scholar]
- 18.Quist, A., I. Doudevski, H. Lin, R. Azimova, D. Ng, B. Frangione, B. Kagan, J. Ghiso, and R. Lal. 2005. Amyloid ion channels: a common structural link for protein-misfolding disease. Proc. Natl. Acad. Sci. USA. 102:10427–10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glabe, C. G., and R. Kayed. 2006. Common structure and toxic function of amyloid oligomers implies a common mechanism of pathogenesis. Neurology. 66:S74–S78. [DOI] [PubMed] [Google Scholar]
- 20.Kreplak, L., and U. Aebi. 2006. From the polymorphism of amyloid fibrils to their assembly mechanism and cytotoxicity. Adv. Protein Chem. 73:217–233. [DOI] [PubMed] [Google Scholar]
- 21.Goldsbury, C. S., G. J. S. Cooper, K. N. Goldie, S. A. Muller, E. L. Saafi, W. T. M. Gruijters, and M. P. Misur. 1997. Polymorphic fibrillar assembly of human amylin. J. Struct. Biol. 119:17–27. [DOI] [PubMed] [Google Scholar]
- 22.Makin, O. S., and L. C. Serpell. 2004. Structural characterization of islet amyloid polypeptide fibrils. J. Mol. Biol. 335:1279–1288. [DOI] [PubMed] [Google Scholar]
- 23.Smith, A. M., T. R. Jahn, A. E. Ashcroft, and S. E. Radford. 2006. Direct observation of oligomeric species formed in the early stages of amyloid fibril formation using electrospray ionization mass spectrometry. J. Mol. Biol. 364:9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soreghan, B., J. Kosmoski, and C. Glabe. 1994. Surfactant properties of Alzheimer's Aβ peptides and the mechanism of amyloid aggregation. J. Biol. Chem. 269:28551–28554. [PubMed] [Google Scholar]
- 25.Roher, A. E., M. O. Chaney, Y. M. Kuo, S. D. Webster, W. B. Stine, L. J. Haverkamp, A. S. Woods, R. J. Cotter, J. M. Tuohy, G. A. Krafft, B. S. Bonnell, and M. R. Emmerling. 1996. Morphology and toxicity of Aβ-(1-42) dimer derived from neuritic and vascular amyloid deposits of Alzheimer's disease. J. Biol. Chem. 271:20631–20635. [DOI] [PubMed] [Google Scholar]
- 26.Kayed, R., E. Head, J. L. Thompson, T. M. McIntire, S. C. Milton, C. W. Cotman, and C. G. Glabe. 2003. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 300:486–489. [DOI] [PubMed] [Google Scholar]
- 27.Kayed, R., and C. G. Glabe. 2006. Conformation-dependent anti-amyloid oligomer antibodies. Methods Enzymol. 413:326–344. [DOI] [PubMed] [Google Scholar]