Abstract
We designed a micromanipulation device that allows the local application of a constant force on living cells, and the measurement of their stiffness. The force is applied through an Arg-Gly-Asp-coated bead adhering on the cell and trapped in optical tweezers controlled by a feedback loop. Epifluorescence observations of green fluorescent protein-actin in the cells are made during force application. We observe a stiffening of cells submitted to a constant force within a few minutes, coupled to actin recruitment both at the bead-cell contact and up to several micrometers from the stress application zone. Moreover, kinetics of stiffening and actin recruitment exhibit a strong correlation. This work presents the first quantification of the dynamics of cell mechanical reinforcement under stress, which is a novel insight into the elucidation of the more general phenomenon of cell adaptation to stress.
INTRODUCTION
Adherent cells are highly sensitive to their mechanical environment which they feel through adhesion molecules embedded in the cell membrane. Among these various binding proteins, integrins link the extracellular matrix to the cell actin cytoskeleton via a dynamic complex of proteins (1,2). These clusters, named focal adhesions (FA), are several micrometers in size and act as mechanosensors (3).
Cellular response to the mechanical environment is based on the mechanotransduction that occurs within FA, and implies processes that happen on very different timescales. Long-term responses include, for example, adaptation of the traction force to the substrate rigidity (4), spreading (5), motility (6–8), or even control of the cell cycle (differentiation, apoptosis) (9–11). Shorter-term responses involve both physiological and mechanical phenomena: mechanical strengthening (12), growth and modification of the biological composition of the existing contacts (13,14), assembly of new focal complexes (15), and enhanced contractility of the actin cytoskeleton (16). Yet the broad cascade of biochemical signaling (17–19) involved in mechanotransduction at the cell membrane has not been totally elucidated.
Cells mechanical integrity is crucial for these adhesion-dependent mechanisms. Hence viscoelastic behaviors of the cells have been widely studied in the aim of establishing a link between their mechanical properties and the biological state of their cytoskeleton (20–23).These investigations lead to identifying actin filaments, and the acto-myosin contractility in particular, as major components of the cells viscoelastic response (24,25) and of their adaptation to stress (4,26).
We set out to focus on the process of cell stiffening under stress that has already been reported previously. It has been shown, for instance, that focal contacts are strengthened by the application of an external force (12) and grow in the direction of the applied force (18). The application of a fluid shear stress on adherent cells causes a dramatic increase in their viscosity, and the actomyosin contractility has been shown to be involved in this process (27). By applying a local force it has also been shown that cell rigidity increases (28) and actin remodels in the vicinity of the applied force (29,30).
To investigate the link between these phenomena and to quantify their dynamics, we designed a method that allows us to both locally apply a constant force, and make epifluorescence observations. Force application is performed via an optically trapped silica microbead. The bead position is measured on a quadrant photodiode detector (see Supplementary Material Fig. S1). A feedback loop (31) is implemented to keep the force on the bead constant. We measure the local creep function and retrieve the cell viscoelastic parameters. We apply a temporal series of step forces so as to have access to the evolution of cell stiffness, and we compare it to the reorganization of green fluorescent protein(GFP)-actin in the cell. We show that the rigidity and the quantity of actin in the vicinity of the force application zone both increase, and that the kinetics of these two phenomena are very well correlated. We propose a typical timescale for actin recruitment and cell stiffening.
MATERIALS AND METHODS
Cell preparation
Both A549 human alveolar epithelial cells (American Type Culture Collection, Rockville, MD) and C2C12 mice myoblastic cells, kindly provided by M. Lambert (Institut du Fer à Moulin, Paris, France), were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 2 mM glutamine, 100 U/mL penicillin, and 50 mg/mL streptomycin (Invitrogen, Carlsbad, CA). Twenty-four hours before manipulation, cells were detached from culture flasks with a trypsin-EDTA solution and plated on a glass coverslip coated with 5 μg/mL fibronectin (Sigma, St. Louis, MO). For epifluorescence observations, GFP-actin plasmid transfection was performed with nanofectin (PAA Laboratories, Pasching, Austria), according to the manufacturer's procedure, ∼15 h before experiment. The plasmid was kindly provided by M. Coppey (Institut Jacques Monod, Paris, France).
Bead coating
Carboxylated silica beads (3.47 μm diameter; Bangs Laboratories, Fishers, IN) were coated with a polypeptide containing the Arg-Gly-Asp (RGD) sequence (PepTide 2000; Telios Pharmaceuticals, San Diego, CA), according to the manufacturer's procedure. It ensured a specific binding to integrin receptors. Beads were then incubated on cells at 37°C for 15 min before manipulation (1–3 beads per cell).
Force application
Our optical tweezers setup, represented in Supplementary Material Fig. S1, is based on a 1064 nm Nd:YAG laser, 600 mW intensity (Spectra Physics, Mountain View, CA), which is focused through the objective of an inverted microscope (model No. DM IRB, ×100 oil immersion objective, 1.25 NA; Leica, Wetzlar, Germany). Galvanometric mirrors are used to deflect the laser beam, and thus displace the optical trap in the XY plane. The position of the bead in the XY plane, rbead, is measured on a quadrant photodiode (S1557; Hamamatsu, Hamamatsu City, Japan). Its spatial resolution is limited by several noise sources: light intensity fluctuations, amplification, and sampling devices noises. The raw acquired signal yields the bead position with a 50 nm resolution. The use of a quadrant detector is essential to achieve a high temporal resolution (much higher than could be achieved through the use of a charge-coupled device camera with an image analysis software), and allows an averaging of the signal over four running data points. This eventually decreases the error on the position down to 25 nm.
The quadrant diode electrical signal is amplified as described by Simmons et al. (31), yielding the analogical value Vdiode = (VX, VY) = (A Xbead, A′ Ybead). The proportional coefficients A and A′ are calibrated before each experiment. The experimental chamber is mounted on an XYZ piezoelectric nanopositioning device (NanoCube, Physik Intrumente, Karlsruhe, Germany), fixed to the microscope stage to allow precise movements of the experimental chamber in three dimensions.
Before any experiment, trap force calibration is performed using the hydrodynamic flow method described in more detail by Balland et al. (24). It gives Ftrap as a nearly linear function of the distance δr between the center of the bead and the center of the trap: ‖δr‖ = ‖rbead − r0‖ for 0 ≤ δr ≤ 1.5 μm. Trap stiffness, depending on the laser power, lies within the range 35–120 pN/μm.
To apply a constant force on the bead, in modulus and direction, δr(t) is kept constant by moving the piezoelectric stage on which the chamber is mounted. This is achieved numerically by a feedback loop under LabVIEW (National Instruments, Austin, TX). The program samples Vdiode at 180 Hz and commands the piezo stage movement rpiezo(t) with a 45 Hz bandwidth. The piezo stage response time is of ∼0.02 s, but the response time of the whole feedback loop depends on the cell rigidity. For very weak cells, large instantaneous displacements are required at t = 0. The force clamp reaches its full efficiency within a maximum time Tmax = 0.1 s. Hence, we measure the instantaneous displacement of the bead with respect to the cell r(t) = −rpiezo(t) in response to a constant force F0.
Measurement of the creep function
When a constant stress is applied to a material (σ = 0 when t < 0, σ = σ0 when t ≥ 0), the creep function is defined as
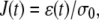 |
(1) |
where ɛ(t) is the induced strain. Here we apply a constant force step F0 and measure the bead displacement r(t). The relationships between force and stress, respectively, and displacement and strain, in a simple linear elasticity model described previously (32), yields
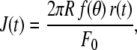 |
(2) |
where R is the bead radius (1.735 μm) and θ is half the angle of the immersion cone of the bead into the cell. The value θ is roughly estimated for each cell on transmission images (refer to (23,32) and Supplementary Material Fig. S2 for more details). The geometrical factor f(θ) is given by
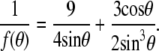 |
(3) |
Fluorescence detection
To follow the evolution of actin density around the bead, we visualize GFP-tagged actin in the cell using the same objective as for trapping. Before experiment, and after each step force application and measurement of the creep function, at time tk (k = 1…N) a stack of epifluorescence images along the z axis is captured with a charge-coupled device camera (Coolsnap ES; Roper Scientific, Trenton, NJ) using an ImageJ plugin. The images are then processed using another ImageJ procedure: for each time tk we define a set of four planes along the z axis (located around the plane of force application, and separated by ∼1 μm) over which we average the fluorescence intensity. In this averaged image, we choose different regions of interest (ROI): ROIbleach, and ROIi, i = 1 to 9.
ROIbleach is a control region, taken in the cell far enough from the bead center (∼6–8 μm) not to be modified by the local force application. The average intensity in this zone, Ibleach, is used as a reference to check the degree of bleaching during the course of the experiment. ROIi are disks of radii R1 = 2.25 to R9 = 6.25 μm (Ri+1 − Ri = 0.5 μm) centered on the bead.
ROI2, which is approximately twice the area of the bead itself, is named ROIbead, and contains the information about short-range actin recruitment around the bead. In this region, bright patches of actin appear at the bead-cell contact during the experiments (see Fig. 3). To evaluate the quantity of actin they contain, we identify and label the patches by indices p using an image treatment that is similar to the one described by Zamir et al. (33). We calculate the area Ap, and average intensity 〈Ip〉 of each patch p and the quantity of actin recruited at the bead-cell contact at time tk, is eventually estimated as
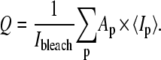 |
(4) |
These patches are fitted by ellipses, from which we retrieve the major and minor axes and the orientation angle (see Fig. 4). We also evaluate the actin present in the network around the bead, by the average fluorescence intensity in ROIbead, still taking into account the bleaching factor:
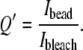 |
(5) |
We perform the same measurements, Q′i, in the disks ROIi. Then, we calculate the average intensity in successive rings around the bead, of radii δRi = (Ri+1 + Ri)/2, by retrieving the differential quantities δQ′i:
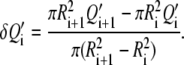 |
(6) |
The values of Q′i and Q are related to actin quantities but are not directly proportional to them. This is why they are only used relatively to the values measured at different times tk within a given experiment.
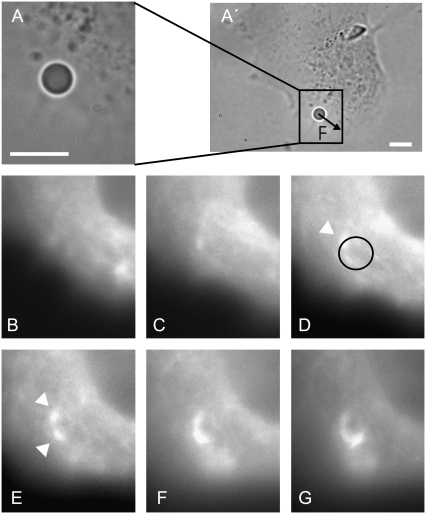
FIGURE 3.
A C2C12 cell during force application: transmission image with an indication of the force direction (A′); a zoom around the bead (A); fluorescence images of GFP-actin at times t1 = 420 s (B), t3 = 1080 s (C), t5 = 1440 s (D), t7 = 1920 s (E), t9 = 2400 s (F), and t10 = 2640 s (G). Arrowheads in images D and E point at adhesion patches. A circle representing the bead in size and position is superimposed on image D. (Bars: 5 μm.)
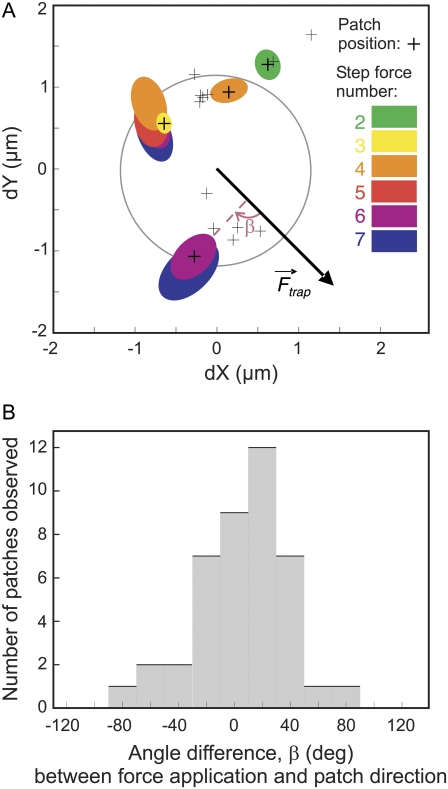
FIGURE 4.
(A) Graphic of the actin recruited in FA around the bead during one experiment. The arrow gives the force direction. The ellipses are the best fits for the actin patches detected (crosses indicate their centers), and are labeled according to the image number on which they are measured. For patches present on step forces number-8 and more, only the centers are displayed. (B) Distribution of the actin patches orientations. Angles are calculated relatively to the direction of force application, as displayed on the top panel.
RESULTS AND DISCUSSION
Creep function and cell viscoelastic parameters
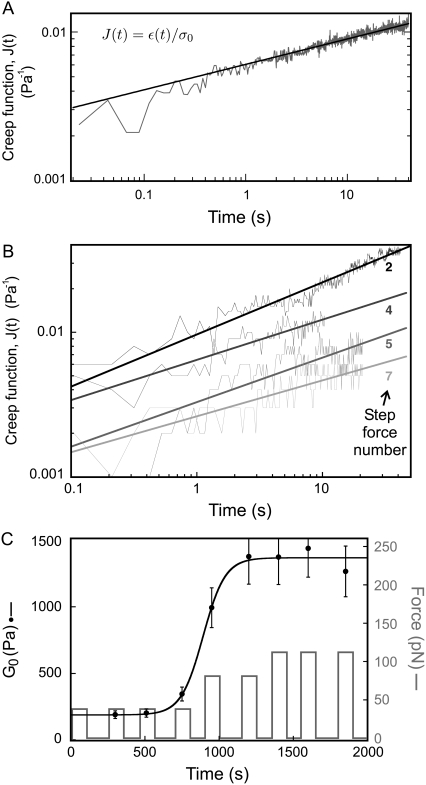
We performed measurements on several cells of two different cell lines: A549 human alveolar epithelial cells and C2C12 mice myoblastic cells. By applying a constant stress on a cell through a RGD-coated bead, and measuring the resulting strain, we retrieved the local creep function (see Materials and Methods). Fig. 1 A shows a typical example of creep function measured on a C2C12 cell. It is well fitted by a power law over more than two time decades: J(t) = A (t/t0)α, where t0 is a reference time, chosen here equal to 1 s. The fit yields α = 0.18 ± 0.01 and A = 5.9 ± 0.8 × 10−3 Pa−1 on this particular example (see Supplementary Material Note S4).
FIGURE 1.
Mechanical measurements on single cells. (A) Creep function of a single C2C12 cell as a function of force application time, fitted by a power law J(t) = 5.9.10−3 t0.18 Pa−1 (in log-log scale). (B) Successive creep functions on a single A549 cell: a series of step forces is applied. For each step, the creep function is measured. For better readability, J(t) is displayed for only four of the nine-step force applications: number 2, 4, 5, and 7. (C) The corresponding viscoelastic modulus G0 versus time, fitted by a sigmoid with a rising time τG = 75 s. Mechanical saturation is reached in this example.
This weak power law, with an exponent of 0.1 to 0.5, is characteristic of the rheology of cells and more generally of materials having a large number of relaxation times distributed over a broad timescale (20,22,23,34).
To interpret our results in terms of cell rigidity, and to compare them with previous measurements, we retrieve the equivalent viscoelastic modulus at 1 Hz, G0. For a creep function that is a power law, G0 is calculated from A and α using the relation
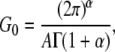 |
(7) |
which has been demonstrated previously (23) (Γ is the Euler γ-function). For example, the creep measurement displayed on Fig. 1 A yields G0 = 257 ± 38 Pa (see Supplementary Material Fig. S3 and Note S4 for comments on the error estimates).
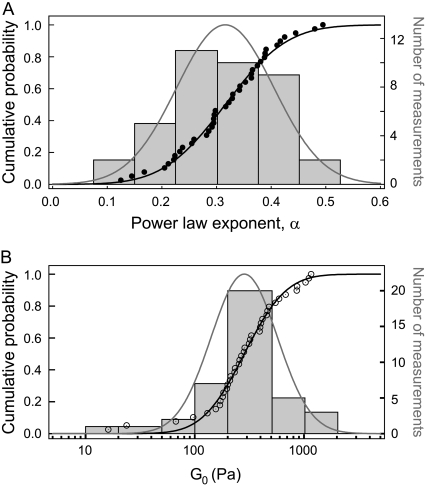
Statistical repartitions show a Gaussian (or normal) distribution for the power-law exponent α, and a log-normal distribution for the prefactor G0, as evidenced on Fig. 2. The mean values and widths of the distributions are obtained by fitting the cumulative probabilities of both α and log(G0) by error functions:
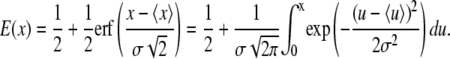 |
(8) |
The average values and distribution widths measured for the power-law exponent α, and the median value of the viscoelastic modulus G0 are 〈α〉 = 0.300, σα = 0.100, and G0, med = 305 Pa on A549 cells; and 〈α〉 = 0.316, σα = 0.089, and G0, med = 285 Pa on C2C12 cells. The mechanical parameters obtained for all the tested cells are summarized in Table 1. These results are consistent with previous measurements on cells probed locally or globally, for harmonic or quasistatic forcings (see data reported in (20–24,34)) and with passive microrheology measurements (35,36). Our data corroborate a large set of mechanical measurements performed on different cell types, which leads us to assert that our estimate of the viscoelastic modulus G0 is a reliable measurement of the local cell stiffness.
FIGURE 2.
Distribution histograms and cumulative probability functions of the power-law parameters: exponent-α (A) and viscoelastic modulus G0 (B), measured on 39 different C2C12 cells.
TABLE 1.
Mechanical parameters calculated from creep measurements performed on A549 cells and C2C12 cells with RGD-coated beads
| Cell type | Nb of cells | 〈α〉 | σα | 〈log G0〉 | σ log Go | G0, med (Pa) | 〈G0〉 (Pa) |
|---|---|---|---|---|---|---|---|
| A549 | 22 | 0.300 | 0.100 | 2.48 | 0.375 | 305 (−175/+420) | 480 |
| C2C12 | 39 | 0.316 | 0.089 | 2.45 | 0.475 | 285 (−140/+570) | 375 |
The “〈〉s” denote average values, σ the width of the distribution, and G0,med is the median value of the viscoelastic modulus G0, given by 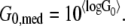
Rigidity evolution on long timescales
To investigate the dynamics of mechanical response under external stress, we followed the evolution of cell rigidity during force application. The magnitude of the applied force was set between 40 and 180 pN. We applied a series of 150 s step forces and measured the creep function during each step. Each step force was followed by an equal time-lapse left for mechanical relaxation of the cell. The main reason for applying a series of force steps rather than a continuous force was to keep the cell deformation small, to remain within the linear regime for strain. The creep function measurements are performed over the maximum time range 0.02–50 s, during which major cell movements or mechanical shifts in the setup can be considered negligible. The reason for using nonmotile cells (C2C12 myoblastic cell line) and for avoiding beads attached to a lamellipodium-like region was to limit the disturbing effects due to membrane ruffling and actin retrograde flow.
A typical series of measurements performed on a single A549 cell is shown in Fig. 1 B. The creep function clearly decreases with the step force application number, meaning that the cell deforms less and less as a force is exerted on it. The cell still exhibits a power law rheology in the time range of 0.1 to 10–20 s, with an exponent α that almost remains the same, but a prefactor A that gets smaller. Hence the modulus G0, which gives the value of the local cell rigidity, increases. We can quantify the stiffening dynamics by plotting the variations of G0 versus time (see Fig. 1 C). For approximately two thirds of the cells tested, we observed such an increase in the cell rigidity in response to force application. All the stiffening curves exhibit a sigmoidlike behavior, even if the saturation in the value of G0 is not always reached. To restrain to a small number of fitting parameters and get results that can easily be displayed as a whole, we use a dimensionless viscoelastic modulus g(t):
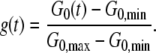 |
(9) |
G0, min and G0, max are, respectively, the minimum and maximum values measured during the course of the experiment. The value g(t) can then be fitted by a sigmoid function,
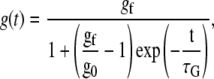 |
(10) |
with a set of three fitting parameters: gf, g0, and τG. The value τG is the stiffening time; g0, which is close to zero, takes into account the fact that the first measured value G0(t0) is not exactly equal to the modulus of the cell at rest, since we exert a perturbation as soon as we start measuring the cell viscoelasticity. The parameter gf is approximately equal to 1 if mechanical saturation is reached (that is, if the last measured value G0(tN) is the cell maximum response to stress), otherwise gf ≥ 1. In Fig. 1 C, for example, G0, min = 190 Pa, G0, max = 1355 Pa, g0 = 7.3.10−7, gf = 1.08, and τG = 90 s for this A549 cell.
Table 2 summarizes the values obtained for C2C12 cells. Between the first and the last step force application, the value of the viscoelastic modulus G0 increases by almost one order of magnitude: G0, min is typically of ∼250 Pa while G0, max is ∼900 Pa. Average, minimum, and maximum values of the parameter τG are also displayed. In these myoblastic cells, the stiffening phenomenon takes place on a timescale of 600 s on average, with some variability from cell to cell. The value τG seems to be greater for C2C12 cells than for A549 cells, but the statistics on A549 cells is too poor to conclude about this point.
TABLE 2.
Mechanical and actin data for C2C12 cells
| G0, ini (Pa) | G0, max (Pa) | τG (s) | τQ (s) | τ′Q (s) | |
|---|---|---|---|---|---|
| mean | 250 | 900 | 630 | 385 | 555 |
| min | 100 | 420 | 90 | 115 | 225 |
| max | 1000 | 2100 | 2000 | 835 | 875 |
Data from 11 experiments in which stiffening and actin recruitment were observed: mean, minimum, and maximum values of G0,ini and G0,max, and of the fitting parameters τG, τQ, and τ′Q.
Actin recruitment around the bead
Actin is known to be one of the key elements in the regulation of cell rigidity (18,37,38). To investigate its role in the phenomenon of stiffening under stress that we observed, we transfected C2C12 cells with a plasmid coding for GFP-actin and followed the actin quantity around the bead.
In more than half of the experiments, such as the one shown in Fig. 3, bright actin patches appeared at the bead-cell contact. Such patches appeared around beads submitted to a force, while they were never present on other cell-bound beads on which we did not apply any force.
When such actin patches could be detected in an experiment, they appeared after the second or third force step. They started as two or three small elongated dots, ∼0.2 × 0.5 μm2 in size, then grew, and eventually merged into an actin ring that surrounded the bead (see Fig. 3, B–G). These observations are consistent with previous observations of actin recruitment within focal adhesions under force application (4,18). These actin-containing patches, which we observed to develop at the bead-cell contact when a force is applied, are thus probably FA.
In the image analysis procedure (see Materials and Methods), the shape of each actin patch was fitted by an ellipsis, defined by its major and minor axes and its orientation angle. This allowed us to follow their direction and size during force application (see Fig. 4 A). The ellipses were elongated in a direction close to the direction of force application (see Fig. 4 B): 70% of their orientations differ by less than 20° from the force direction.
Additional important information was the position of these patches around the bead, with regard to the force direction. They appeared preferentially ahead of and behind the bead, which are the regions of highest strain and stress fields (39), both in compression and elongation. This growth of FA in the direction of the applied force is consistent with previous experimental observations (18) and theoretical predictions (40).
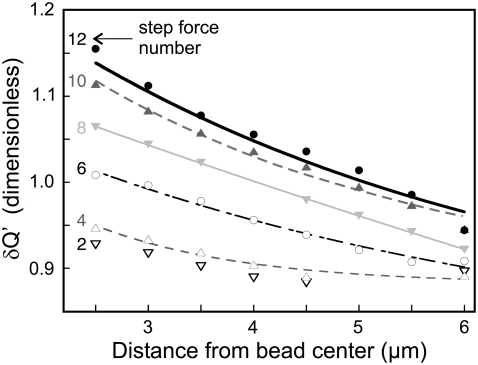
A quantification of the actin contained in these contacts is given by the value Q(t). We also measured the global actin density in the network around the bead: it is given by Q′(t), the average fluorescence intensity in a disk approximately twice the area of the bead. Both calculations are described in Materials and Methods. Most experiments exhibited an increase in Q′ with time, even when actin dots could not be detected. To test whether this could be attributed to a reinforcement in the actin network around the bead, we measured the average fluorescence intensity δQ′i in rings of increasing radii δRi around the bead: from δR1 = 2.5 to δR8 = 6.0 μm (see Materials and Methods and Fig. 5). This is an estimate of the intermediate-range actin recruitment within the cell. We observed that the actin quantity in those rings increased during the course of the experiments, and that the increase was substantial even far from the bead. Fig. 5 shows the variation of this actin density with the distance r from the bead center, for different force-application times. The observed decrease of δQ′ with r is fitted by an exponential decay, which yields a cutoff radius of ∼3–4 μm. The actin network is thus reinforced not only at the bead-cell contact, but also up to a distance of several microns from the force-application zone. This length scale is of the same order of magnitude as the characteristic length for stress and strain field decay (39,41).
FIGURE 5.
Plot of the actin density δQ′i in successive rings around the bead (of radii δR1 = 2.5 to δR8 = 6.0 μm), at different step force application times tk, k = 2 (open down-triangles), 4 (open up-triangles), 6 (open circles), 8 (shaded down-triangles), 10 (shaded up-triangles), and 12 (solid circles). During force application, the actin quantity increases even far from the bead. An exponential decay fit δQ′(r) = A + B exp(– r/Rc) yields a cutoff radius Rc of ∼3.5 μm (1.4, 5.2, 3.25, and 4.55 μm, respectively, for t4, t6, t10, and t12).
On the positive experiments during which we could observe actin recruitment, we eventually stopped the force application to let the cell relax. During this relaxation time, we kept acquiring images, to see whether the fluorescence would decrease. Such a decrease, down to the intensity level observed before force application, was observed in more than half of the cases, but not all of them. Due to this variability in the cells relaxation behavior, we cannot conclude on any relaxation timescale.
Control experiments were performed to ensure that the actin recruitment was not due to attraction of cell material toward the optical trap. We performed the same temporal series of tweezers application as in the real experiments, but without trapping any bead on the cell. We did not see any increase in the actin quantity around the laser application zone.
Correlation between the rigidity increase and actin recruitment
To compare the kinetics of actin recruitment observed in all the experiments, we plotted the time evolution of both dimensionless actin quantities, 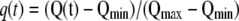 and
and 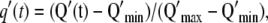 that are defined in the same way as g(t).
that are defined in the same way as g(t).
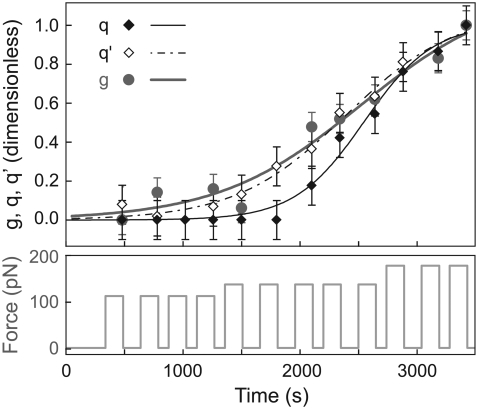
We fitted these data by sigmoids with a similar set of three parameters as for g: (q0, qf, τQ) and (q′0, q′f, τ′Q). Fig. 6 shows the result of an experiment in which g, q, and q′ increase with very similar behaviors. The most interesting parameters of the fit, the timescales, are τG = 612 s, τQ = 300 s, and τ′Q = 470 s, in this example. As summarized in Table 2, for all the cells tested, we observed typical recruitment times of a few hundreds of seconds. It should be noted that, despite cell-to-cell variability, scattering of the timescales τG, τQ, and τ′Q turns out to be rather small.
FIGURE 6.
Normalized cell viscoelastic modulus g(t) (shaded dots), actin quantities q(t) (solid diamonds), and q′(t) (open diamonds) versus time, during a series of step-force applications. Fitting this experiment by a sigmoid yields the parameters {q0 = 1.9.10−4, qf = 1.01, τQ = 300 s}; {q′0 = 6.2.10−3, q′f = 1.03, τ′Q = 471 s}; and {g0 = 1.6.10−3, gf = 1.2, τG = 612 s}.
We must also take into account the waiting time that is left between two successive force steps, which is approximately equal to the force application duration itself. We can estimate that the force is applied during an effective time teff, which is half the total time t. Effective timescales for stiffening and actin recruitment can also be roughly estimated as half the mean measured times: 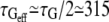 s,
s, 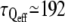 s, and
s, and 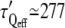 s.
s.
The increase in G0 appears highly correlated to the actin recruitment around the bead. The bead-cell contact strengthening exhibits a sigmoidlike dynamics, with an effective rising time τQeff of ∼200 s. However, in most of the experiments, the effective rigidification time τGeff is closer to the rising time for the overall actin network density τ′Qeff than to τQeff. This is also evident in the average values: both τGeff and τ′Qeff are very close to 300 s. This shows that the reinforcement of the actin network results in a greater cell rigidity. The increase in the measured rigidity is thus very dependent on the actin network densification, and not only on the strengthening of the contacts as previously reported (12).
CONCLUSION
This work is the first attempt to quantify the dynamics of both cell stiffening and actin recruitment in response to a controlled external stress. We evidence a mechanical strengthening of cells in response to force application, and a reinforcement both of the cell-substrate contact and of the local actin network in the vicinity of the force application zone, and up to several micrometers from it. The rising times for cell rigidity and actin quantity are ∼300 s for C2C12 cells.
In this study, we use mere wide-field epifluorescence images. This allows us to perform a semiquantitative analysis of the local actin density. We do not provide results regarding the dynamics of single actin filaments or the mechanisms underlying actin network reorganization. The increase in the actin quantity may be due to a densification or a broadening of the actin cortex in this zone, and/or to a more global reorganization of the filaments within the cell, allowing for instance new actin bundles to grow from or toward the force application zone. Addressing these issues will require the use of more sophisticated fluorescence observation techniques like confocal microscopy, fluorescence recovery after photobleaching (42), or fluorescence speckle microscopy (43). This could help elucidate the precise cytoskeleton dynamics underlying the recruitment that we evidence here.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Acknowledgments
The authors thank François Gallet (laboratoire Matière et Systèmes Complexes) for very helpful assistance and advice, and Myriam Allioux and Maïté Coppey (Institut Jacques Monod, Paris, France) for interesting discussions and the GFP-actin plasmid.
This work was supported by the Association pour la Recherche contre le Cancer.
Editor: Gaudenz Danuser.
References
- 1.Felsenfeld, D. P., D. Choquet, and M. P. Sheetz. 1996. Ligand binding regulates the directed movement of beta1 integrins on fibroblasts. Nature. 383:438–440. [DOI] [PubMed] [Google Scholar]
- 2.Zamir, E., and B. Geiger. 2001. Molecular complexity and dynamics of cell-matrix adhesions. J. Cell Sci. 114:3583–3590. [DOI] [PubMed] [Google Scholar]
- 3.Geiger, B., A. Bershadsky, R. Pankov, and K. M. Yamada. 2001. Transmembrane extracellular matrix-cytoskeleton crosstalk. Nat. Rev. Mol. Cell Biol. 2:793–805. [DOI] [PubMed] [Google Scholar]
- 4.Saez, A., A. Buguin, P. Silberzan, and B. Ladoux. 2005. Is the mechanical activity of epithelial cells controlled by deformations or forces? Biophys. J. 89:L52–L54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engler, A., L. Bacakova, C. Newman, A. Hategan, M. Griffin, and D. Discher. 2004. Substrate compliance versus ligand density in cell on gel responses. Biophys. J. 86:617–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pelham, R. J., and Y.-L. Wang. 1997. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc. Natl. Acad. Sci. USA. 94:13661–13665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lo, C.-M., H.-B. Wang, M. Dembo, and Y.-L. Wang. 2000. Cell movement is guided by the rigidity of the substrate. Biophys. J. 79:144–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.du Roure, O., A. Saez, A. Buguin, R. H. Austin, P. Chavrier, P. Silberzan, and B. Ladoux. 2005. Force mapping in epithelial cell migration. Proc. Natl. Acad. Sci. USA. 102:2390–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giancotti, F. G., and E. Ruhoslahti. 1999. Integrin signaling. Science. 285:1028–1032. [DOI] [PubMed] [Google Scholar]
- 10.Wang, H.-B., M. Dembo, and Y.-L. Wang. 2000. Substrate flexibility regulates growth and apoptosis of normal but not transformed cells. Am. J. Physiol. Cell Physiol. 279:C1345–C1350. [DOI] [PubMed] [Google Scholar]
- 11.Saez, A., M. Ghibaudo, A. Buguin, P. Silberzan, and B. Ladoux. 2007. Rigidity driven growth and differentiation of epithelial cells on microstructured anisotropic substrates. Proc. Natl. Acad. Sci. USA. 104:8281–8286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choquet, D., D. Felsenfeld, and M. P. Sheetz. 1997. Extracellular matrix rigidity causes strengthening of the integrin-cytoskeleton linkages. Cell. 88:39–48. [DOI] [PubMed] [Google Scholar]
- 13.Balaban, N. Q., U. S. Schwarz, D. Riveline, P. Goichberg, G. Tzur, I. Sabanay, D. Mahalu, S. Safran, A. Bershadsky, L. Addadi, and B. Geiger. 2001. Force and focal adhesion assembly, a close relationship studied using elastic micropatterned substrates. Nat. Cell Biol. 3:466–472. [DOI] [PubMed] [Google Scholar]
- 14.Katz, B.-Z., E. Zamir, A. Bershadsky, Z. Kam, K. M. Yamada, and B. Geiger. 2000. Physical state of the extracellular matrix regulates the structure and molecular composition of cell-matrix adhesions. Mol. Biol. Cell. 11:1047–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galbraith, C. G., K. M. Yamada, and M. Sheetz. 2002. The relationship between force and focal complex development. J. Cell Biol. 159:695–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delanoë-Ayari, H., R. al Kurdi, D. Gulino-Debrac, and D. Riveline. 2004. Membrane and acto-myosin tension promote clustering of adhesion proteins. Proc. Natl. Acad. Sci. USA. 101:2229–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felsenfeld, D. P., P. L. Schwartzbberg, A. Venegas, R. Tse, and M. P. Sheetz. 1999. Selective regulation of integrin-cytoskeleton interactions by the tyrosine kinase SRC. Nat. Cell Biol. 1:200–206. [DOI] [PubMed] [Google Scholar]
- 18.Riveline, D., E. Zamir, N. Q. Balaban, U. S. Schwarz, T. Ishizaki, S. Narumiya, Z. Kam, B. Geiger, and A. Bershadsky. 2001. Focal contacts as mechanosensors, externally applied local mechanical force induces growth of focal contacts in an mDia1-dependent and ROCK-independent mechanism. J. Cell Biol. 153:1175–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang, Y., E. L. Botvinick, Y. Zhao, M. Berns, S. Usami, R. Y. Tsien, and S. Chien. 2005. Visualizing the mechanical activation of SRC. Nature. 434:1040–1045. [DOI] [PubMed] [Google Scholar]
- 20.Fabry, B., G. N. Maksym, J. P. Butler, M. Glogauer, D. Navajas, and J. J. Fredberg. 2001. Scaling the microrheology of living cells. Phys. Rev. Lett. 87:148102. [DOI] [PubMed] [Google Scholar]
- 21.Fabry, B., G. N. Maksym, S. A. Shore, P. E. Moore, R. A. Panettieri, Jr., J. P. Butler, and J. J. Fredberg. 2001. Selected contribution, time course and heterogeneity of contractile responses in cultured human airway smooth muscle cells. J. Appl. Physiol. 91:986–994. [DOI] [PubMed] [Google Scholar]
- 22.Desprat, N., A. Richert, J. Simeon, and A. Asnacios. 2005. Creep function of a single living cell. Biophys. J. 88:2224–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balland, M., N. Desprat, D. Icard, S. Féréol, A. Asnacios, J. Browaeys, S. Hénon, and F. Gallet. 2006. Power laws in microrheology experiments on living cells, comparative analysis and modeling. Phys. Rev. E. 74:021911. [DOI] [PubMed] [Google Scholar]
- 24.Balland, M., A. Richert, and F. Gallet. 2005. The dissipative contribution of myosin II in the cytoskeleton dynamics of myoblasts. Eur. Biophys. J. 34:255–261. [DOI] [PubMed] [Google Scholar]
- 25.Smith, B. A., B. Tolloczko, J. Martin, and P. Grütter. 2005. Probing the viscoelastic behavior of cultured airway smooth muscle cells with atomic force microscopy, stiffening induced by contractile agonist. Biophys. J. 88:2994–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang, N., J. P. Butler, and D. E. Ingber. 1993. Mechanotransduction across the cell surface and through the cytoskeleton. Science. 260:1124–1127. [DOI] [PubMed] [Google Scholar]
- 27.Lee, J. S. H., P. Panorchan, C. M. Hale, S. B. Khatau, T. P. Kole, Y. Tseng, and D. Wirtz. 2006. Ballistic intracellular nanorheology reveals ROCK-hard cytoplasmic stiffening response to fluid flow. J. Cell Sci. 119:1760–1768. [DOI] [PubMed] [Google Scholar]
- 28.Wang, N., and D. E. Ingber. 1994. Control of cytoskeletal mechanics by extracellular matrix, cell shape and mechanical tension. Biophys. J. 66:2181–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deng, L., N. J. Fairbank, B. Fabry, P. Smith, and G. N. Maksym. 2004. Localized mechanical stress induces time-dependent actin cytoskeletal remodeling and stiffening in cultured airway smooth muscle cells. Am. J. Physiol. Cell Physiol. 287:C440–C448. [DOI] [PubMed] [Google Scholar]
- 30.Matthews, B. D., D. R. Overby, R. Mannix, and D. E. Ingber. 2006. Cellular adaptation to mechanical stress, role of integrins, Rho, cytoskeletal tension and mechanosensitive ion channels. J. Cell Sci. 119:508–519. [DOI] [PubMed] [Google Scholar]
- 31.Simmons, R. M., J. T. Finer, S. Chu, and J. Spudich. 1996. Quantitative measurements of force and displacement using an optical trap. Biophys. J. 70:1813–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laurent, V. M., S. Henon, E. Planus, R. Fodil, M. Balland, D. Isabey, and F. Gallet. 2002. Assessment of mechanical properties of adherent living cells by bead micromanipulation, comparison of magnetic twisting cytometry vs. optical tweezers. J. Biomech. Eng. 124:408–421. [DOI] [PubMed] [Google Scholar]
- 33.Zamir, E., B.-Z. Katz, S. i. Aota, K. M. Yamada, B. Geiger, and Z. Kam. 1999. Molecular diversity of cell-matrix adhesions. J. Cell Sci. 112:1655–1669. [DOI] [PubMed] [Google Scholar]
- 34.Lenormand, G., E. Millet, B. Fabry, J. P. Butler, and J. J. Fredberg. 2004. Linearity and time scale invariance of the creep function in living cells. J. R. Soc. Interface. 1:91–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kole, T. P., Y. Tseng, I. Jiang, J. L. Katz, and D. Wirtz. 2005. Intracellular mechanics of migrating fibroblasts. Mol. Biol. Cell. 16:328–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoffman, B. D., G. Massiera, K. M. Van Citters, and J. C. Crocker. 2006. The consensus mechanics of cultured mammalian cells. Proc. Natl. Acad. Sci. USA. 103:10259–10264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chrzanowska-Wodnicka, M., and K. Burridge. 1996. Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. J. Cell Biol. 133:1403–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trepat, X., M. Grabulosa, L. Buscemi, F. Rico, R. Farré, and D. Navajas. 2005. Thrombin and histamin induce stiffening of alveolar epithelial cells. J. Appl. Physiol. 98:1567–1574. [DOI] [PubMed] [Google Scholar]
- 39.Karcher, H., J. Lammerding, H. Huang, R. T. Lee, R. D. Kamm, and M. Kaazempur-Mofrad. 2003. A three-dimensional viscoelastic model for cell deformation with experimental verification. Biophys. J. 85:3336–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nicolas, A., and S. Safran. 2006. Limitation of cell adhesion by the elasticity of the extracellular matrix. Biophys. J. 91:61–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bausch, A., F. Ziemann, A. A. Boulbitch, K. Jacobson, and E. Sackmann. 1998. Local measurements of viscoelastic parameters of adherent cell surfaces by magnetic bead microrheometry. Biophys. J. 75:2038–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Osborn, E., A. Rabodzey, C. Dewey, and J. Hartwig. 2006. Endothelial actin cytoskeleton remodeling during mechanostimulation with fluid shear stress. Am. J. Physiol. Cell Physiol. 290:444–452. [DOI] [PubMed] [Google Scholar]
- 43.Vallotton, P., S. L. Gupton, C. M. Waterman-Storer, and G. Danuser. 2004. Simultaneous mapping of filamentous actin flow and turnover in migrating cells by quantitative fluorescent speckle microscopy. Proc. Natl. Acad. Sci. USA. 101:9660–9665. [DOI] [PMC free article] [PubMed] [Google Scholar]