Abstract
β-Barrel proteins are present in the outer membranes of Gram-negative bacteria, mitochondria and chloroplasts. The central component of their assembly machinery is called Omp85 in bacteria. Omp85 is predicted to consist of an integral membrane domain and an amino-terminal periplasmic extension containing five polypeptide-transport-associated (POTRA) domains. We have addressed the function of these domains by creating POTRA domain deletions in Omp85 of Neisseria meningitidis. Four POTRA domains could be deleted with only slight defects in Omp85 function. Only the most carboxy-terminal POTRA domain was essential, as was the membrane domain. Thus, similar to the mitochondrial Omp85 homologue, the functional core of bacterial Omp85 consists of its membrane domain and a single POTRA domain, that is, POTRA5.
Keywords: β-barrel, Omp85, outer membrane, POTRA domain
Introduction
Membrane-embedded β-barrel proteins are found in the outer membranes of Gram-negative bacteria and in those of mitochondria and chloroplasts, probably reflecting the endosymbiont origin of these organelles. Some of the assembly machinery for outer membrane proteins (OMPs), in particular the assembly factor Omp85, is also evolutionarily highly conserved (Gentle et al, 2005). Omp85 is an essential OMP the function of which was first identified in Neisseria meningitidis (Voulhoux et al, 2003) and subsequently confirmed in Escherichia coli (Doerrler & Raetz, 2005; Wu et al, 2005). Depletion of Omp85 resulted in assembly defects in all OMPs tested. Omp85 is part of a multisubunit complex (Voulhoux et al, 2003) that in E. coli at least, contains four different lipoproteins (Wu et al, 2005; Sklar et al, 2007). The mitochondrial Omp85 homologue, called Sam50 or Tob55, was also shown to be required for the assembly of β-barrel proteins into the outer membrane (Kozjak et al, 2003; Paschen et al, 2003; Gentle et al, 2004). In chloroplasts, it seems that during endosymbiont evolution the Omp85 family has expanded into having many members. The best-studied member, translocon outer membrane complex 75-III (Toc75-III), is involved in protein transport across rather than into the membrane, whereas another family member, Toc75-V or AtOEP80, might be dedicated to the insertion of β-barrel proteins into the outer membrane (Inoue & Potter, 2004).
We predicted that the bacterial Omp85 consists of a carboxy-terminal membrane-embedded β-barrel of 315 amino-acid residues and a 461-residue-long amino-terminal extension residing in the periplasm (Voulhoux et al, 2003). Within the periplasmic moiety, five so-called polypeptide-transport-associated (POTRA) domains, suggested to have chaperone-like qualities, were identified using hidden Markov model searches (Sánchez-Pulido et al, 2003). POTRA domains were also identified in other members of the Omp85 superfamily of polypeptide-transporting β-barrels, including Sam50, Toc75 and the outer-membrane-located TpsB component of the two-partner secretion systems of Gram-negative bacteria. However, in these other proteins, the number of POTRA domains is restricted to three or fewer. In the present study, we have addressed the possible redundancy of POTRA domains in Omp85 of N. meningitidis by mutational analysis and found that only one specific domain is essential for Omp85 function. This domain, together with the β-barrel, constitutes the functional core of Omp85.
Results
POTRA domains 1–4 are dispensable for viability
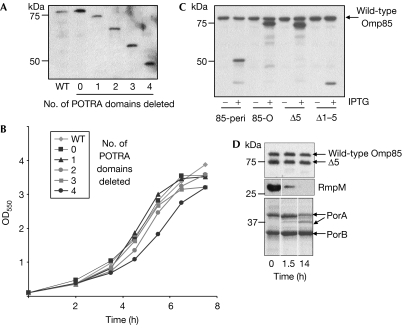
To address the role of the different POTRA domains, we generated 5′ truncations in the omp85 gene (Fig 1). The mutant alleles were cloned under the control of an isopropyl-β-D-thiogalactopyranoside (IPTG)-inducible promoter on a neisserial replicative plasmid and, as a result of the cloning strategy, the signal sequence of the recombinant gene product was substituted by that of the lactoferrin receptor LbpA. The plasmids were introduced into N. meningitidis strain HB-1, and the ability of the Omp85 variants to substitute for the wild-type Omp85 was assessed by testing whether the chromosomal copy of omp85 could be inactivated. To this end, we transformed the mutant Omp85-expressing strains with an omp85 allele disrupted with a kanamycin resistance cassette and constructed in such a way that it can only recombine into the chromosomal omp85 locus. Successful allelic exchange was tested by PCR. In strains producing complete Omp85 or variants lacking 1, 2, 3 or 4 POTRA domains from plasmids, the chromosomal omp85 gene could be inactivated. Immunoblots confirmed that these strains no longer expressed wild-type Omp85, but only the plasmid-encoded proteins at levels comparable with wild-type Omp85 (Fig 2A). By contrast, we were not able to inactivate the chromosomal omp85 allele when an Omp85 protein lacking all five POTRA domains (Δ1–5) was expressed from plasmid (Fig 2C). Thus, at least one POTRA domain is required for Omp85 function. The absence of up to four POTRA domains hardly affected growth rates: only the strain expressing Omp85 that lacked four POTRA domains grew slightly more slowly than the others (Fig 2B).
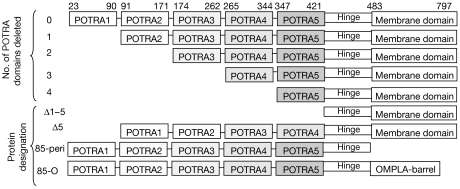
Figure 1.
Domain organization of Omp85 and constructed Omp85 variants. POTRA domains were defined according to Sánchez-Pulido et al (2003) and the membrane domain according to Voulhoux et al (2003). The numbers at the top indicate the positions of the amino- and carboxy-terminal amino acids of each domain in the Omp85 sequence. Protein designations are shown on the left. OMPLA, outer membrane phospholipase A; POTRA, polypeptide-transport-associated.
Figure 2.
Analysis of Omp85 mutants. Wild type (WT) refers to strain HB-1. Molecular-weight markers are indicated on the left side of the blots. (A) Immunoblots containing cell lysates from bacteria expressing functional Omp85 variants were probed with an Omp85 carboxy-terminal antiserum. Loadings were equalized on the basis of optical density of the cultures. (B) Growth curves of bacteria expressing functional Omp85 variants in tryptic soy broth plus IPTG. (C) Cell lysates of bacteria expressing Omp85 variants that did not allow disruption of the chromosomal omp85 gene were probed with Omp85 amino-terminal antiserum. Protein designations are as indicated in Fig 1. (D) Cell envelopes of bacteria expressing Δ5 were treated at room temperature with 50 μg/ml trypsin for the time periods indicated below the lanes, subjected to denaturing SDS–PAGE, blotted and probed with Omp85 N-terminal antiserum (top panel), RmpM antibody (middle panel) or stained with Coomassie brilliant blue (lower panel). IPTG, isopropyl-β-D-thiogalactopyranoside; OD, optical density; Omp, outer membrane protein; PorA, porin A; POTRA, polypeptide-transport-associated; SDS–PAGE, SDS–polyacrylamide gel electrophoresis.
POTRA5 is essential for Omp85 function
To examine whether POTRA5 specifically is required for Omp85 function, we constructed an Omp85 variant lacking this domain while retaining the other four POTRA domains (Δ5; Figs 1, 2C). The chromosomal wild-type omp85 allele could not be inactivated in a strain expressing this mutant protein. Thus, the POTRA5 domain is essential. We tested whether Δ5 was correctly assembled in the outer membrane, by trypsin treatment of cell envelopes. Misassembled OMPs are sensitive to trypsin in this type of assay (Voulhoux et al, 2003). Similar to the native Omp85, the Δ5 protein was not degraded, indicating that it was correctly assembled (Fig 2D). As controls, digestion of porin PorA and the outer-membrane-associated protein RmpM was evaluated. PorA contains a trypsin-cleavage site in an extracellular loop, resulting in a distinct degradation product on prolonged trypsin treatment, whereas RmpM was completely degraded (Fig 2D). Furthermore, Δ5 was not extractable with urea (data not shown), confirming that it is assembled in the outer membrane. Thus, Omp85 lacking only POTRA5 is assembled in the outer membrane but is not functional.
The Omp85 membrane domain is essential
To examine whether the predicted membrane domain of Omp85 is required for function, we constructed an Omp85 variant lacking this domain (85-peri) and a chimeric protein (85-O) in which the barrel of Omp85 is replaced by the 12-stranded β-barrel of outer membrane phospholipase A (OMPLA) of N. meningitidis (Figs 1, 2C). The chromosomal omp85 copy could not be inactivated in strains expressing either of these proteins, indicating that the membrane domain of Omp85 is essential and not only acts as a membrane anchor for the periplasmic domain, but is also specifically required for function. None of the mutant Omp85 proteins described exerted dominant-negative effects: expression of these proteins did not affect growth rates (data not shown).
OMP assembly in viable POTRA-deletion mutants
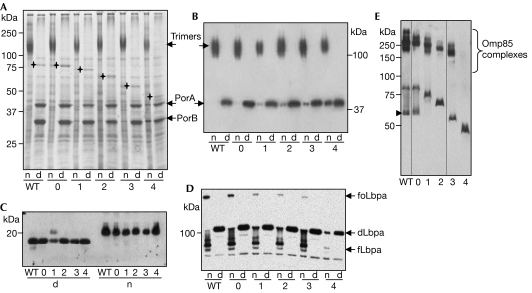
Although deletions of POTRA1–4 did not, or only marginally, diminish cellular viability, they still might partly perturb Omp85 function. To examine this possibility, cell envelopes of the strains expressing mutant Omp85 proteins were analysed by semi-native SDS–polyacrylamide gel electrophoresis (SDS–PAGE). The most-abundant OMPs in N. meningitidis, porins PorA and PorB, remain in their native trimeric conformation during this procedure. Porin trimers were detected in the cell envelopes of all mutants, although also some monomeric porins were detected in the native samples, particularly in that of the mutant lacking four POTRA domains (Fig 3A). Immunoblot analysis showed that the amount of monomeric PorA accumulated was inversely correlated with the number of POTRA domains retained in the Omp85 derivatives (Fig 3B).
Figure 3.
Assessment of outer membrane protein folding and assembly in POTRA deletion mutants. Cell envelopes were denatured by boiling in SDS (d) or left on ice (n). The number of deleted POTRA domains is indicated below the lanes. Molecular-weight markers are indicated next to the blots. (A) Coomassie brilliant blue-stained gel. The positions of the Omp85 proteins are indicated on the gel with crosses. (B,C) Immunoblots of similar samples as shown in (A) were probed with antibodies against PorA (B) or NspA (C). (D) Cells were grown under iron limitation and the blot was probed with antibodies against LbpA. The positions of folded monomer (fLbpa), denatured monomer (dLbpa) and folded oligomers (foLbpa) of LbpA are indicated. (E) Blot containing only native samples probed with Omp85 amino-terminal antiserum. The arrowhead indicates folded monomers of intact Omp85. The lanes are from one single blot, but different exposure time durations are shown (separated by the black lines) to optimize visualization of Omp85 reactivity for all samples. Omp, outer membrane protein; PorA, porin A; POTRA, polypeptide-transport-associated.
Omp85 was shown to interact directly with its substrate proteins (Robert et al, 2006), presumably through its periplasmic domain. Furthermore, Omp85 is thought to exist as a tetramer (Robert et al, 2006), resulting in the presence of 20 POTRA domains in such a complex. It could be that larger OMPs are more sensitive to deletion of POTRA domains in Omp85 than the smaller ones. To investigate this possibility, we examined the folding of the monomeric protein NspA, which, as an eight-stranded β-barrel, is considerably smaller than the 16-stranded β-barrels of porins (Vandeputte-Rutten et al, 2003). In semi-native SDS–PAGE, folded NspA migrates more slowly than the denatured form. The non-denatured samples of all deletion mutants showed only the folded form of NspA (Fig 3C), indicating that the folding of this small β-barrel protein is indeed less sensitive to deletion of POTRA domains in Omp85 than the folding of porins.
The largest β-barrels known are the iron-limitation-inducible receptors, which have 22 β-strands (Chimento et al, 2005), matching closely the number of POTRA domains in the tetrameric Omp85 complex. To induce the synthesis of these receptors in the POTRA deletion mutants, the bacteria were grown under iron limitation. Under these conditions, all mutants showed a slightly reduced growth rate compared with the strains expressing the wild-type Omp85 either from the chromosome or from plasmid (in the latter case, with the LbpA signal sequence): the generation time was approximately 15% higher (data not shown). In semi-native SDS–PAGE, two folded forms of the lactoferrin receptor LbpA can be detected, that is, an oligomer and a folded monomer, which migrate slower and faster, respectively, than the denatured form (Pettersson et al, 2006). The assembly of LbpA was affected to some extent by deletion of Omp85 POTRA domains, most severely on deletion of four domains (Fig 3D). Moreover, the total levels of LbpA seemed to be reduced, presumably by proteolysis of unassembled proteins. Similar effects were found for the transferrin receptor TbpA (data not shown). The folding status of the enterobactin receptor FrpB is not easily assessable in semi-native SDS–PAGE (Kortekaas et al, 2006), but, also in this case, the amounts of protein detected were correlated with the number of POTRA domains in the Omp85 derivatives (data not shown). Thus, assembly defects in larger OMPs become more pronounced on deletion of multiple POTRA domains, although at least a proportion of the LbpA and TbpA proteins produced was correctly assembled even in the Omp85 mutant lacking four POTRA domains.
The secretin PilQ forms a large dodecameric complex and is also dependent on Omp85 for its assembly (Voulhoux et al, 2003). The number of β-strands in each PilQ protomer is not known. The high-molecular-weight PilQ complex was detected in all strains, with some unassembled monomers detected only in the mutant lacking four POTRA domains (data not shown).
Omp85 complex formation can also be assessed by semi-native SDS–PAGE (Voulhoux et al, 2003). High-molecular-weight Omp85 complexes were detected in all POTRA deletion variants, except for the one lacking four POTRA domains (Fig 3E). Of the wild-type Omp85, but not of the mutants, also a folded monomeric form was detected. Apparently, the folded monomers of the mutants are more sensitive to SDS. In conclusion, POTRA1–3 are not required for complex formation. Omp85 lacking four POTRA domains is, although functional, either not present in a complex or forms an instable complex that falls apart during semi-native SDS–PAGE.
Phylogenetic analysis
Our data indicate an essential role for POTRA5 in Omp85 functioning in N. meningitidis. To investigate whether the importance of this domain would be reflected in its positional conservation, we carried out a phylogenetic analysis of Omp85 POTRA domains from 59 bacterial species (supplementary Fig 1 online). This analysis showed that the POTRA domains almost entirely cluster by their positional number, suggesting that their arrangement might indeed be functionally important. A previous analysis with fewer proteins including three Sam50 homologues (Sánchez-Pulido et al, 2003) indicated that the single Sam50 POTRA domain was most similar to Omp85 POTRA5. We included 22 mitochondrial Omp85 homologues in our analysis and found that the Sam50 POTRA domains cluster with both POTRA1 and POTRA5 (supplementary Fig 1 online). Thus, the single POTRA domain of Sam50 might have evolved from POTRA1 or, more likely considering the importance of POTRA5 shown in this study, from POTRA5 of Omp85.
Discussion
The bacterial OMP assembly factor Omp85 consists of an integral membrane domain and an N-terminal periplasmic moiety containing five POTRA domains. Here, we show that the functional core of Omp85 consists of the integral membrane domain plus POTRA domain 5. The importance of the membrane domain was inferred from the observation that its substitution by an unrelated β-barrel did not result in a functional Omp85. The importance of the POTRA domains was assessed in a deletion analysis, which showed that POTRA1–4 could be deleted resulting in only mild OMP assembly defects, whereas POTRA5 was essential.
The function of the N terminus of Omp85 family members has been addressed in a few studies. In contrast to POTRA5 of bacterial Omp85, the single POTRA domain of mitochondrial Sam50 was not required for viability, at least not in yeast, although its absence reduced growth and impaired import of β-barrel OMPs (Habib et al, 2007). It was not required for assembly of the Sam50 complex, but it was shown to bind to precursors of β-barrel proteins. A similar function was postulated for the N terminus of bacterial Omp85, as purified Omp85 was shown to interact directly with its substrate OMPs (Robert et al, 2006). Also the N terminus of Toc75-III of the chloroplast protein import machinery, which contains two POTRA domains, was reported to bind to substrate proteins and, additionally, to mediate binding of accessory proteins of the Toc complex (Bredemeier et al, 2007). The N terminus of the cyanobacterial counterpart also binds to substrate proteins and, additionally, mediates homo-oligomerization and interacts with the C-terminal domain, which is not the case for the plastidic member (Bredemeier et al, 2007). Thus, it seems that the N termini of different members of the Omp85 superfamily have divergent functions. This notion fits with phylogenetic analyses of the separate N and C termini, which suggest that these domains have evolved independently (Bredemeier et al, 2007).
In a recent paper, the structure of a fragment of E. coli Omp85 encompassing POTRA1–4 was described, and a series of mutants in which the five POTRA domains were deleted, one at a time, was analysed (Kim et al, 2007). In sharp contrast to our results, all mutations severely infringed OMP assembly and not only POTRA5 but also POTRA3 and 4 seemed to be essential for the viability of E. coli. This discrepancy might be due to differences in the OMP assembly pathway between the two organisms. Alternatively, the relative position of the individual POTRA domains might be important, as suggested by the strong positional conservation of these domains in our phylogenetic analysis. Thus, taking one POTRA domain out juxtaposes the two domains, which are not adjacent in wild-type Omp85 and might be more deleterious to Omp85 function than completely removing multiple domains.
Our result shows that POTRA5 is essential in bacterial Omp85, and we speculate that it might interact directly with the substrate proteins, particularly with the C-terminal signature motif that is present in these proteins (Robert et al, 2006). Alternatively, or in addition, POTRA5 might be required for the binding of the essential accessory lipoprotein YfiO as was indicated in the analysis of the POTRA5 deletion in E. coli Omp85 (Kim et al, 2007). The nonessential POTRA1–4 domains might enhance the efficiency of OMP assembly, perhaps by forming a broader platform for OMP binding, which might be particularly relevant for the larger β-barrels. Consistently, the assembly of a small β-barrel protein, NspA, seemed totally unaffected by the deletion of POTRA1–4 in Omp85, but even the largest β-barrels, the receptors, could still assemble in such mutants. Also, POTRA4 is possibly involved in stabilization of the assembly complex, as this complex could no longer be detected in semi-native SDS–PAGE in the POTRA1–4 deletion mutant.
Methods
Bacterial strains and growth conditions. Strain HB-1, an unencapsulated derivative of N. meningitidis serogroup B strain H44/76 (Bos & Tommassen, 2005), and derivatives were grown on GC agar (Becton Dickinson, Sparks, MD, USA) plates containing Vitox (Oxoid, Basingstoke, UK) and antibiotics when appropriate (kanamycin 100 μg/ml; chloramphenicol 10 μg/ml) in candle jars at 37 °C. Cultures were grown in tryptic soy broth or, to impose iron limitation, in RPMI (Sigma-Aldrich, St Louis, MO, USA) in plastic flasks at 37 °C with shaking. Expression of omp85 alleles from plasmids was induced with 0.1 mM IPTG. E. coli strains DH5α and TOP10F' (Invitrogen, Carlsbad, CA, USA) were used for routine cloning. E. coli was propagated on LB plates containing antibiotics when appropriate (kanamycin 50 μg/ml; chloramphenicol 25 μg/ml).
Constructions. Relevant segments of the omp85 gene were amplified by PCR from H44/76 genomic DNA with 5′ primers containing an NdeI restriction site and 3′ primers containing a stop codon followed by an AatII restriction site. The PCR products were cloned into pCRII-TOPO (Invitrogen) and sequenced. Subsequently, they were subcloned using NdeI/AatII restriction into pFP10-c-LbpA, a Neisseria replicative plasmid containing the lbpA gene plus 37 bp upstream sequence behind a tandem lac-tac promoter (Pettersson et al, 2006). In the plasmid-encoded mutant Omp85 proteins, the authentic signal sequence is substituted by the signal sequence plus four N-terminal amino acids from LbpA (that is, sequence MNKKHSFPLTLTALAIATAFPSYAANPE). Strain HB-1 was transformed with the plasmids as described previously (Pettersson et al, 2006).
The chromosomal omp85 gene was disrupted through a gene replacement strategy as described previously (Voulhoux et al, 2003). Briefly, using pRV1300 as the template, a PCR product was generated containing a part of the gene upstream from omp85 (to allow for homologous recombination), a kanamycin resistance cassette replacing 1,200 bp at the 5′ end of omp85 and the 3′ end of omp85. After gel purification, the PCR product was used to transform HB-1 derivatives containing a mutant omp85 allele on a plasmid in the presence of 1 mM IPTG. Bacteria were plated on GC plates containing kanamycin and IPTG. Replacement of the chromosomal copy of omp85 was tested by PCR using primers that anneal in the 3′ end of omp85 and in the upstream gene (Voulhoux et al, 2003). When replacement of the chromosomal omp85 copy failed, the transformation was repeated at least once to substantiate the conclusion that disruption of the chromosomal gene was not possible. In parallel, a strain carrying a wild-type omp85 allele (albeit with the substitution of the signal-sequence-encoding segment) on the plasmid was used as a positive control. The omp85–OMPLA gene fusion and the omp85 allele lacking the POTRA5-encoding segment were constructed by overlap PCR. From H44/76 genomic DNA, two PCR products were generated containing 20-bp overlapping sequences in the fusion region. The two PCR products were purified and combined in a second PCR. The resulting product was cloned into pCRII-TOPO, sequenced and subcloned into pFP10-c-LbpA using NdeI/AatII sites, which were incorporated in the primers. The OMPLA part of the protein starts with amino-acid residues TrpTyrAsnAsn, corresponding to the beginning of the β-barrel part of neisserial OMPLA. This part was identified by alignment with E. coli OMPLA the crystal structure of which has been solved (Snijder & Dijkstra, 2000).
Cell envelopes. Cell envelopes were prepared and treated with trypsin as described previously (Voulhoux et al, 2003).
Gel electrophoresis and immunoblotting. SDS–PAGE under denaturing or semi-native conditions and immunoblotting were carried out as described previously (Voulhoux et al, 2003). For detection of Omp85 complexes in semi-native PAGE, conditions were slightly adjusted: the running buffer contained 0.025% SDS and the sample buffer 1% SDS.
Antibodies. Rabbit antisera raised against amino-acid residues 22–464 (anti-Omp85 N-terminal serum) or residues 455–797 (anti-Omp85 C-terminal serum) of Omp85 were generous gifts from R. Judd (University of Montana, Missoula, MT, USA). Antibodies against PorA and RmpM came from the Netherlands Vaccine Institute (Bilthoven, The Netherlands). TbpA antiserum and NspA-specific monoclonal antibody AL4 were generously provided by A. Schryvers (University of Calgary, Canada) and D. Granoff (Children's Hospital Oakland Research Institute, Oakland, CA, USA), respectively. LbpA antiserum was described previously (Pettersson et al, 2006).
Phylogenetic analysis. From the recently identified 213 Omp85 homologues in the databases (Bredemeier et al, 2007), we took 59 bacterial and 22 mitochondrial proteins from different genera. Bacterial POTRA domains were defined using the Pfam database (http://www.sanger.ac.uk/Software/Pfam/). As mitochondrial POTRA domains are not always detected by Pfam, we used secondary structure predictions (http://bioinf.cs.ucl.ac.uk/psipred) and defined POTRA domains as having strand–helix–helix–strand–strand structure. The sequences are supplied as supplementary Tables 1,2 online. A dendrogram was constructed using ClustalW1.83 at http://clustalw.genome.jp.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Figure and Tables
Acknowledgments
M.P.B and V.R. were funded by the Research Councils for Earth and Life Sciences (ALW) and Chemical Sciences (CW), respectively, with financial aid from the Netherlands Organization for Scientific Research (NWO).
References
- Bos MP, Tommassen J (2005) Viability of a capsule- and lipopolysaccharide-deficient mutant of Neisseria meningitidis. Infect Immun 73: 6194–6197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredemeier R, Schlegel T, Ertel F, Vojta A, Borissenko L, Bohnsack MT, Groll M, von Haeseler A, Schleiff E (2007) Functional and phylogenetic properties of the pore-forming β-barrel transporters of the Omp85 family. J Biol Chem 282: 1882–1890 [DOI] [PubMed] [Google Scholar]
- Chimento DP, Kadner RJ, Wiener MC (2005) Comparative structural analysis of TonB-dependent outer membrane transporters: implications for the transport cycle. Proteins 59: 240–251 [DOI] [PubMed] [Google Scholar]
- Doerrler W, Raetz CR (2005) Loss of outer membrane proteins without inhibition of lipid export in an Escherichia coli YaeT mutant. J Biol Chem 280: 27679–27687 [DOI] [PubMed] [Google Scholar]
- Gentle I, Gabriel K, Beech P, Waller R, Lithgow T (2004) The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J Cell Biol 164: 19–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentle IE, Burri L, Lithgow T (2005) Molecular architecture and function of the Omp85 family of proteins. Mol Microbiol 58: 1216–1225 [DOI] [PubMed] [Google Scholar]
- Habib SJ, Waizenegger T, Niewienda A, Paschen SA, Neupert W, Rapaport D (2007) The N-terminal domain of Tob55 has a receptor-like function in the biogenesis of mitochondrial β-barrel proteins. J Cell Biol 176: 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Potter D (2004) The chloroplastic protein translocation channel Toc75 and its paralog OEP80 represent two distinct protein families and are targeted to the chloroplastic outer envelope by different mechanisms. Plant J 39: 354–365 [DOI] [PubMed] [Google Scholar]
- Kim S, Malinverni JC, Sliz P, Silhavy TJ, Harrison SC, Kahne D (2007) Structure and function of an essential component of the outer membrane protein assembly machinery. Science 317: 961–964 [DOI] [PubMed] [Google Scholar]
- Kortekaas J, Müller SA, Ringler P, Gregorini M, Weynants VE, Rutten L, Bos MP, Tommassen J (2006) Immunogenicity and structural characterisation of an in vitro folded meningococcal siderophore receptor (FrpB, FetA). Microbes Infect 8: 2145–2153 [DOI] [PubMed] [Google Scholar]
- Kozjak V, Wiedemann N, Milenkovic D, Lohaus C, Meyer HE, Guiard B, Meisinger C, Pfanner N (2003) An essential role of Sam50 in the protein sorting and assembly machinery of the mitochondrial outer membrane. J Biol Chem 278: 48520–48523 [DOI] [PubMed] [Google Scholar]
- Paschen SA, Waizenegger T, Stan T, Preuss M, Cyrklaff M, Hell K, Rapaport D, Neupert W (2003) Evolutionary conservation of biogenesis of β-barrel membrane proteins. Nature 426: 862–866 [DOI] [PubMed] [Google Scholar]
- Pettersson A, Kortekaas J, Weynants VE, Voet P, Poolman J, Bos MP, Tommassen J (2006) Vaccine potential of the Neisseria meningitidis lactoferrin-binding proteins LbpA and LbpB. Vaccine 24: 3545–3557 [DOI] [PubMed] [Google Scholar]
- Robert V, Volokhina EB, Senf F, Bos MP, Van Gelder P, Tommassen J (2006) Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol 4: e377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Pulido L, Devos D, Genevrois S, Vicente M, Valencia A (2003) POTRA: a conserved domain in the FtsQ family and a class of β-barrel outer membrane proteins. Trends Biochem Sci 28: 523–526 [DOI] [PubMed] [Google Scholar]
- Sklar JG, Wu T, Gronenberg LS, Malinverni JC, Kahne D, Silhavy TJ (2007) Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc Natl Acad Sci USA 104: 6400–6405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder HJ, Dijkstra BW (2000) Bacterial phospholipase A: structure and function of an integral membrane phospholipase. Biochim Biophys Acta 1488: 91–101 [DOI] [PubMed] [Google Scholar]
- Vandeputte-Rutten L, Bos MP, Tommassen J, Gros P (2003) Crystal structure of neisserial surface protein A (NspA), a conserved outer membrane protein with vaccine potential. J Biol Chem 278: 24825–24830 [DOI] [PubMed] [Google Scholar]
- Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J (2003) Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299: 262–265 [DOI] [PubMed] [Google Scholar]
- Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Kahne D (2005) Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121: 235–245 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure and Tables