Abstract
The effect of cotranslationally active chaperones on the conformation of incomplete protein chains is poorly understood. The secondary structure of a 77-residue chaperone-bound N-terminal protein fragment corresponding to the first five helices (A–E) of apomyoglobin (apoMb1-77) is investigated here at the residue-specific level by multidimensional NMR. The substrate-binding domain of DnaK, DnaK-β, is employed as a chaperone model. By taking advantage of the improved spectral quality resulting from chaperone deuteration, we find that DnaK-β-bound apoMb1-77 displays a region of nonnative helicity at residues away from the main chaperone binding site. The nonnative structural motif comprises portions of the native D and E helices and has similar characteristics to the reported nonnative DE helical region of acid-unfolded full-length apoMb. Upon incorporation of the missing C-terminal amino acids, a structural kink develops between residues 56 and 57, and two separate native D and E helices are generated. This work highlights, for the first time to our knowledge, the presence of a nonnative helical motif in a large chaperone-bound protein fragment under physiologically relevant solution conditions.
Our understanding of how proteins achieve their native structure in buffered solutions has progressed considerably over the past few decades (1). However, we know very little about how proteins fold in the cell, where the presence of additional interacting and noninteracting molecules has the potential of affecting both mechanisms and yields.
A number of recent studies utilizing model systems have targeted the effect of physiologically relevant factors on protein folding, including vectorial chain elongation (2,3), ribosomal tethering (4), molecular chaperones (5), and molecular crowding (6). Biophysical investigations addressing cotranslational protein folding, however, are still scarce. We showed that the folding behavior of N-terminal fragments derived from apomyoglobin (apoMb) is fundamentally different from the in vitro refolding of the full-length protein. The N-terminal apoMb polypeptides misfold and self-associate under physiologically relevant conditions (2), suggesting that chaperones and/or the ribosome are needed to prevent the aggregation of apoMb in the cell during translation. Indeed, self-association of the protein fragments is prevented in the presence of DnaK-β, the substrate-binding domain of the cotranslationally active bacterial chaperone Hsp70 (5). More recent multidimensional NMR investigations revealed that misfolding-prone apoMb fragments are held in a globally unfolded conformation retaining some residual secondary structure (7). However, several important conformational details could not be elucidated. Given the incomplete level of information and the fact that related studies only address very small substrates (8,9), it is fair to state that the structural properties of chaperone-bound polypeptides and the effect of chaperones on protein folding (both during and after protein biosynthesis) are still poorly understood.
Here, we provide the first detailed description, to our knowledge, of the residual secondary structure of a chaperone-bound protein fragment whose length largely exceeds that of the known core residues directly in contact with the chaperone (10). Surprisingly, we detect nonnative helical secondary structure for some of the residues away from the chaperone binding site.
The substrate of interest is the 77-residue N-terminal apoMb fragment apoMb1-77, which includes amino acids spanning the first five helices of the full-length native protein. ApoMb1-77 misfolds and self-associates in isolation, resulting in barely detectable NMR signals (5). In the presence of chaperone, however, most of the expected NMR resonances are retrieved. We selected the substrate-binding domain of DnaK, DnaK-β (5), as model chaperone. At 1:1 molar ratios of apoMb1-77:DnaK-β, the protein fragment is in a nearly completely chaperone-bound state (5). The moderate HN chemical shift dispersion of DnaK-β-bound apoMb1-77 shows lack of an overall tertiary structure. Previously reported initial assignments and corresponding Cα secondary chemical shifts for DnaK-β-bound apoMb1-77 lacked information on a significant portion of the polypeptide (7). Practical challenges associated with sample conditions included: 1), mandatory use of low temperature (needed to prevent substrate aggregation); and 2), spin-diffusion across the macromolecular complex. The above challenges led to broad lineshapes, which prevented detailed conformational analysis and hindered the assignment process.
Here, we resort to deuteration of the DnaK-β chaperone, which partially solves the spin diffusion problem and leads to improved NMR spectral quality. Most importantly, chaperone deuteration enables elucidating novel key structural details on the chaperone-bound substrate.
Despite its improved spectral features, the 13C,15N-apoMb1-77-2H-DnaK-β substrate-chaperone complex defies substrate backbone assignments via the classical suite of triple-resonance experiments. Even TROSY-type strategies at 800 MHz did not lead to higher quality data. Therefore, we resorted to a combination of HNCA and a series of 1H, 15N HSQC-detected pH titrations to maximize the number of assigned resonances. The HNCA and the starting spectrum of the pH titrations were acquired under the conditions noted in Fig. 1. The pH was then gradually decreased, and multiple spectra were collected at several progressively lower pH values, down to pH 2.5. At the pH titration endpoint, both the chaperone and the fragment are unfolded and no longer bound. Assignments at low pH (S. Rajagopalan, C. Chow, E. C. Fulmer, D. Fedyukina, Y.-J. Eun, and S. Cavagnero, unpublished), where NMR resonances are significantly more intense, were done via conventional NMR methods and then back-traced to higher pH to assign the chaperone-bound apoMb1-77 resonances at pH 5.8. The backbone assignments resulting from pH titration were confirmed by the detectable connectivities and partial assignments from the pH 5.8 HNCA, which also provided Cα chemical shifts for the assigned residues.
FIGURE 1.
1H, 15N -HSQC NMR spectrum of apoMb1-77 in the presence of deuterated DnaK-β chaperone. Both species were 100 μM in 10 mM sodium acetate at pH 5.8. Data were acquired on a Varian INOVA 600 MHz NMR spectrometer at 4°C. The assigned resonances are labeled.
The above strategy led to backbone HN, N, and Cα assignments for most of the detectable resonances, corresponding to 46 out of 77 residues (12), as shown in Fig. 1. The unassigned residues belong to either amino acids 26–35, previously identified as composing the core of the chaperone-binding site (10), or the highly nonpolar residues belonging to the native A helix. The former set of resonances are likely broadened beyond detection because of lack of local mobility at the binding site core of the 23.5 kDa complex at 4°C (see also Supplementary Material). The amino acids belonging to the native A helix are likely exchange-broadened since they are nearly-undetectable at 4°C even in the acid-unfolded state of apoMb1-77.
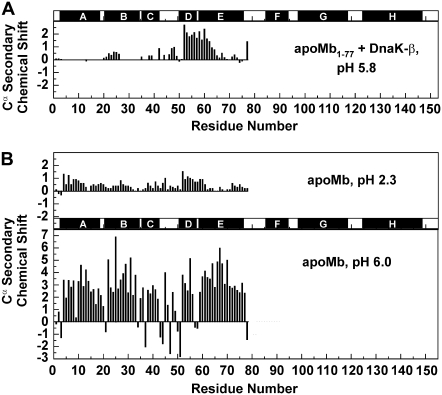
Secondary chemical shifts for the assigned Cα nuclei (Fig. 2 A) were calculated by subtracting residue-specific random coil chemical shifts (13) from the experimentally determined chemical shifts of the relevant nuclei. Backbone chemical shifts are very sensitive to electronic environment and are effective reporters of secondary structure at atomic resolution (13). The helices of full-length native apoMb show large and positive Cα secondary chemical shifts ((14); Fig. 2 B, lower panel).
FIGURE 2.
(A) Cα secondary chemical shifts of apoMb1-77 N-terminal fragment in the presence of deuterated DnaK-β (1:1 molar ratio) at pH 5.8 and 4°C. Chemical shifts were obtained from resonance assignments performed in the presence of deuterated chaperone. NMR experimental details are provided in Supplementary Material. (B) Known Cα secondary chemical shifts of the 1-77 region of acid-unfolded (pH 2.3, 25°C) (15) and native (pH 6.0, 35°C) (14) full-length apoMb.
Remarkably, chaperone-bound apoMb1-77 displays nonnative helicity across the region comprising the native D and E helices. In addition, the detectable helical profile of chaperone-bound apoMb1-77 is similar to that of acid-unfolded apoMb ((15); Fig. 2 B, top panel) except for the approximately twofold larger helicity in the DE region. Either the lower temperature and/or differences in the carboxyl group protonation state of the four acidic residues (Glu-52, Glu-54, Glu-59 and Asp-60) of the apoMb1-77 DE region are likely responsible for the increased helicity at pH 5.8.
The short peptide (T51-K63) corresponding to the DE portion of the sequence was reported to be partially helical in isolation (16). In addition, the presence of the DE helical motif away from the chaperone binding site and the fact that this motif is also present at low pH in the absence of chaperone suggest that the nonnative structure results from a local intrinsic conformational trend of the DE sequence in the context of apoMb1-77.
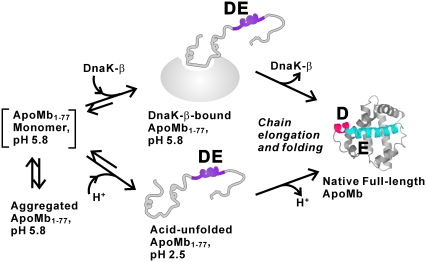
Fig. 3 provides a schematic representation of the chaperone-bound apoMb1-77 conformation supported by this work and a view of apoMb1-77 under different experimental conditions. At pH 5.8, apoMb1-77 has a tendency to self-associate and aggregate, which can be averted either by a decrease in pH or by complex formation with DnaK-β (5). In both the chaperone-bound and low pH cases, a globally unfolded conformation lacking significant stable tertiary contacts and comprising the nonnative DE residual helical region is generated. We propose that the chaperone-bound state of N-terminal protein fragments may effectively serve as an unfolded state under physiologically relevant conditions, before protein synthesis is complete. The presence of a chaperone-bound complex is important primarily because this state enables the full-length protein to undergo folding and become bioactive within the same medium that ensures minimal aggregation via chaperone-binding while protein synthesis is in progress.
FIGURE 3.
Proposed cartoon representation of apoMb1-77 conformations populated under different conditions. The species enclosed in square brackets may be metastable and is not detectable in solution. The residual nonnative helicity of DnaK-β-bound apoMb1-77 is shown in purple. Upon substrate chain elongation and folding, the DE helical region is replaced by individual D (red) and E (blue) helices.
In summary, this work identifies, for the first time to our knowledge, the presence of nonnative helical conformation in a chaperone-bound substrate lacking the C-terminal region. Our previous studies with a small peptide substrate corresponding to the apoMb1-77 binding site core (7) revealed that the peptide undergoes local helix unwinding and switches to a more extended conformation upon chaperone binding. Therefore, chaperone-bound apoMb1-77 is characterized by two types of nonnative motifs: helical and extended. On incorporation of the missing C-terminal residues, apoMb acquires the ability to reach its folded state, and native D and E helices develop.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Acknowledgments
We thank Margarita Santiago and Ashok Sekhar for their critical reading of the manuscript.
This work was supported by National Institutes of Health (NIH) grant GM068538. This study made use of the National Magnetic Resonance Facility at Madison, which is supported by NIH grants P41RR02301 (BRTP/ NCRR) and P41GM66326 (NIGMS). Additional equipment was purchased with funds from the University of Wisconsin, the NIH (RR02781, RR08438), the National Science Foundation (DMB-8415048, OIA-9977486, BIR-9214394), and the United States Department of Agriculture.
Editor: Kathleen B. Hall.
References
- 1.Laurents, D. V., and R. L. Baldwin. 1998. Protein folding: matching theory and experiment. Biophys. J. 75:428–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chow, C. C., C. Chow, V. Raghunathan, T. J. Huppert, E. B. Kimball, and S. Cavagnero. 2003. Chain length dependence of apomyoglobin folding: structural evolution from misfolded sheets to native helices. Biochemistry. 42:7090–7099. [DOI] [PubMed] [Google Scholar]
- 3.Neira, J. L., and A. R. Fersht. 1999. Exploring the folding funnel of a polypeptide chain by biophysical studies on protein fragments. J. Mol. Biol. 285:1309–1333. [DOI] [PubMed] [Google Scholar]
- 4.Hsu, S. T. D., P. Fucini, L. D. Cabrita, H. Launay, C. M. Dobson, and J. Christodoulou. 2007. Structure and dynamics of a ribosome-bound nascent chain by NMR spectroscopy. Proc. Natl. Acad. Sci. USA. 104:16516–16521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kurt, N., S. Rajagopalan, and S. Cavagnero. 2006. Effect of Hsp70 chaperone on the folding and misfolding of polypeptides modeling an elongating protein chain. J. Mol. Biol. 355:809–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Minton, A. P. 2001. The influence of macromolecular crowding and macromolecular confinement on biochemical reactions in physiological media. J. Biol. Chem. 276:10577–10580. [DOI] [PubMed] [Google Scholar]
- 7.Chen, Z. J., N. Kurt, S. Rajagopalan, and S. Cavagnero. 2006. Secondary structure mapping of DnaK-bound protein fragments: chain helicity and local helix unwinding at the binding site. Biochemistry. 45:12325–12333. [DOI] [PubMed] [Google Scholar]
- 8.Landry, S. J., R. Jordan, R. McMacken, and L. M. Gierasch. 1992. Different conformations for the same polypeptide bound to chaperones DnaK and Groel. Nature. 355:455–457. [DOI] [PubMed] [Google Scholar]
- 9.Zhu, X. T., X. Zhao, W. F. Burkholder, A. Gragerov, C. M. Ogata, M. E. Gottesman, and W. A. Hendrickson. 1996. Structural analysis of substrate binding by the molecular chaperone DnaK. Science. 272:1606–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vega, C., N. Kurt, Z. Chen, S. Rudiger, and S. Cavagnero. 2006. Binding specificity of an α-helical protein sequence to a full length Hsp70 chaperone and its minimal substrate binding domain. Biochemistry. 45:13835–13846. [DOI] [PubMed] [Google Scholar]
- 11.Reference deleted in proof.
- 12.Backbone chemical shift assignments for chaperone-bound apoMb1–77 were deposited in the BioMagResBank (BMRB ID 15589).
- 13.Wishart, D., C. Bigam, A. Holm, R. Hodges, and B. Sykes. 1995. 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J. Biomol. NMR. 5:67–81. [DOI] [PubMed] [Google Scholar]
- 14.Eliezer, D., and P. E. Wright. 1996. Is apomyoglobin a molten globule? Structural characterization by NMR. J. Mol. Biol. 263:531–538. [DOI] [PubMed] [Google Scholar]
- 15.Eliezer, D., J. Yao, H. J. Dyson, and P. E. Wright. 1998. Structural and dynamic characterization of partially folded states of apomyoglobin and implications for protein folding. Nat. Struct. Biol. 5:148–155. [DOI] [PubMed] [Google Scholar]
- 16.Yao, J., J. Chung, D. Eliezer, P. E. Wright, and H. J. Dyson. 2001. NMR structural and dynamic characterization of the acid-unfolded state of apomyoglobin provides insights into the early events in protein folding. Biochemistry. 40:3561–3571. [DOI] [PubMed] [Google Scholar]