Abstract
Background
Late blight is the most serious potato disease world-wide. The most effective and environmentally sound way for controlling late blight is to incorporate natural resistance into potato cultivars. Several late blight resistance genes have been cloned recently. However, there is almost no information available about the resistance pathways mediated by any of those genes.
Results
We previously cloned a late blight resistance gene, RB, from a diploid wild potato species Solanum bulbocastanum. Transgenic potato lines containing a single RB gene showed a rate-limiting resistance against all known races of Phytophthora infestans, the late blight pathogen. To better understand the RB-mediated resistance we silenced the potato Rar1 and Sgt1 genes that have been implicated in mediating disease resistance responses against various plant pathogens and pests. The Rar1 and Sgt1 genes of a RB-containing potato clone were silenced using a RNA interference (RNAi)-based approach. All of the silenced potato plants displayed phenotypically normal growth. The late blight resistance of the Rar1 and Sgt1 silenced lines were evaluated by a traditional greenhouse inoculation method and quantified using a GFP-tagged P. infestans strain. The resistance of the Rar1-silenced plants was not affected. However, silencing of the Sgt1 gene abolished the RB-mediated resistance.
Conclusion
Our study shows that silencing of the Sgt1 gene in potato does not result in lethality. However, the Sgt1 gene is essential for the RB-mediated late blight resistance. In contrast, the Rar1 gene is not required for RB-mediated resistance. These results provide additional evidence for the universal role of the Sgt1 gene in various R gene-mediated plant defense responses.
Background
Potato late blight, a disease caused by the oomycete pathogen Phytophthora infestans, is one of the world's most devastating crop diseases. World-wide losses due to late blight exceed several billion dollars annually [1]. Most of the potato cultivars currently grown in the United States are highly susceptible to late blight and control of this disease relies almost exclusively on fungicide applications. The most effective and environmentally sound way for controlling late blight is to incorporate natural resistance into potato cultivars. The pedigrees of many potato cultivars currently used in different countries include late blight resistant germplasm derived from Solanum demissum, Solanum andigena, and other wild species. However, most of the resistance derived from these wild species is controlled by single dominant resistance genes (R genes). These R genes are only effective in preventing the development of late blight if the invading P. infestans race contains the corresponding avirulence genes. This R gene-mediated resistance is often short-lived and is rapidly overcome by new races of the late blight pathogen.
Solanum bulbocastanum (2n = 2x = 24) is a diploid species that has adapted in the same environment as the late blight pathogen. This wild species was characterized as possessing durable resistance against P. infestans, even under high disease pressure [2,3]. Two resistance genes, RB (Rpi-blb1) and Rpi-blb2, have been cloned from S. bulbocastanum [4-6]. Both genes confer broad-spectrum resistance against a wide range of known P. infestans races. Transgenic potato lines containing a single RB gene showed a high-level resistance in the Toluca Valley, Mexico, where the potato fields are naturally intensively infested with the most diversified P. infestans populations [7]. Most interestingly, transgenic RB plants did not show total immunity to late blight, but instead showed a marked delay in both onset of symptoms and development of lesions. Such rate-limiting resistance may put less selection pressure on the P. infestans populations and protect the durability of this resistance gene. The RB gene therefore provides an excellent model to study the mechanism of broad-spectrum and rate-limiting disease resistances. An understanding of the underlying mechanism of this type of resistance is important for developing strategies to breed durable and sustainable disease resistance.
Several genes have been implicated in the regulation of R gene function. Of these genes, Rar1 and Sgt1 are among the most extensively studied genes. The Rar1 (required for Mla12 resistance) gene was first identified for its essential role in the function of a subset of Mla genes that confer resistance to barley powdery mildew [8]. The RAR1 protein contains two highly similar but distinct cysteine- and histidine-rich (CHORD) Zn2+-binding domains and was proposed to play a role in stabilizing R proteins in a confirmation that is implicated in receiving pathogen signals [9]. The Sgt1 gene (suppressor of the G2 allele of skp1) is an essential gene with multiple functions in yeast. SGT1 protein was initially identified as a RAR1-interacting partner in a yeast two-hybrid screen [10]. SGT1 may play a role in R protein accumulation [11]. Rar1 and Sgt1 genes are required in various R-gene mediated resistance against viral, bacterial, oomycete or fungal pathogens [12]. However, none of the previously studied R genes showed a race-non-specific and rate-limiting resistance phenotype as the RB gene. In addition, the role the Rar1 and Sgt1 genes are not universal and these genes are not essential for resistance involving some R genes [12,13].
Besides the two broad-spectrum resistance genes RB and Rpi-blb2, several race-specific late blight resistance genes have also been cloned [14-16]. Numerous late blight resistance genes have recently been mapped in various potato species or populations [17-25]. However, there is almost no information available about the resistance pathways mediated by any of these genes. As an initial effort to understand the RB-mediated late blight resistance pathway, we silenced the Rar1 and Sgt1 genes using an RNAi-based approach in a potato line containing the RB gene. We demonstrated that SGT1, but not RAR1, is essential for the RB-mediated broad-spectrum resistance to potato late blight.
Results
Identification of the potato Rar1 and Sgt1 genes
A search of the Institute for Genomic Research (TIGR) potato database [26] using a sequence from the tobacco Rar1 gene (AF480487) identified a potato EST, TC121848 (1187 bp), which showed 96% sequence similarity to the tobacco and tomato Rar1 transcripts. Similarly, a search using a sequence from the tomato Sgt1 gene (TC85297) identified a potato EST, TC112395 (1461 bp), which showed 98% sequence similarity to the tobacco and tomato Sgt1 genes and 90% sequence similarity to the Arabidopsis thaliana Sgt1b gene. Since the full-length cDNAs for the A. thaliana Rar1 and Sgt1b genes are 901 and 1290 bp, respectively [27,28], the identified potato ESTs almost cover each of the complete potato genes. Southern blot hybridization was performed to determine the copy numbers of the Rar1 and Sgt1 genes in the potato genome. Genomic DNA was isolated from potato clone K41, which contains the RB gene introgressed from S. bulbocastanum, and was hybridized with the potato Rar1 and Sgt1 gene probes. The Southern hybridization results showed that the haploid potato genome contains only one copy of the Rar1 gene and two copies of the Sgt1 gene (data not shown), which agree with a similar conclusion reported by Pajerowska et al. (2005).
RNAi-based silencing of the potato Rar1 and Sgt1 genes
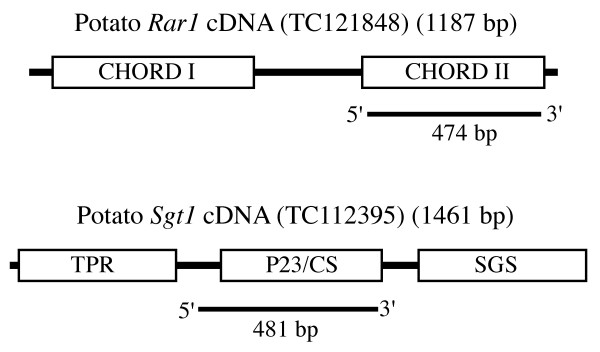
We developed RNAi constructs for the potato Rar1 and Sgt1 genes. The Rar1 silencing construct contained a 474-bp fragment covering the CHORD II domain. The Sgt1 construct contained a 481-bp fragment that targets the P23/CS domain (Figure 1). These constructs were used for Agrobacterium-mediated transformation of the potato line K41. We obtained 65 and 58 independent transgenic lines for the Rar1 and Sgt1 genes, respectively.
Figure 1.
Schematic representation of the potato Rar1 and Sgt1 genes and regions used for RNAi construct development.
The expression of the Rar1 and Sgt1 genes in the transgenic lines was analyzed by Northern blot hybridization using the Rar1 (1187 bp) and Sgt1 (1461 bp) genes as probes. A significant reduction of the Rar1 transcripts was observed in 47 of the 65 Rar1-RNAi transgenic lines analyzed (Table 1). Sgt1 transcript reduction was also observed in 35 of the 58 Sgt1-RNAi lines (Table 1). Three Rar1-RNAi lines and one Sgt1-RNAi line showed no detectable Northern hybridization signals after exposure for over one month using intensifying screens (Figure 2). Only these four lines were used in the late blight resistance evaluation. We observed no distinguishable morphological features associated with RNAi-induced silencing of the Rar1 and Sgt1 genes in these transgenic lines.
Table 1.
RNAi Silencing efficiencies of the Rar1 and Sgt1 genes based on Northern blot hybridization
| Gene | Transgenic lines screened | Transgenic lines with non-detectable transcript | Transgenic lines with normal transcript level | Transgenic lines with partial transcript level | Silencing efficiency | |
| >50% | <50% | |||||
| Rar1 | 65 | 32 | 18 | 7 | 8 | 72% |
| Sgt1 | 58 | 1 | 23 | 20 | 14 | 60% |
>50% indicates that transcript loss was less than 50%
<50% indicates that transcript loss was more than 50%
Figure 2.
Transcription analysis of Rar1 and Sgt1 silenced transgenic lines. (A) Northern blot analysis of transcription of the Rar1 gene. Lane 1: a transgenic plant (clone 3007) containing the Rar1-RNAi construct but not silenced; Lane 2–4: three independent Rar1-silenced lines (clones 3128, 2998, 3028) showing no transcript; Lane 5: untransformed K41 control. (B) Northern blot analysis of transcription of the Sgt1 gene. Lane 1: a transgenic plant (clone 3061) containing the Sgt1-RNAi construct but not silenced; Lane 2: Sgt1-silenced line (clone 3095) showing no transcript; Lane 3: untransformed K41 control. Both blots were re-probed with an actin gene probe to ensure equal loading of mRNA in each lane.
Late blight resistance evaluation of the Rar1- and Sgt1-silenced potato lines
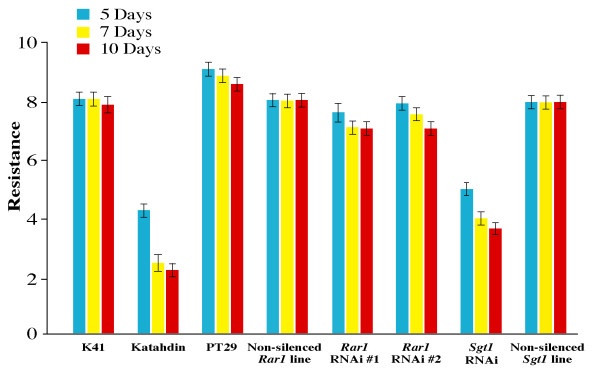
We evaluated the late blight resistance of the Rar1- and Sgt1-silenced lines together with several controls that either contain or do not contain the RB gene, including transformed but not silenced Rar1-RNAi and Sgt1-RNAi lines (no transcript reduction was found in these RNAi lines). Triplicates of each line were used in each of two independent inoculation experiments. All variation within replicates and experiments were taken into account in determining resistance scores. The average score of the late blight infection on the Rar1-silenced plants was 7.3 (± 0.6) after 7 days post inoculation (dpi) and 6.9 (± 0.0) after 10 dpi, which represents ~13% and 18% foliage infection, respectively (Figures 3, 4). The untransformed K41 plants and the non-silenced Rar1-RNAi line showed an average score of 8.0 (± 0.0), representing less than 10% infection. All susceptible controls showed an average score of 2.1 (± 0.6), representing ~81% infection. ANOVA showed that there was a significant difference in mean resistance scores among the tested plants (P-val = 2.2e-16) after seven days. Fisher's Least Significant Difference (LSD) test as a comparison of resistance score means revealed no significant differences between the Rar1-silenced plants and other resistant controls but revealed significant differences with the susceptible control, Katahdin. These results show that the RB-mediated resistance was not affected in the Rar1 silenced lines.
Figure 3.
Late blight resistance phenotype of Rar1- and Sgt1-silenced lines. Controls and silenced plants were inoculated with P. infestans and photographs were taken seven days after inoculation. (A, F) Untransformed K41 control (RB+); (B) A transformed but not silenced Rar1-RNAi line (clone 3007); (C, D) Two independent Rar1-silenced lines (clones 3128, 2998); (E, I) Susceptible control Katahdin (RB-); (G) A transformed but not silenced Sgt1-RNAi line (clone 3061); (H) A Sgt1-silenced line (clone 3095).
Figure 4.
Late blight resistance evaluation of Rar1- and Sgt1-silenced lines. Resistance readings were taken 5, 7 and 10 days post inoculation (dpi). K41: untransformed control (RB+); Katahdin: susceptible control (RB-); PT29: a S. bulbocastanum clone as the resistant control (RB+); Non-silenced Rar1 line (RB+): a transgenic, but non-silenced Rar1-RNAi line (clone 3007); Rar1 RNAi line #1 (Rar1 silenced, RB+, clone 3128); Rar1 RNAi line #2 (Rar1 silenced, RB+, clone 2998); Sgt1 RNAi line (Sgt1 silenced, RB+, clone 3095); Non-silenced Sgt1 line (RB+): a non-silenced Sgt1-RNAi line (clone 3061). Error bars represent the standard deviation from the means.
The Sgt1-silenced plants showed an average late blight infection of 4.0 (± 0.0) after 7 dpi and 3.7 (± 0.7) after 10 dpi, representing ~70% and ~73% infection, respectively (Figures 3, 4). Significant differences among the mean resistance scores were observed among the tested plants starting from day seven using ANOVA with P-val = 2.2e-16. Fisher's LSD test revealed that the Sgt1-silenced plant had a significantly different resistance score compared to the resistant control (untransformed K41) and non-silenced Sgt1 plants. These results suggested that silencing of the Sgt1 gene compromised the RB-mediated late blight resistance.
The late blight resistance of the Rar1- and Sgt1-silenced lines were also evaluated using the GFP-tagged P. infestans isolate 208m2 [29]. The Phytophthora growth was quantified by counting the fluorescing sporangia within each area; more P. infestans growth is indicated by larger counts. One and three days after inoculation, no significant differences were observed between the sporangial counts on the silenced and non-silenced controls. Six days after inoculation, however, there were significant differences among the sporangial counts of the tested plants (Figure 5) based on a one-way ANOVA with unequal variance (F = 3.627, P-val = 0.03142). Fisher's LSD, using an alpha of 0.01, indicated that the Sgt1-silenced plant had significantly more P. infestans growth, as indicated by sporangial count, than the K41 control, the transgenic but not silenced Rar1-RNAi and Sgt1-RNAi lines and the Rar1-silenced plants (Figure 6). These results are consistent with those from the conventional greenhouse evaluations.
Figure 5.
Late blight resistance evaluation using a GFP-tagged P. infestans strain. Leaves were inoculated on plants with GFP-tagged isolate 208m2 and were removed 6 days post inoculation and photographed on a dissecting microscope under a narrow-band GFP filter. (A) Untransformed K41 control. (B) Katahdin. (C) A silenced Rar1-RNAi line (clone 3128). (D) A Sgt1-silenced line (clone 3095). The red circles outline the original location of the 10 μl inoculation drops. Green fluorescence is only observed within the circles in A and C but spreads out of the original inoculation sites in B and D. Bars = 2.5 mm.
Figure 6.
Quantitative analysis of GFP Phytophthora lesions. The average GFP sporangial counts (particle count) are shown from K41: untransformed control (RB+); Non-silenced Rar1 line (RB+): a transgenic, but non-silenced Rar1-RNAi line (clone 3007); A Rar1-RNAi line (clone 3128); Non-silenced Sgt1 line (RB+): a non-silenced Sgt1-RNAi line (clone 3061); A Sgt1-RNAi line (clone 3095). Using a one-way ANOVA, with unequal variance, and Fisher's LSD at an alpha of 0.01, the only group that has significantly higher sporangial growth is the Sgt1-silenced line.
Discussion
The Rar1 and Sgt1 genes in potato
Rar1 is a single copy gene in barley [10] and A. thaliana [28,30]. Our data from Southern blot hybridizations confirm the previous report that only one copy of Rar1 exists in the haploid potato genome [31]. Plant rar1 mutants show no visible growth defects [32], although silencing of the Rar1 homolog in C. elegans resulted in semi sterility and embryo lethality [8]. The Rar1-silenced potato plants did not show any visible growth phenotypes in our study.
SGT1 is an essential protein for proper kinetochore function in yeast and null mutations of the single copy Sgt1 gene are lethal [33]. Most plant species analyzed appear to contain two Sgt1 genes [11,31,34,35]. The Sgt1a and Sgt1b double mutant in A. thaliana is embryo lethal [11]. Similarly, virus-induced gene silencing (VIGS) of the Sgt1 genes (NbSgt1.1 and NbSgt1.2) in N. benthamiana caused a stunt phenotype [34]. In tomato, silencing of the Sgt1-1 gene, but not Sgt1-2, was lethal. This result was possibly caused by silencing of both the Sgt1-1 and Sgt1-2 genes using the VIGS construct designed for Sgt1-1 [35]. In contrast, silencing of the wheat and barley SGT1 genes by VIGS did not result in a growth phenotype [36,37]. However, the copy number and functional redundancy of the SGT1 genes in wheat and barley are not clear, this may be caused by either partial silencing of the gene and/or compensated by partially homologous and non-silenced SGT genes in these species.
We did not observe any abnormal growth phenotype in the Sgt1 silenced line. The complete silencing of the Sgt1 gene was confirmed by Northern blot hybridization (Figure 2B). The available sequences of the two potato Sgt1 genes (StSgt1-1: AY615272; StSgt1-2: AY615274) are 100% identical. The Sgt1 cDNA fragment used in our RNAi contruct also shares 100% homology to these sequences. These results suggest that both copies of the potato Sgt1 gene are likely silenced in the RNAi silencing line. Therefore, it appears that silencing both of the Sgt1 genes is not lethal in potato. This result will need to be confirmed by multiple completely silenced Sgt1 lines.
Requirement of the RAR1 and SGT1 proteins in disease resistance
A role of the RAR1 protein in disease resistance signaling was first recognized in barley through Mla12-mediated resistance against powdery mildew [8]. The same role for RAR1 has since been identified in the resistance pathways conferred by several genes that belong to the NB-LRR family [12,36-38]. Interestingly, there were also reports that the Rar1 gene appears to not play a role in the resistance pathway. The RAR1-independent cases include the Mla1-mediated resistance against powdery mildew in barley [10,39], Bs2/AvrBs2-mediated resistance against bacterial spot disease in N. benthamiana [40], and Mi-1-mediated resistance against root-knot nematodes in tomato [35]. We demonstrated that silencing of the single Rar1 gene in potato does not affect the RB-mediated late blight resistance.
The previous RAR1-independent conclusions were based on gene silencing techniques by biolistic delivery of double stranded RNA constructs into single cells [10,39] or by VIGS [35,40]. These techniques may not result in complete suppression of a target gene and uniform silencing of the gene throughout the infected plants. False negative results have to be carefully sorted out because a low level of the transcripts resulting from incomplete silencing may be sufficient to facilitate the function of RAR1. The RNAi-silenced Rar1 potato lines showed no transcripts based on stringent Northern blot hybridizations (Figure 2), suggesting a near complete silencing of the Rar1 gene. These stable silenced lines allowed repeated resistance evaluations. Thus, our results from multiple RNAi silencing lines provide conclusive evidence that RB-mediated resistance is RAR1 independent.
The RAR1 protein has been proposed to be involved in forming or stabilizing the recognition complexes associated with R proteins or in assisting conformational changes of the recognition complexes [12]. Such functions may not be required in the complex assembly of a subset of R proteins. Bieri et al (2004) showed that in barley rar1 mutants that are compromised for MLA6 but not MLA1 resistance, the steady state level of both MLA isoforms is reduced. However, MLA6 accumulated to about a four-fold lower level than MLA1 in transgenic lines. Interestingly, Mla1 functions independently of Rar1 only where MLA1 abundance exceeds a threshold level [39]. A number of previous studies suggest that RAR1 may control the abundance of the NB-LRR type R proteins [39]. We propose that the RB protein level in the Rar1 silenced potato lines may be sufficient to trigger the resistance pathway.
Many previous studies confirmed the requirement of the SGT1 protein in resistance mediated by both NB-LRR and Pto-kinase type resistance genes, as well as in non-host resistance [34,35,37,40,41]. Several resistance genes in A. thaliana were SGT1b independent in the sgt1b mutant background [27,28,30]. However, SGT1a may complement the loss of SGT1b [11]. The MLA1-triggered resistance in barley, unlike other MLA variants, was largely unaffected when the HvSgt1 gene was silenced [10]. The transient silencing method used in this system may not have been complete, leaving some levels of HvSGT1 for MLA1 to function. Alternatively, MLA1 requires low levels of HvSGT1 to operate as compared to other MLA variants [10]. Bhattarai et al. (2007) recently showed that partial silencing of the Sgt1-1 gene in tomato resulted in attenuation of Mi-1-mediated potato aphid resistance, but the same plants still held the Mi-1-mediated root-knot nematode resistance. These results support the hypothesis that plant R proteins differ in the amounts of SGT1 needed to trigger effective resistance [11]. We demonstrate that silencing of Sgt1 clearly compromised the RB-mediated late blight resistance (Figures 3, 4, 5), indicating that the RNAi approach most likely silenced both copies of the Sgt1 gene and the Sgt1 gene plays the key role in the pathway. Our results provide further evidence for the universal role of the Sgt1 gene in various R gene-mediated plant defense responses.
Conclusion
We developed RNAi-based silencing lines of genes Rar1 and Sgt1 in a potato line that contains the late blight resistance gene RB, which confers broad-spectrum resistance against all known races of P. infestans. Silencing of the Rar1 and Sgt1 genes in potato does not result in phenotypic changes. Late blight resistance evaluation of the Rar1 and Sgt1 silenced lines showed that the Sgt1 gene is essential for the RB-mediated late blight resistance. In contrast, silencing of the Rar1 gene does not abolish the RB-mediated resistance. These results provide additional evidence for the universal role of the Sgt1 gene in various R gene-mediated plant defense responses.
Methods
Plant materials
Potato line J101K6A6K41 (K41) is a tetraploid clone and was developed from the somatic hybrid J101 between potato and S. bulbocastanum (clone PT29) [42]. J101 was used as the female parent to backcross with Katahdin (BC1), Atlantic (BC2) and Katahdin (BC3). K41 is a late blight resistant BC3 clone. Presence of the RB gene in K41 was confirmed by polymerase chain reaction (PCR) using RB-specific primers [43] and this clonal line was used in transformation experiments designed to silence the Rar1 and Sgt1 genes. S. bulbocastanum (PT29) and potato cultivar 'Katahdin' were used as controls for late blight resistance evaluation.
RNAi construct design and potato transformation
Total RNA was extracted from leaf tissue of K41 using the RNeasy Plant Mini Kit (Qiagen, Valencia, California) and treated with TURBO DNA-free (Ambion, Austin, Texas) to remove DNA contamination. First strand cDNA was synthesized using 1 μg of total RNA, oligo d(T) primer and superscript reverse transcriptase (Invitrogen, Carlsbad, California). The cDNA fragments used to silence Rar1 and Sgt1 were amplified by PCR. A 474-bp cDNA fragment from Rar1, corresponding to the TIGR potato EST TC121848 (nt 346 through 820), was amplified from K41 cDNA using Platinum Taq DNA polymerase (Invitrogen). The primers used for amplification included 5' CACC CAA CAC CAT CTG CTA CCA AAA A 3'(forward) and 5' GAC ACT GGG TCA GCG TTG TG 3'(reverse). A 481-bp cDNA fragment from Sgt1, corresponding to the TIGR potato EST TC112395 (nt 359 through 840 bp) (this EST is recently split into TC133190 and TC159283) was amplified from K41 cDNA using primers 5' CACC GGC CTG TAT GAA GCT TGA AGA A 3'(forward) and 5' TCT GCA TTT TGC AGG TGT TAT C 3' (reverse). The Rar1 and Sgt1 amplicons were successively purified using QIAquick PCR purification kit (Qiagen), gel verified and then cloned into pENTR/D-TOPO vector using the pENTR Directional TOPO Cloning kit (Invitrogen). The Rar1 and Sgt1 DNA fragments were then transferred into the pHellsGate8 vector using the LR Clonase recombination method according to Helliwell et al. (2002) [44]. The sequence-verified pHellsgate8-Rar1 and pHellsgate8-Sgt1 constructs were then transformed into K41 using standard Agrobacterium-mediated transformation protocols [45].
Gel-blot hybridizations
Southern blot hybridization was performed according to Stupar et al. (2002) [46]. Genomic DNA was isolated from leaf tissues of K41 and digested with restriction enzymes. The DNA blots were probed with a 1-kb genomic DNA fragment of the Rar1 gene and a 1.1-kb genomic DNA fragment of the Sgt1 gene to evaluate the copy numbers of these two genes in the potato genome. RNA blots were prepared using 10–15 μg of total RNA isolated from leaf tissue using the TRIzol protocol (Invitrogen). To verify the transcription of the Rar1 and Sgt1 genes in the RNAi lines, RNA blots were hybridized with the complete cDNA fragments of Rar1 (TC121848) and Sgt1 (TC112395) amplified from K41 cDNAs. Hybridizations were performed as described previously [47]. Intensifying screens were used to detect the Northern blot hybridization signals and the blots were exposed for at least one month to reveal if any transcripts were detectable in the Rar1- and Sgt1-RNAi lines.
Late blight resistance evaluation
Rar1 and Sgt1 silenced plants along with several controls were evaluated for late blight resistance in environmentally controlled greenhouses at the University of Wisconsin-Madison Biotron facility. Controls include the susceptible potato cultivar Katahdin (without RB gene), S. bulbocastanum (PT29), non-transformed K41 lines and K41 lines that were transformed with the construct containing either Rar1 or Sgt1, but not silenced. The inoculation and resistance evaluation were performed as described previously [43]. Briefly, the selected lines were grown in triplicates and were randomly placed in a mist chamber eight hours prior to inoculation. The mist chamber held a 24-hour relative humidity of 100%, an 8-hour light period, a daytime temperature between 17–19°C and a nighttime temperature at 13–15°C. The plants were inoculated with 76,000–80,000 sporangia/ml of sporangial suspensions of P. infestans isolate US930287 (US-8 genotype, A-2 mating type). Measurements of the foliage blight were interpreted and scored according to the Malcolmson scale [48]. The scale was based on percent of foliage infected and scores were as follows: 9 – no visible infection; 8 – <10% infection; 7 – 11–25%; 6 – 26–40%; 5 – 41–60%; 4 – 61–70%; 3 – 71–80%; 2 – 81–90%; 1 – >90%; 0 – 100% infection. Blight scores were recorded 5, 7 and 10 days after inoculation. An average score for the resistance was determined using the three replicates of each clone in each inoculation experiment.
Resistance evaluation using a GFP-tagged P. infestans strain
A quantitative method was employed using a strain of P. infestans, which contains GFP, to better quantify pathogen growth. All silenced lines, as well as the transgenic but not silenced Rar1-RNAi and Sgt1-RNAi lines, were inoculated by the GFP-tagged P. infestans isolate 208m2 provided by Dr. Felix Mauch (University of Friberg, Switzerland) [29]. The final average sporangial concentration was 64,259 sporangia/mL, using a hemocytometer. The sporangial suspension was placed at 15°C for three hours before inoculation to release the zoospores. One 10 μl drop of the suspension was placed on both sides of the midvein, on the bottom of the leaf, in approximately the same location. Two leaves were sampled from each plant 24 and 72 hours after inoculation and four leaves were sampled 144 hours (six days) after inoculation. Each leaf was examined for the presence of actively growing Phytophthora using an Olympus SZX12 dissecting scope. Each area was photographed with an Olympus DP70 digital camera using the 41020 Chroma narrow-band GFP filter with an excitation of 425/75 nm and emission of 500/50 nm.
The spreading Phytophthora on each leaf surface were quantified by first converting the GFP images to black and white. The color conversion highlights the GFP Phytophthora sporangia and mycelium growth within each area. The area was then outlined and analyzed using the "analyze particle" tool in the ImageJ software [49]. This tool scans the image selection until it finds the edge of an object, corresponding to fluorescing sporangia or mycelium on the leaf surface. The tool outlines each object measures it and fills it in. It counts this small area as one particle and resumes scanning until it reaches another particle, where it repeats the process until the end of the image selection. The data report details the total number of counted particles, representing the total number of fluorescing sporangia on the leaf surface. As the Phytophthora grows, more sporangia and mycelium are present, which translates into a higher particle count. We measure the pathogen growth directly, not just the lesion, or area of dead tissue present on the leaf surface, which may not correspond to the actual pathogen spread. This experiment was designed as a Complete Randomized Design (CRD) with a total of 56 observations for days one and three and 50 observations for day six. Data quality tests and assessments were performed, such as residual and qq-plots. A one-way ANOVA with unequal variance was performed to compare the particle counts for each of the silenced lines with the empty vector controls and Fisher's LSD tested each group individually.
Authors' contributions
PBB and JJ conceived the project. PBB developed RNAi constructs. JAR and SA developed transgenic lines. PBB and PN characterized transgenic lines. PBB, LCK, and SMW conducted disease resistance evaluation. PBB and JJ drafted the manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We thank CSIRO Plant Industry(Australia) for providing the pHellsGate 8 silencing vector. This research was supported by a grant from the National Science Foundation Plant Genome Research Program (DBI-0218166) to S.A.P. and J.J. and Hatch funds to J.J.
Contributor Information
Pudota B Bhaskar, Email: pudota1@wisc.edu.
John A Raasch, Email: jaraasch@wisc.edu.
Lara C Kramer, Email: lmcolton@wisc.edu.
Pavel Neumann, Email: neumann@umbr.cas.cz.
Susan M Wielgus, Email: swielgus@wisc.edu.
Sandra Austin-Phillips, Email: sandra@biotech.wisc.edu.
Jiming Jiang, Email: jjiang1@wisc.edu.
References
- Kamoun S. Nonhost resistance to Phytophthora: novel prospects for a classical problem. Curr Opin Plant Biol. 2001;4:295–300. doi: 10.1016/S1369-5266(00)00176-X. [DOI] [PubMed] [Google Scholar]
- Niederhauser JS, Millis WR. Resistance of Solanum species to Phytophthora infestans in Mexico. Phytopathol. 1953;43:456–457. [Google Scholar]
- Vansoest LJM, Schober B, Tazelaar MF. Resistance to Phytophthora infestans in tuber-bearing species of Solanum and its geographical distribution. Potato Res. 1984;27:393–411. doi: 10.1007/BF02357427. [DOI] [Google Scholar]
- Song J, Bradeen JM, Naess SK, Raasch JA, Wielgus SM, Haberlach GT, Liu J, Kuang H, Austin-Phillips S, Buell CR, et al. Gene RB cloned from Solanum bulbocastanum confers broad spectrum resistance to potato late blight. Proc Natl Acad Sci USA. 2003;100:9128–9133. doi: 10.1073/pnas.1533501100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vossen E, Sikkema A, Hekkert BL, Gros J, Stevens P, Muskens M, Wouters D, Pereira A, Stiekema W, Allefs S. An ancient R gene from the wild potato species Solanum bulbocastanum confers broad-spectrum resistance to Phytophthora infestans in cultivated potato and tomato. Plant J. 2003;36:867–882. doi: 10.1046/j.1365-313X.2003.01934.x. [DOI] [PubMed] [Google Scholar]
- van der Vossen EAG, Gros J, Sikkema A, Muskens M, Wouters D, Wolters P, Pereira A, Allefs S. The Rpi-blb2 gene from Solanum bulbocastanum is an Mi-1 gene homolog conferring broad-spectrum late blight resistance in potato. Plant J. 2005;44:208–222. doi: 10.1111/j.1365-313X.2005.02527.x. [DOI] [PubMed] [Google Scholar]
- Lozoya-Saldana H, Belmar-Diaz C, Bradeen JM, Helgeson JP. Characterization of Phytophthora infestans isolates infecting transgenic and somatic hybrid potatoes resistant to the pathogen in the Toluca Valley, Mexico. Am J Potato Res. 2005;82:79. [Google Scholar]
- Shirasu K, Lahaye T, Tan MW, Zhou FS, Azevedo C, Schulze-Lefert P. A novel class of eukaryotic zinc-binding proteins is required for disease resistance signaling in barley and development in C. elegans. Cell. 1999;99:355–366. doi: 10.1016/S0092-8674(00)81522-6. [DOI] [PubMed] [Google Scholar]
- Hubert DA, Tornero P, Belkhadir Y, Krishna P, Takahashi A, Shirasu K, Dangl JL. Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO J. 2003;22:5679–5689. doi: 10.1093/emboj/cdg547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo C, Sadanandom A, Kitagawa K, Freialdenhoven A, Shirasu K, Schulze-Lefert P. The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science. 2002;295:2073–2076. doi: 10.1126/science.1067554. [DOI] [PubMed] [Google Scholar]
- Azevedo C, Betsuyaku S, Peart J, Takahashi A, Noel L, Sadanandom A, Casais C, Parker J, Shirasu K. Role of SGT1 in resistance protein accumulation in plant immunity. EMBO J. 2006;25:2007–2016. doi: 10.1038/sj.emboj.7601084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu K, Schulze-Lefert P. Complex formation, promiscuity and multi-functionality: protein interactions in disease-resistance pathways. Trends Plant Sci. 2003;8:252–258. doi: 10.1016/S1360-1385(03)00104-3. [DOI] [PubMed] [Google Scholar]
- Muskett P, Parker J. Role of SGT1 in the regulation of plant R gene signalling. Microbes Infect. 2003;5:969–976. doi: 10.1016/S1286-4579(03)00183-7. [DOI] [PubMed] [Google Scholar]
- Ballvora A, Ercolano MR, Weiss J, Meksem K, Bormann CA, Oberhagemann P, Salamini F, Gebhardt C. The R1 gene for potato resistance to late blight (Phytophthora infestans) belongs to the leucine zipper/NBS/LRR class of plant resistance genes. Plant J. 2002;30:361–371. doi: 10.1046/j.1365-313X.2001.01292.x. [DOI] [PubMed] [Google Scholar]
- Huang SW, Vleeshouwers VGAA, Werij JS, Hutten RCB, van Eck HJ, Visser RGF, Jacobsen E. The R3 resistance to Phytophthora infestans in potato is conferred by two closely linked R genes with distinct specificities. MPMI. 2004;17:428–435. doi: 10.1094/MPMI.2004.17.4.428. [DOI] [PubMed] [Google Scholar]
- Huang SW, van der Vossen EAG, Kuang HH, Vleeshouwers V, Zhang NW, Borm TJA, van Eck HJ, Baker B, Jacobsen E, Visser RGF. Comparative genomics enabled the isolation of the R3a late blight resistance gene in potato. Plant J. 2005;42:251–261. doi: 10.1111/j.1365-313X.2005.02365.x. [DOI] [PubMed] [Google Scholar]
- Gebhardt C, Valkonen JPT. Organization of genes controlling disease resistance in the potato genome. Annu Rev Phytopathol. 2001;39:79–102. doi: 10.1146/annurev.phyto.39.1.79. [DOI] [PubMed] [Google Scholar]
- Gebhardt C, Ballvora A, Walkemeier B, Oberhagemann P, Schuler K. Assessing genetic potential in germplasm collections of crop plants by marker-trait association: a case study for potatoes with quantitative variation of resistance to late blight and maturity type. Mol Breed. 2004;13:93–102. doi: 10.1023/B:MOLB.0000012878.89855.df. [DOI] [Google Scholar]
- Park TH, Gros J, Sikkema A, Vleeshouwers V, Muskens M, Allefs S, Jacobsen E, Visser RGF, van der Vossen EAG. The late blight resistance locus Rpi-blb3 from Solanum bulbocastanum belongs to a major late blight R gene cluster on chromosome 4 of potato. MPMI. 2005;18:722–729. doi: 10.1094/MPMI-18-0722. [DOI] [PubMed] [Google Scholar]
- Park TH, Vleeshouwers V, Huigen DJ, van der Vossen EAG, van Eck HJ, Visser RGF. Characterization and high-resolution mapping of a late blight resistance locus similar to R2 in potato. Theor Appl Genet. 2005;111:591–597. doi: 10.1007/s00122-005-2050-4. [DOI] [PubMed] [Google Scholar]
- Villamon FG, Spooner DM, Orrillo M, Mihovilovich E, Perez W, Bonierbale M. Late blight resistance linkages in a novel cross of the wild potato species Solanum paucissectum (series Piurana) Theor Appl Genet. 2005;111:1201–1214. doi: 10.1007/s00122-005-0053-9. [DOI] [PubMed] [Google Scholar]
- Bradshaw JE, Bryan GJ, Lees AK, McLean K, Solomon-Blackburn RM. Mapping the R10 and R11 genes for resistance to late blight (Phytophthora infestans) present in the potato (Solanum tuberosum) R-gene differentials of Black. Theor Appl Genet. 2006;112:744–751. doi: 10.1007/s00122-005-0179-9. [DOI] [PubMed] [Google Scholar]
- Bradshaw JE, Hackett CA, Lowe R, McLean K, Stewart HE, Tierney I, Vilaro MDR, Bryan GJ. Detection of a quantitative trait locus for both foliage and tuber resistance to late blight [Phytophthora infestans (Mont.) de Bary] on chromosome 4 of a dihaploid potato clone (Solanum tuberosum subsp. tuberosum) Theor Appl Genet. 2006;113:943–951. doi: 10.1007/s00122-006-0353-8. [DOI] [PubMed] [Google Scholar]
- Rauscher GM, Smart CD, Simko I, Bonierbale M, Mayton H, Greenland A, Fry WE. Characterization and mapping of RPi-ber, a novel potato late blight resistance gene from Solanum berthaultii. Theor Appl Genet. 2006;112:674–687. doi: 10.1007/s00122-005-0171-4. [DOI] [PubMed] [Google Scholar]
- Simko I, Costanzo S, Ramanjulu V, Christ BJ, Haynes KG. Mapping polygenes for tuber resistance to late blight in a diploid Solanum phureja × S. stenotomum hybrid population. Plant Breed. 2006;125:385–389. doi: 10.1111/j.1439-0523.2006.01232.x. [DOI] [Google Scholar]
- http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=potato
- Austin MJ, Muskett P, Kahn K, Feys BJ, Jones JDG, Parker JE. Regulatory role of SGT1 in early R gene-mediated plant defenses. Science. 2002;295:2077–2080. doi: 10.1126/science.1067747. [DOI] [PubMed] [Google Scholar]
- Tornero P, Merritt P, Sadanandom A, Shirasu K, Innes RW, Dangl JL. RAR1 and NDR1 contribute quantitatively to disease resistance in Arabidopsis, and their relative contributions are dependent on the R gene assayed. Plant Cell. 2002;14:1005–1015. doi: 10.1105/tpc.001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si-Ammour A, Mauch-Mani B, Mauch F. Quantification of induced resistance against Phytophthora species expressing GFP as a vital marker: Beta-aminobutyric acid but not BTH protects potato and Arabidopsis from infection. Mol Plant Pathol. 2003;4:237–248. doi: 10.1046/j.1364-3703.2003.00168.x. [DOI] [PubMed] [Google Scholar]
- Muskett PR, Kahn K, Austin MJ, Moisan LJ, Sadanandom A, Shirasu K, Jones JDG, Parker JE. Arabidopsis RAR1 exerts rate-limiting control of R gene-mediated defenses against multiple pathogens. Plant Cell. 2002;14:979–992. doi: 10.1105/tpc.001040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajerowska KM, Parker JE, Gebhardt C. Potato homologs of Arabidopsis thaliana genes functional in defense signaling – Identification, genetic mapping, and molecular cloning. MPMI. 2005;18:1107–1119. doi: 10.1094/MPMI-18-1107. [DOI] [PubMed] [Google Scholar]
- Takahashi A, Casais C, Ichimura K, Shirasu K. HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc Natl Acad Sci USA. 2003;100:11777–11782. doi: 10.1073/pnas.2033934100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa K, Skowyra D, Elledge SJ, Harper JW, Hieter P. SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol Cell. 1999;4:21–33. doi: 10.1016/S1097-2765(00)80184-7. [DOI] [PubMed] [Google Scholar]
- Peart JR, Lu R, Sadanandom A, Malcuit I, Moffett P, Brice DC, Schauser L, Jaggard DAW, Xiao SY, Coleman MJ, et al. Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc Natl Acad Sci USA. 2002;99:10865–10869. doi: 10.1073/pnas.152330599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattarai KK, Li Q, Liu Y, Dinesh-Kumar SP, Kaloshian I. The Mi-1-mediated pest resistance requires Hsp90 and Sgt1. Plant Physiol. 2007;144:312–323. doi: 10.1104/pp.107.097246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein I, Pacak MB, Hrubikova K, Williamson S, Dinesen M, Soenderby IE, Sundar S, Jarmolowski A, Shirasu K, Lacomme C. Virus-induced gene silencing-based functional characterization of genes associated with powdery mildew resistance in barley. Plant Physiol. 2005;138:2155–2164. doi: 10.1104/pp.105.062810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield SR, Huang L, Brandt AS, Gill BS. Development of a virus-induced gene-silencing system for hexaploid wheat and its use in functional analysis of the Lr21-mediated leaf rust resistance pathway. Plant Physiol. 2005;138:2165–2173. doi: 10.1104/pp.105.061861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YL, Schiff M, Marathe R, Dinesh-Kumar SP. Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 2002;30:415–429. doi: 10.1046/j.1365-313X.2002.01297.x. [DOI] [PubMed] [Google Scholar]
- Bieri S, Mauch S, Shen QH, Peart J, Devoto A, Casais C, Ceron F, Schulze S, Steinbiss HH, Shirasu K, et al. RAR1 positively controls steady state levels of barley MLA resistance proteins and enables sufficient MLA6 accumulation for effective resistance. Plant Cell. 2004;16:3480–3495. doi: 10.1105/tpc.104.026682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister RT, Dahlbeck D, Day B, Li Y, Chesnokova O, Staskawicz BJ. Molecular genetic evidence for the role of SGT1 in the intramolecular complementation of Bs2 protein activity in Nicotiana benthamiana. Plant Cell. 2005;17:1268–1278. doi: 10.1105/tpc.104.029637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tor M, Gordon P, Cuzick A, Eulgem T, Sinapidou E, Mert-Turk F, Can C, Dangl JL, Holub EB. Arabidopsis SGT1b is required for defense signaling conferred by several downy mildew resistance genes. Plant Cell. 2002;14:993–1003. doi: 10.1105/tpc.001123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgeson JP, Pohlman JD, Austin S, Haberlach GT, Wielgus SM, Ronis D, Zambolim L, Tooley P, McGrath JM, James RV, et al. Somatic hybrids between Solanum bulbocastanum and potato: a new source of resistance to late blight. Theor Appl Genet. 1998;96:738–742. doi: 10.1007/s001220050796. [DOI] [Google Scholar]
- Colton LM, Groza HI, Wielgus SM, Jiang JM. Marker-assisted selection for the broad-spectrum potato late blight resistance conferred by gene RB derived from a wild potato species. Crop Sci. 2006;46:589–594. doi: 10.2135/cropsci2005.0112. [DOI] [Google Scholar]
- Helliwell CA, Wesley SV, Wielopolska AJ, Waterhouse PM. High-throughput vectors for efficient gene silencing in plants. Funct Plant Biol. 2002;29:1217–1225. doi: 10.1071/FP02033. [DOI] [PubMed] [Google Scholar]
- Ziegelhoffer T, Will J, Austin-Phillips S. Expression of bacterial cellulase genes in transgenic alfalfa (Medicago sativa L.), potato (Solanum tuberosum L.) and tobacco (Nicotiana tabacum L.) Mol Breed. 1999;5:309–318. doi: 10.1023/A:1009646830403. [DOI] [Google Scholar]
- Stupar RM, Song JQ, Tek AL, Cheng ZK, Dong FG, Jiang JM. Highly condensed potato pericentromeric heterochromatin contains rDNA-related tandem repeats. Genetics. 2002;162:1435–1444. doi: 10.1093/genetics/162.3.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupar RM, Beaubien KA, Jin WW, Song JQ, Lee MK, Wu CC, Zhang HB, Han B, Jiang JM. Structural diversity and differential transcription of the patatin multicopy gene family during potato tuber development. Genetics. 2006;172:1263–1275. doi: 10.1534/genetics.105.051219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruickshank G, Stewart HE, Wastie RL. An illustrated assessment key for foliage blight of potatoes. Potato Res. 1982;25:213–214. doi: 10.1007/BF02359807. [DOI] [Google Scholar]
- http://rsb.info.nih.gov/ij/