Abstract
The stalk of Saccharomyces cerevisiae ribosomes contains, on average, five distinct proteins, namely P0 and four acidic proteins, P1α, P1β, P2α, and P2β. Each ribosome contains only one copy of P0, but the distribution of the acidic proteins among the ribosome population in vivo has not been determined. Using two-photon fluorescence correlation spectroscopy and scanning FCS, on cells expressing EGFP-tagged P0, P1, and P2 proteins, we show, with brightness analysis, that individual yeast ribosomes in vivo are compositionally heterogeneous in regard to P1α, P1β, P2α, and P2β. These results are relevant to the hypothesis, based on in vitro studies, that the overall cellular pattern of expressed proteins can be determined by the distribution of the stalk proteins among the ribosome population.
INTRODUCTION
The bacterial ribosomal stalk has a relatively simple structure composed of either two or three dimers of the acidic 12-kDa protein L7/L12, which interact through their N-terminal domains with the 23-kDa protein L10 (1). This protein complex attaches to the highly conserved 23S rRNA GTPase Associated Region. The globular L7/L12 C-domain is exposed to the cytoplasm and is joined to the rest of the molecule through a flexible hinge region (2). The two C-domains of the L7/L12 dimer are highly mobile and, on average, far apart (3,4). The high mobility of the stalk has hindered its resolution in the available prokaryotic ribosome crystal structures (5,6).
Evolution has notably increased the complexity of the ribosomal stalk. The eukaryotic protein L10 counterpart, phosphoprotein P0, is larger because of its extended carboxyl domain, which shows significant sequence similarity with the corresponding L7/L12-like proteins, which have evolved into two phosphoprotein families, P1 and P2. Each of these families of proteins has a variable number of members depending on the organism (see Ballesta and Remacha (7) for a review). In vitro studies showing exchange of P1 and P2 proteins between the ribosome and a cytoplasmic pool of free proteins during translation (8–10) suggest that the composition of the eukaryotic stalk structure is variable.
In Saccharomyces cerevisiae, the ribosomal stalk is made of protein P0 and four 12-kDa acidic proteins, P1α, P1β, P2α, and P2β. The formation of acidic protein homodimers as well as heterodimers of different types, e.g., P1α/P2β and P1β/P2α, has been shown by cross-linking studies in vitro (11–17) on ribosomes isolated from various eukaryotic species. However, the composition of the stalk, i.e., homogeneous versus heterogeneous, and the functional significance of different types of stalks inside the cell are still open questions.
For example, ribosomes with different stalk compositions as a result of either the presence of different acidic proteins (18) or different phosphorylation states (19) have been isolated from cells in different metabolic conditions (20). The various subpopulations are active in protein synthesis, but their efficiencies vary depending on the specific mRNA being translated (21). In this way, the overall cellular pattern of expressed proteins can be affected by the relative proportion of the different ribosomes. In fact, the regulatory activity of the eukaryotic stalk can be considered as a paradigmatic example of the so-called ribosome filter hypothesis (22).
We note that information regarding ribosome composition and structure has been obtained almost exclusively from purified particles, after cell fractionation and washing, which might result in the loss of elements sensitive to the purification procedures. Two-photon fluorescence correlation spectroscopy (FCS) and scanning FCS have been applied to oligomeric protein systems (23,24). More recently, analysis of the FCS data using the photon counting histogram (PCH) method has permitted the study of the oligomerization state of proteins in vivo (25,26). We have applied these techniques to study the ribosome in S. cerevisiae cells.
METHODS
Strains and growth conditions
All S. cerevisiae strains used in this work, described in Table 1, are derived from strain BJ5458 (27). For ribosome preparation, yeast were grown in rich YEP medium (2% bactopeptone, 1% yeast extracts). For fluorescence experiments, cells were grown in minimal YNB medium (0.67% yeast nitrogen base) containing the nutritional requirements of the strains. Both media were supplemented with either 2% glucose (YEPD, YNBD) or 2% galactose (YEPG, YNBG) as required. E. coli DH5α was used for plasmid constructions and was grown in LB medium.
TABLE 1.
Strains expressing EGFP-tagged ribosomal stalk proteins used in this report
| Strain | Native protein missing | Transforming plasmid | Expressed EGFP-tagged protein |
|---|---|---|---|
| DpGP0-P0t | P0 | pFL36-P0t | P0-EGFP |
| Dp4GP0-P0t,P2αt | P0, P2α | pFL36-P0t, pUG35-P2αt | P0-EGFP, P2α-EGFP |
| Dp6GP0-P0t,P1βt | P0, P1β | pFL36-P0t, pFL39-P1βt | P0-EGFP, P1β-EGFP |
| Dp4-P2αt | P2α | pUG35-P2αt | P2α-EGFP |
| Dp5-P2βt | P2β | pUG35-P2βt | P2β-EGFP |
| Dp7-P1αt | P1α | pUG35TRP-P1αt | P1α-EGFP |
| Dp6-P1βt | P1β | pFL39-P1βt | P1β-EGFP |
| Dp46-P2αt,P1βt | P2α, P1β | pUG35-P2αt, pFL39-P1βt, | P2α-EGFP, P1β-EGFP |
All the strains were derived from S. cerevisiae BJ5458 (Dp) and obtained as described in Methods.
Disrupted strains
Gene disruption was performed by recombination using cassettes containing the KanMXLoxP selection marker flanked by the appropriate sequences complementary to the ends of the DNA fragment to be deleted according to Güldener et al. (28). In this way, genes RPP2A and RPP1B were either individually or consecutively disrupted to obtain strains Dp4, Dp6, and Dp46. When required, the KanMX marker was eliminated by recombination using the flanking LoxP sequences (28). The absence of the deleted protein in disrupted strains was checked by SDS-PAGE and Western blot using specific antibodies.
Protein P0 null conditional mutants DpGP0, Dp4GP0, and Dp6GP0 were obtained using a substitution cassette containing the same KanMX marker and a copy of the RPP0 gene coding sequence under the control of the GAL1 promoter (29). In these strains, the wild-type P0 protein is expressed only when cells are grown in galactose. For growing in glucose, the strains depend on exogenous RPP0 genes encoded in transforming plasmids.
Plasmids
pUG35-P2αt
Using appropriate oligonucleotides as primers, a 334-bp DNA fragment containing the RPP2A ORF flanked by BamHI and EcoRI restriction sites was directly obtained by PCR from the S. cerevisiae genomic DNA. The fragment was cloned in the corresponding sites of centromeric vector pUG35, which carries the URA3 genetic marker (Güldener and Hegemann, unpublished data). In this construct the enhanced green fluorescent protein (EGFP) is fused in phase to the C-end of protein P2α, which is under the control of the MET25 promoter.
pFL39-P1βt
As a first step, a pUG35-P1βt plasmid was constructed as described for pUG35-P2αt. Then, a 2042-bp KpnI-SacI fragment containing the MET25 controlled RPP1B-EGFP and the corresponding terminator was obtained from this construct and subcloned into the corresponding sites of centromeric TRP1 pFL39 vector (30).
pFL36-P0t
Similarly, a 952 bp BamHI-EcoRI PCR fragment containing the RPP0 ORF was first cloned into vector pUG23, similar to pUG35 but carrying a HIS3 marker (Güldener and Hegemann, unpublished data). A 3.3-kbp SacI-PstI fragment encoding the EGFP fused P0 protein under the MET25 promoter was subcloned in the centromeric LEU2 pFL36 centromeric vector (30).
Cell transformations
Bacterial transformations were performed according to the method described by Hanahan (31). Yeasts were transformed using lithium acetate as described previously (32).
Cell fractionation and ribosome purification
Cells grown in YEPD medium up to A600 = 0.8 were broken with glass beads, and the ribosomes purified from the cell extracts as previously described (33).
Sample preparation for fluorescence microscopy
For in vitro studies, the purified ribosomes from the EGFP-labeled strains were diluted to a final concentration of 0.25 mg/ml in 100 mM Tris-HCl, pH 7.4, 20 mM KCl, 12.5 mM MgCl2, 5 mM DTT, and single-point FCS measurements were performed.
For in vivo studies, 10 μl of cells growing in minimal medium up to midexponential phase were mixed with 10 μl of melted 2% low-gelling-temperature agarose (Sigma Type VII) in the same medium, deposited in a microscope slide, pressed with a slide cover, and the slide borders were sealed with nail varnish. The samples were immediately taken for fluorescence determination in the two-photon microscope at room temperature. From each strain, 9 to 10 cells were measured at three different orbit centers using the scanning FCS method. At least two preparations from different cell cultures were tested for each strain.
Electrophoretic methods
Proteins were resolved either by 12% SDS-PAGE or by isoelectrofocusing (34) and detected by Western blotting using specific antibodies (35).
Two-photon point and scanning FCS
The two-photon excitation scanning fluorescence microscope used in these experiments was assembled at the Laboratory for Fluorescence Dynamics and has been described previously (36). A mode-locked titanium-sapphire laser with 80-MHz, 100-fs pulse width (Tsunami; Spectra-Physics, Mountain View, CA) was used as the excitation light source at 920 nm. The laser was guided into the microscope by x-y galvanoscanner mirrors (Model 6350; Cambridge Technology, Watertown, MA) to achieve beam scanning in both x and y directions and to direct the laser beam to the desired X-Y position. A photomultiplier tube (Hamamatsu R7400P, Hamamatsu City, Japan) was used for light detection in the photon counting mode. A BG39 optical filter (Chroma Technologies, Brattleboro, VT) was placed before the photomultiplier for suppression of IR excitation light. An Olympus 60× (1.2 N.A.) water immersion objective was used for all measurements.
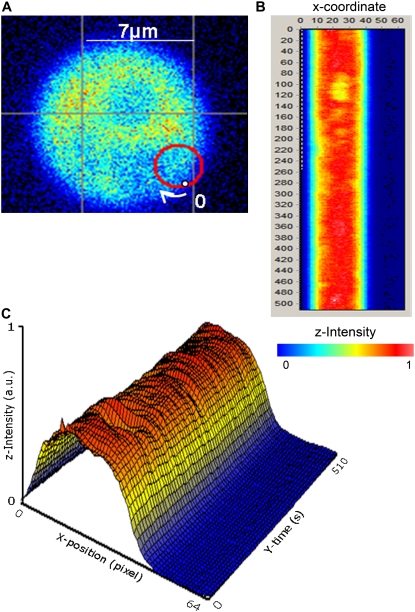
For the single-point FCS measurements a 50-kHz or a 64-kHz sampling frequency was used. For the scanning FCS measurement, the center of the circular scanning path was selected from the fluorescence image. The data acquisition Frequency was set at 64 kHz, with a 1-ms orbit period and radius of 1.52 μm (see Fig. 3 A). Therefore, 64 data points, corresponding to 64 locations in the cell, were collected in each scanning orbit (see Fig. 3 B).
FIGURE 3.
Scanning FCS analysis. (A) Intensity image of a DpGP0-P0t cell showing the scanning orbit (red circle) of 1.52 μm radius used in the scanning FCS data acquisition. The point labeled “0” corresponds to the beginning of the scan (point 0 in the X-position column in the “carpet”) and the end of the scan (Point 63 in the X-position column in the “carpet”). (B) XY transformation of the raw scanning FCS. The X-position columns represent points along one circular scan, and the Y-position rows represent successive scans with each scan taking 1 ms. The color scale indicates the relative intensities of the sections, with orange being the most intense and blue corresponding to intensities outside of the cell. Data were acquired at 64 KHz and 1 ms period. (C) The time-intensity-position data of the “carpet” shown in B are replotted as a surface. The intensity-color scale is the same.
DATA ANALYSIS
Point spread function scaling analysis
In this type of analysis, the point spread function (PSF) is artificially changed by binning adjacent pixels and then calculating the characteristic correlation time of the data averaged. The XY representation of the scanning data is shown (see Fig. 3 B). In this representation, one position inside the cell (orange areas) is chosen and the program (SimFCS) is set to bin three, five, seven, or nine adjacent columns to calculate the average diffusion coefficients after each binning operation. Because the orbit spans regions inside and outside the cell, data can be analyzed at different locations along one orbit as controls.
RESULTS
Expression of EGFP-labeled ribosomal stalk components in S. cerevisiae
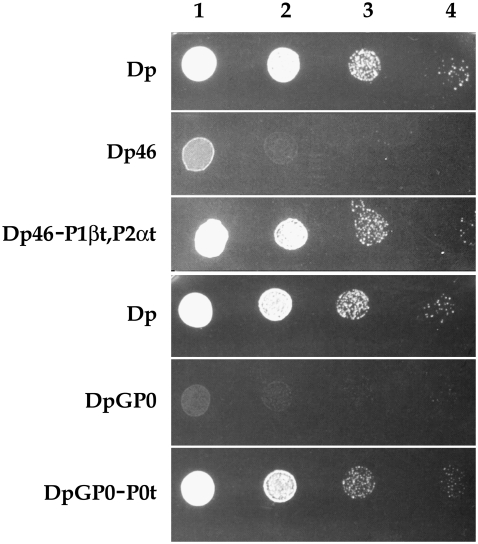
Proteins P0, P1β, and P2α fused to EGFP were expressed in strains lacking the corresponding native proteins derived from S. cerevisiae BJ5458 (Dp) (summarized in Table 1). Tagged proteins complement the absence of the native components in the transformed strains with little or no effect on cell growth (Fig. 1).
FIGURE 1.
Cells (∼104, 103, 102, 10) from the indicated yeast strains growing in rich medium at A600 = 0.8, were spotted in positions 1, 2, 3, and 4, respectively, on a YEPD agar plate and allowed to grow at 30° for 48 h.
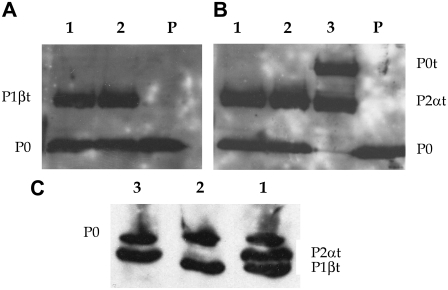
Ribosomes purified from the different strains and the tagged proteins were resolved either by SDS-PAGE and Western blotting (Fig. 2, A and B) or by isoelectrofocusing (Fig. 2 C). The results showed that the amounts of the tagged components are similar whether they are expressed alone in strains Dp6-P1βt and Dp4-P2αt or in pairs in strains Dp46-P2αt, P1βt, and Dp4GP0-P0t, P2αt. Therefore, the presence of the EGFP tag does not appear to affect the capacity of the proteins to complement the absence of the native component or to be a major steric impediment for their interaction with the ribosome.
FIGURE 2.
Estimation of tagged stalk proteins bound to purified ribosomes. Ribosomes from strains Dp46-P2αt,P1βt (1), Dp6-P1βt (2), Dp4-P2αt (3), Dp4GP0-P0t,P2αt (4), and parental Dp (P) were resolved by either SDS-PAGE (A and B) or isoelectrofocusing (C). In A and B, proteins were detected by Western blotting using first specific monoclonal antibodies to protein P2α (A) and P1β (B) and then rabbit antibody to protein P0. In C, proteins were detected using monoclonal antibody 3BH5 to all stalk P proteins.
Fluorescence measurements
FCS data analysis
We carried out two-photon FCS and scanning FCS using strains in which different stalk proteins were tagged with either one or two green fluorescent proteins (EGFP) and then used PCH analysis (25) to distinguish between ribosomes carrying one or two tagged proteins. For point FCS measurement, the fluorescence intensity of the sample at the laser focus was collected as a function of time. The autocorrelation curves of the FCS measurements were calculated using the normalized autocorrelation function (37). For scanning FCS measurements, data points were taken along an orbit (Fig. 3) and saved as a long data string in the same way as the FCS measurements (37). Data segments were transposed to form a fluorescence intensity matrix as a function of time (Fig. 3 B). Thus, each vertical column contains information on the fluorescence intensity fluctuation as a function of time at a particular sample position. Autocorrelation curves and PCHs were calculated from each vertical column, each reflecting a specific position in the cell.
Experimental autocorrelation functions and PCHs were fit using a Gaussian-Lorentzian intensity profile model (25). A beam waist of 0.4 μm calibrated using fluorescein in buffer at pH 8, which has a known diffusion coefficient of 300 μm2/s, was used in the autocorrelation analysis as in previous work (23,38). The SimFCS software (Laboratory for Fluorescence Dynamics, Irvine, CA) was used for all analysis.
Characterization of intracellular diffusion coefficients
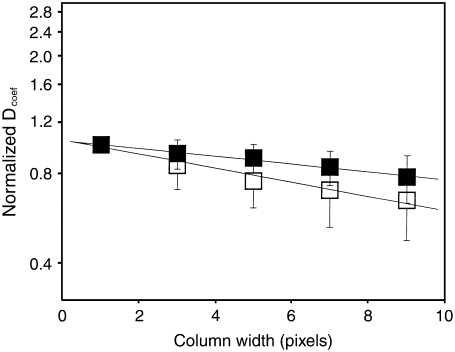
For the purified ribosomes in solution, values in the range of 15 ± 2 μm2/s were found, close to the expected value according to the Stokes-Einstein relation for a 6-MDa molecule diffusing in water (24). For the cellular studies, autocorrelation analysis of all strains showed an average diffusion coefficient around 0.02–0.05 μm2/s. The diffusion coefficients of some proteins inside the cells have been reported to be three to five times slower than in solution (36). The diffusion processes we are measuring in the cells are clearly too slow to be free diffusion of ribosomes in a medium with a viscosity three to five times that of water. On consideration, one might suppose that this slow diffusion could correspond to ribosomes attached to polysomes. However, under the conditions of our experiments (anaerobic and 25°C), a significant portion of the ribosomes are not expected to be associated with polysomes. Moreover, if we were observing diffusion of polysome-associated ribosomes, we would expect to have significantly higher brightness values. One possibility is that the slow diffusion we observe results from binding of the ribosomes to immobile or slowly moving structures. To distinguish binding from diffusion, we analyzed the scanning FCS data by binning adjacent pixels and then calculating the characteristic correlation time of the averaged data. If fluctuations are caused by diffusion, then the characteristic autocorrelation time should become larger as more adjacent pixels are averaged because it will take longer to cross this artificially constructed larger volume. Instead, if there are binding and unbinding processes, the fluctuation rate should remain unchanged, but the amplitude of the signal should decrease. The analysis of several scanning FCS runs using this method is shown in Fig. 4. One notes a continuous decrease of the apparent diffusion coefficient as more columns are averaged, a result compatible with diffusion of very large structures.
FIGURE 4.
Point spread function (PSF) scaling analysis. The horizontal axis is the binning of adjacent (columns) pixels. In the vertical axis the diffusion coefficients are normalized to the average of seven measurements. Solid squares correspond to DpGP0-P0t, and open squares to Dp46-P1βt,P2αt.
Determination of ribosome molecular brightness standards
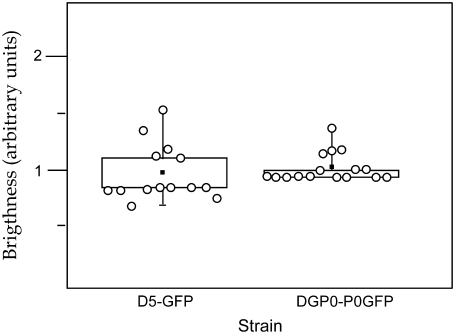
PCH analysis gives the molecular brightness of the ribosomes inside the cells. A comparison of strain Dp5-EGFP, which expresses free EGFP, and strain DpGP0-P0t, which expresses EGFP-tagged P0, showed that the average brightness per particle is similar in both samples (Fig. 5). Therefore the brightness value obtained in the DpGP0-P0t strain was considered as the brightness unit corresponding to one tagged protein bound per ribosome. Similarly, measurements on DpGP0-P0t served to estimate the standard deviation of the technique.
FIGURE 5.
Fluorescence brightness values from strains expressing free EGFP and EGFP-tagged protein P0. The mean value of the determinations is indicated by a black square. The interquartile corresponding to 50% of the determinations in the intermediate range is boxed. The vertical line marks the complete range of values.
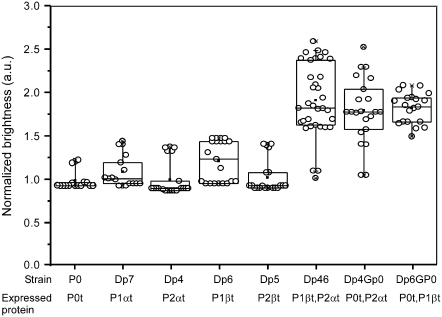
Estimation of tagged stalk components
FCS measurements on strains expressing the tagged proteins P1βt and P2αt, alone or in different combinations (Table 1), were carried out, and the average brightness values per particle are summarized in Fig. 6. When only one tagged acidic protein is expressed, namely in strains Dp4-P2αt and Dp6-P1βt, the average brightness per particle and the corresponding standard deviation are similar to those of the reference DpGP0-P0t strain. When two labeled proteins are simultaneously expressed, either P1βt and P2αt (strain Dp46-P2αt,P1βt), or P0t and one acidic protein (strains Dp4GP0-P0t,P2αt and Dp6GP0-P0t,P1βt), the brightness is about doubled (Fig. 6).
FIGURE 6.
Fluorescence in cells from strains expressing tagged stalk components. The plots are as expressed in the legend to Fig. 5. The number of measurements is represented by the open circles, and the mean value by the solid square.
DISCUSSION
The estimated diffusion coefficients strongly indicate that the tagged ribosomes move very slowly inside the cell. This result suggests either that there are diffusional barriers for particles of this size or that the ribosomes are attached to large structures that diffuse slowly. Nevertheless, most ribosomes have been found free in the cytoplasm and not bound to endoplasmic reticulum in yeast (39). It is possible, that ribosomes are actually loosely associated with either membranes or other slowly moving cellular structures and are released on cell disruption. Further investigation is required to clarify this issue, but it seems that our results provide a new picture of the intracellular ribosome mobility that must be taken into account to understand how protein synthesis takes place in the cell.
The scanning FCS data from the transformed strains expressing one tagged protein indicate that most of the free ribosomes inside the yeast cells carry only one copy of each stalk component. Similarly, when two tagged components are simultaneously expressed, the results show that, on average, close to two fluorescent molecules are bound per particle. Although we have not investigated the full stalk complex but only complexes in which one or two EGFP-labeled proteins are present at any given time, the results presented here are consistent with a model wherein the yeast ribosomal stalk, in vivo, is formed by a complex made of one molecule of each of the acidic proteins, P1α, P1β, P2α, and P2β, and protein P0 as in vitro data suggest (18,40).
Analysis of the dispersion (standard deviation) of the brightness values in some strains (Fig. 6) raises some interesting questions. By comparison with the DpGP0-P0t sample, the data from strains expressing the tagged proteins P2αt and either P0t or P1βt show a dispersion that is considerably larger than the dispersion in strains expressing one acidic protein or even when protein P0t and P1βt are expressed together. The higher data dispersion cannot be the consequence of steric hindrance caused by the EGFP moiety, which, as previously mentioned, does not seem to affect the simultaneous interaction of two tagged proteins.
Alternatively, the data dispersion can be the result of the compositional variability of the eukaryotic ribosomal stalk. Thus, the bound acidic proteins exchange with a free cytoplasmic pool of these components during protein synthesis (7). Moreover, a small fraction of the yeast ribosome population in exponentially growing cells has been shown to carry only two acidic proteins or to be totally depleted of P1/P2 proteins (18), and these fractions apparently increase in stationary phase (20). Cross-linking techniques have also indicated that a fraction of the ribosomes, studies in vitro, can eventually carry P2 dimers, which are stabilized by S-S bonds (17). With this information, the dispersion of the brightness values can result from changes in the stalk composition in a small and variable fraction of the ribosome population. Some ribosomes may lack acidic proteins or carry a dimer of the same protein, not firmly bound to the particle and consequently difficult to detect in purified wild-type ribosomes. The present lack of knowledge concerning the real mechanism of the stalk function makes it difficult to understand the functional significance of these stalk composition variations, but they support the conclusion that the heterogeneity of the ribosome population detected in vitro (11) may also be present in the cell. By the same reasoning, the differences in the data dispersion found in strains Dp4GP0-P0t,P2αt and Dp6GP0-P0t,P1βt (Fig. 6) suggest that the P2α system is more dynamic than the P1β system. Different functions have been previously reported for the two acidic protein families pointing to a determinant role of the P1 proteins in the stalk assembly (41). Although the two protein types probably bind as P1-P2 heterodimers (12), the P1 type seems to play a leading role in the process (14,42). Our FCS studies support the in vitro results and indicate that the differences between the two acidic protein families are also significant inside the cell. Although many questions remain, our study demonstrates that multiphoton FCS methods can be used to investigate ribosome composition and dynamics in vivo.
Acknowledgments
This work was supported by grants BFU2006-00365BMC from the Ministerio de Ciencia y Tecnología (Spain) and by an institutional grant from the Fundación Ramón Areces (Madrid). D.M.J. acknowledges support from National Science Foundation Grant MCB 9808427. The Laboratory for Fluorescence Dynamics is supported by the National Center for Research Resources of the National Institutes of Health PHS 5 P41 RR00315.
Alberto García-Marcos and Susana A. Sánchez contributed equally to this work.
Alberto García-Marcos' present address is Centro de Investigaciones Científicas, Ramiro de Maeztu 9, 28040 Madrid, Spain.
Pilar Parada's present address is BioSigma S.A., Avenida. General San Martin 16.500, Lote 106, Colina, Santiago, Chile.
Editor: David W. Piston.
References
- 1.Diaconu, M., U. Kothe, F. Schlunzen, N. Fischer, J. M. Harms, A. G. Tonevitsky, H. Stark, M. V. Rodnina, and M. C. Wahl. 2005. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell. 121:991–1004. [DOI] [PubMed] [Google Scholar]
- 2.Bocharov, E. V., A. G. Sobol, K. V. Pavlov, D. M. Korzhnev, V. A. Jaravine, A. T. Gudkov, and A. S. Arseniev. 2004. From structure and dynamics of protein L7/L12 to molecular switching in ribosome. J. Biol. Chem. 279:17697–17706. [DOI] [PubMed] [Google Scholar]
- 3.Hamman, B. D., A. V. Oleinikov, G. G. Jokhadze, R. R. Traut, and D. M. Jameson. 1996. Rotational and conformational dynamics of Escherichia coli ribosomal protein L7/L12. Biochemistry. 35:16672–16679. [DOI] [PubMed] [Google Scholar]
- 4.Hamman, B. D., A. V. Oleinikov, G. G. Jokhadze, R. R. Traut, and D. M. Jameson. 1996. Dimer/monomer equilibrium and domain separations of Escherichia coli ribosomal protein L7/L12. Biochemistry. 35:16680–16686. [DOI] [PubMed] [Google Scholar]
- 5.Ban, N., P. Nissen, J. Hansen, P. B. Moore, and T. A. Steitz. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 289:905–920. [DOI] [PubMed] [Google Scholar]
- 6.Schuwirth, B. S., M. A. Borovinskaya, C. W. Hau, W. Zhang, A. Vila-Sanjurjo, J. M. Holton, and J. H. Cate. 2005. Structures of the bacterial ribosome at 3.5 A resolution. Science. 310:827–834. [DOI] [PubMed] [Google Scholar]
- 7.Ballesta, J. P. G., and M. Remacha. 1996. The large ribosomal subunit stalk as a regulatory element of the eukaryotic translational machinery. Prog. Nucleic Acid Res. Mol. Biol. 55:157–193. [DOI] [PubMed] [Google Scholar]
- 8.Tsurugi, K., and K. Ogata. 1985. Evidence for the exchangeability of acidic ribosomal proteins on cytoplasmic ribosomes in regenerating rat liver. J. Biochem. 98:1427–1431. [DOI] [PubMed] [Google Scholar]
- 9.Scharf, K.-D., and L. Nover. 1987. Control of ribosome biosynthesis in plant cell cultures under heat shock conditions. II. Ribosomal proteins. Biochim. Biophys. Acta. 909:44–57. [DOI] [PubMed] [Google Scholar]
- 10.Remacha, M., A. Jimenez-Diaz, C. Santos, R. Zambrano, E. Briones, M. A. Rodriguez Gabriel, E. Guarinos, and J. P. G. Ballesta. 1995. The proteins P1, P2, and P0, components of the eukaryotic ribosome stalk. New structural and functional aspects. Biochem. Cell Biol. 73:959–968. [DOI] [PubMed] [Google Scholar]
- 11.Guarinos, E., M. Remacha, and J. P. G. Ballesta. 2001. Asymetric interactions between the acidic P1 and P2 proteins in the Saccharomyces cerevisiae ribosomal stalk. J. Biol. Chem. 276:32474–32479. [DOI] [PubMed] [Google Scholar]
- 12.Tchorzewski, M., D. Krokowski, A. Boguszewska, A. Liljas, and N. Grankowski. 2003. Structural characterization of yeast acidic ribosomal P proteins forming the P1A–P2B heterocomplex. Biochemistry. 42:3399–3408. [DOI] [PubMed] [Google Scholar]
- 13.Tchorzewski, M., B. Boldyreff, O. Issinger, and N. Grankowski. 2000. Analysis of the protein-protein interactions between the human acidic ribosomal P-proteins: evaluation by the two hybrid system. Int. J. Biochem. Cell Biol. 32:737–746. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalo, P., J. P. Lavergne, and J. P. Reboud. 2001. Pivotal role of the P1 N-terminal domain in the assembly of the mammalian ribosomal stalk and in the proteosynthetic activity. J. Biol. Chem. 276:19762–19769. [DOI] [PubMed] [Google Scholar]
- 15.Uchiumi, T., A. J. Wahba, and R. R. Traut. 1987. Topography and stoichiometry of acidic proteins in large ribosomal subunits from Artemia salina as determined by crosslinking. Proc. Natl. Acad. Sci. USA. 84:5580–5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zurdo, J., J. M. Sanz, C. Gonzalez, M. Rico, and J. P. G. Ballesta. 1997. The exchangeable yeast ribosomal acidic protein YP2beta shows characteristics of a partly folded state under physiological conditions. Biochemistry. 36:9625–9635. [DOI] [PubMed] [Google Scholar]
- 17.Qiu, D., P. Parada, A. G. Marcos, D. Cardenas, M. Remacha, and J. P. Ballesta. 2006. Different roles of P1 and P2 Saccharomyces cerevisiae ribosomal stalk proteins revealed by cross-linking. Mol. Microbiol. 62:1191–1202. [DOI] [PubMed] [Google Scholar]
- 18.Guarinos, E., C. Santos, A. Sanchez, D. Y. Qiu, M. Remacha, and J. P. Ballesta. 2003. Tag-mediated fractionation of yeast ribosome populations proves the monomeric organization of the eukaryotic ribosomal stalk structure. Mol. Microbiol. 50:703–712. [DOI] [PubMed] [Google Scholar]
- 19.Montoya-Garcia, L., V. Munoz-Ocotero, R. Aguilar, and E. Sanchez de Jimenez. 2002. Regulation of acidic ribosomal protein expression and phosphorylation in maize. Biochemistry. 41:10166–10172. [DOI] [PubMed] [Google Scholar]
- 20.Saenz-Robles, M. T., M. Remacha, M. D. Vilella, S. Zinker, and J. P. G. Ballesta. 1990. The acidic ribosomal proteins as regulators of the eukaryotic ribosomal activity. Biochim. Biophys. Acta. 1050:51–55. [DOI] [PubMed] [Google Scholar]
- 21.Remacha, M., A. Jimenez-Diaz, B. Bermejo, M. A. Rodriguez-Gabriel, E. Guarinos, and J. P. G. Ballesta. 1995. Ribosomal acidic phosphoproteins P1 and P2 are not required for cell viability but regulate the pattern of protein expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 15:4754–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mauro, V. P., and G. M. Edelman. 2002. The ribosome filter hypothesis. Proc. Natl. Acad. Sci. USA. 99:12031–12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berland, K. M., P. T. C. So, and E. Gratton. 1995. Two-photon fluorescence correlation spectroscopy: Method and application to the intracellular environment. Biophys. J. 68:694–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen, Y., J. D. Muller, K. M. Berland, and E. Gratton. 1999. Fluorescence fluctuation spectroscopy. Methods. 19:234–252. [DOI] [PubMed] [Google Scholar]
- 25.Muller, J. D., Y. Chen, and E. Gratton. 2000. Resolving heterogeneity on the single molecular level with the photon-counting histogram. Biophys. J. 78:474–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen, Y., J. D. Muller, Q. Ruan, and E. Gratton. 2002. Molecular brightness characterization of EGFP in vivo by fluorescence fluctuation spectroscopy. Biophys. J. 82:133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez, E., B. Segui-Real, E. Silles, M. J. Mazon, and I. V. Sandoval. 1999. The prepropeptide of vacuolar aminopeptidase I is necessary and sufficient to target the fluorescent reporter protein GFP to the vacuole of yeast by the Ccvt pathway. Mol. Microbiol. 33:52–62. [DOI] [PubMed] [Google Scholar]
- 28.Güldener, U., S. Heck, T. Fielder, J. Beinhauer, and J. H. Hegemann. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24:2519–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santos, C., and J. P. Ballesta. 1994. Ribosomal protein P0, contrary to phosphoproteins P1 and P2, is required for ribosome activity and Saccharomyces cerevisiae viability. J. Biol. Chem. 269:15689–15696. [PubMed] [Google Scholar]
- 30.Bonneaud, N., O. Ozier-Kalogeropoulos, G. Li, M. Labouesse, L. Minvielle-Sebastia, and F. Lacroute. 1991. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast. 7:609–615. [DOI] [PubMed] [Google Scholar]
- 31.Hanahan, D. 1985. Techniques for transformation of E. coli. In DNA Cloning: A Practical Approach. D. M. Glover, editor. IRL Press, Oxford. 109–136.
- 32.Gietz, R. D., R. H. Schiestl, A. R. Willems, and R. A. Woods. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS- DNA/PEG procedure. Yeast. 11:355–360. [DOI] [PubMed] [Google Scholar]
- 33.Perez-Fernandez, J., M. Remacha, and J. P. Ballesta. 2005. The acidic protein binding site is partially hidden in the free Saccharomyces cerevisiae ribosomal stalk protein P0. Biochemistry. 44:5532–5540. [DOI] [PubMed] [Google Scholar]
- 34.Zambrano, R., E. Briones, M. Remacha, and J. P. G. Ballesta. 1997. Phosphorylation of the acidic ribosomal P proteins in Saccharomyces cerevisiae. A reappraisal. Biochemistry. 36:14439–14446. [DOI] [PubMed] [Google Scholar]
- 35.Vilella, M. D., M. Remacha, B. L. Ortiz, E. Mendez, and J. P. G. Ballesta. 1991. Characterization of the yeast acidic ribosomal phosphoproteins using monoclonal antibodies. Proteins L44/L45 and L44′ have different functional roles. Eur. J. Biochem. 196:407–414. [DOI] [PubMed] [Google Scholar]
- 36.Ruan, Q., Y. Chen, E. Gratton, M. Glaser, and W. W. Mantulin. 2002. Cellular characterization of adenylate kinase and its isoform: two-photon excitation fluorescence imaging and fluorescence correlation spectroscopy. Biophys. J. 83:3177–3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruan, Q., M. A. Cheng, M. Levi, E. Gratton, and W. W. Mantulin. 2004. Spatial-temporal studies of membrane dynamics: scanning fluorescence correlation spectroscopy (SFCS). Biophys. J. 87:1260–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanchez, S. A., Y. Chen, J. D. Muller, E. Gratton, and T. L. Hazlett. 2001. Solution and interface aggregation states of Crotalus atrox venom phospholipase A2 by two-photon excitation fluorescence correlation spectroscopy. Biochemistry. 40:6903–6911. [DOI] [PubMed] [Google Scholar]
- 39.Schneider, E., E. R. Lochmann, H. Lother. 1976. Distribution of membrane-bound and free ribosomes in growing yeast. Biochem. Biophys. Acta. 432:92–97. [DOI] [PubMed] [Google Scholar]
- 40.Krokowski, D., A. Boguszewska, D. Abramczyk, A. Liljas, M. Tchorzewski, and N. Grankowski. 2006. Yeast ribosomal P0 protein has two separate binding sites for P1/P2 proteins. Mol. Microbiol. 60:386–400. [DOI] [PubMed] [Google Scholar]
- 41.Ballesta, J. P. G., E. Guarinos, J. Zurdo, P. Parada, G. Nusspaumer, V. S. Lalioti, J. Perez-Fernandez, and M. Remacha. 2000. Structure of the yeast ribosomal stalk. In The Ribosome. Structure, Function, Antibiotics and Cellular Interactions. R. Garrett, S. Douthwaite, A. Matheson, A. Liljas, P. B. Moore, and H. F. Noller, editors. American Society for Microbiology. Washington. 115–125.
- 42.Zurdo, J., C. Gonzalez, J. M. Sanz, M. Rico, M. Remacha, and J. P. G. Ballesta. 2000. Structural differences between Saccharomyces cerevisiae ribosomal stalk proteins P1 and P2 support their functional diversity. Biochemistry. 39:8935–8943. [DOI] [PubMed] [Google Scholar]