Abstract
The self-perpetuating conformational change of the translation termination factor Sup35 is associated with a prion phenomenon of Saccharomyces cerevisiae. In vitro, the prion-determining region (NM) of Sup35 assembles into amyloid-like fibres through a mechanism of nucleated conformational conversion. Here, we describe an alternative assembly pathway of NM that produces filaments that are composed of β-strands and random coiled regions with several-fold smaller diameters than the amyloid fibres. NM filaments are not detectable with either thioflavin T or Congo Red and do not show SDS or protease resistance. As filaments do not self-convert into fibres and do not act as seed, they are not intermediates of amyloid fibre formation. Instead, they represent a stable off-pathway form. Similar to mammalian prion proteins, Sup35 contains oligopeptide repeats located in the NM region. We found that the number of repeats determines the partitioning of the protein between filaments and amyloid-like fibres. Low numbers of repeats favour the formation of the filamentous structure, whereas high numbers of repeats favour the formation of amyloid-like fibres.
Keywords: filament, AFM, FTIR, oligopeptide repeats
Introduction
Prions are a diverse group of unrelated proteins that share one unusual characteristic: they are able to adopt different conformational states. A self-perpetuating conformational switch of the mammalian prion PrP can lead to severe neurological disorders (Scheibel & Buchner, 2006). In fungi, however, prion proteins are not inherently toxic and instead cause epigenetically heritable traits (Wickner, 1994; Shorter & Lindquist, 2005). One of the most intensively studied fungal prion proteins is the translation termination factor Sup35 of Saccharomyces cerevisiae. The prion propagation of Sup35 creates a diverse array of phenotypes, enhancing or reducing growth and survival in distinct environments (True & Lindquist, 2000). These traits are heritable by a ‘protein-only' mechanism because Sup35 proteins in the prion state are transmitted from the cytoplasm of the mother to the daughter cell. The prion form converts new Sup35 protein to the prion state, producing a self-perpetuating epigenetic element of inheritance (Serio et al, 2000; DePace & Weissman, 2002).
Sup35 converts from the normal to the prion state in vivo, whereas soluble Sup35 converts into amyloid-like fibres in vitro. Prion propagation in vivo, as well as amyloid formation in vitro, is mediated by the prion-determining amino-terminal asparagine- and glutamine-rich region (N), and the highly charged middle region (M; Glover et al, 1997). N contains oligopeptide repeats similar to those found in the amino-terminal domain of mammalian prion proteins. The repeats of N are critically involved in both prion propagation (Liu & Lindquist, 1999; Parham et al, 2001) and amyloid formation (Scheibel & Lindquist, 2001). Decreasing the number of oligopeptide repeats reduces the propensity of prion propagation and amyloid formation. Similarly, expansion of the repeats induces the spontaneous appearance of the prion state in vivo and leads to accelerated amyloid fibre assembly in vitro.
In vitro NM amyloid fibre assembly is a two-stage process following a mechanism of nucleated conformational conversion (Serio et al, 2000; Scheibel et al, 2004). The soluble state of NM represents an equilibrium between monomers and oligomers. On formation of a nucleus, further protein is converted into thermodynamically stable amyloid-like fibres. The underlying conformational change mainly occurs in the N-terminal region N, with the oligopeptide repeats probably involved in forming the amyloid core (Krishnan & Lindquist, 2005).
Fibrous intermediates have been described for several amyloidogenic proteins and are apparently on-pathway for amyloid formation (Harper et al, 1997; Walsh et al, 1997; Kelly, 1998; Serpell, 2000). These protofibrils have many common structural characteristics similar to amyloid fibres. Here, we describe a filamentous species of NM that is not amyloidogenic and that is off-pathway for amyloid fibre formation. Its structural features are profoundly different from those of amyloid fibres. Interestingly, the number of oligopeptide repeats determines whether NM spontaneously assembles into the amyloid-like form or into the alternative filamentous form.
Results
NM assembly forms
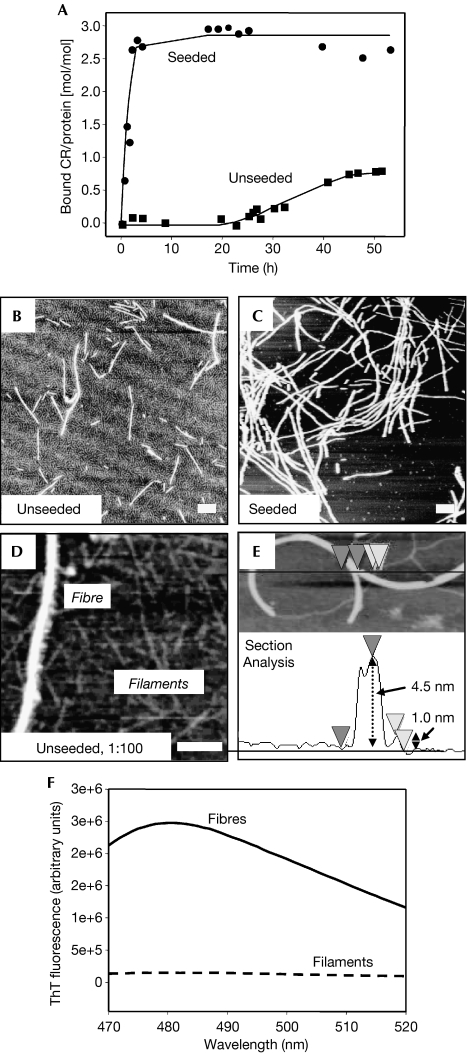
NM amyloid fibre assembly was monitored using congo red (CR). On incubation of soluble NM without agitation, which is normally used to speed up the reactions, CR staining started after a lag phase of approximately 20 h—reflecting nuclei formation—and was completed after a period of approximately 45 h. De novo formation of nuclei can be bypassed by the addition of preformed fibres, which act as seed. In this case, fibre assembly started immediately after the addition of seeds and was completed after approximately 6 h (Fig 1A). The lower CR signal in unseeded reactions indicated a smaller amount of amyloid-like fibres. In these samples, an apparently filamentous protein background layer was detected, in addition to the characteristic fibres, by atomic force microscopy (AFM; Fig 1B,C). At higher magnifications, this protein layer showed a needle-like morphology with a height of 0.8–1.3 nm, with four- to eightfold smaller diameter than that of amyloid-like NM fibres with 4–6 nm height, under the conditions used (Fig 1D,E). It is important to note that although AFM in air does not necessarily measure true heights of proteins, these NM filaments showed several morphological characteristics of protofibrils detected for other amyloid-forming proteins (Harper et al, 1997).
Figure 1.
Amyloid fibre assembly of NM monitored by congo red binding in quiescent conditions. (A) Unseeded (squares) and seeded with 4% w/w sonicated fibres (circles); 150 μg/ml soluble protein each. After completion of the assembly reaction, samples were investigated by AFM: (B) unseeded and (C) seeded; (D) reflects a diluted (1:100) sample of (B). (E) AFM section analysis to exemplarily determine filament and fibre heights. Scale bars, 200 nm. (F) ThT binding (excitation at 450 nm) of fibres and filaments. AFM, atomic force microscopy; CR, congo red; M, highly charged middle region; N, asparagine- and glutamine-rich region; ThT, thioflavin T.
Characterization of NM filaments
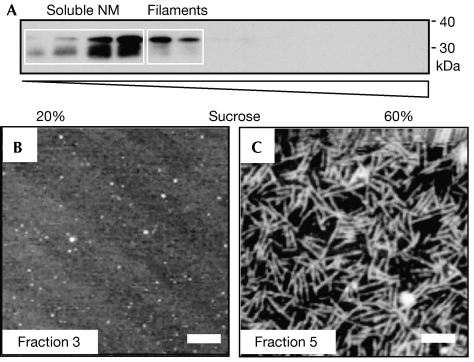
A sedimentation/filtration assay was established to separate filaments from amyloid-like fibres in unseeded assembly reactions after 4 days of incubation. All amyloid-like fibres could be easily sedimented, whereas filaments (and soluble protein) were found only in the supernatant after centrifugation. The fibrous pellet fraction showed the amyloid-characteristic CR spectral shift. In the filament-containing supernatant, no dye binding, either CR or thioflavin T (ThT; Fig 1F), was observed, which indicated the absence of amyloid-like structures. The supernatant was characterized further by centrifugation in a continuous sucrose gradient, and subsequent SDS–polyacrylamide gel electrophoresis (SDS–PAGE) and immunoblotting (Fig 2A). AFM analysis showed soluble protein in the first four gradient fractions and filaments in the next two (Fig 2B,C), indicating that filaments are not artefacts of sample preparation but a separable and stable structural state of NM. Interestingly, degradation bands were only found in fractions of soluble NM and not in filament-containing fractions (Fig 2A). In several independent experiments, densitometric quantification of the non-degraded protein bands at approximately 35 kDa showed 50–80% of NM in the soluble fractions and substantially 20–50% in the filamentous fractions. As it was not possible to separate filaments from the remaining soluble protein quantitatively, further analysis was carried out with the mixture of both species (soluble and filamentous; termed filaments) and compared with freshly dissolved NM (solely reflecting soluble protein (100%)).
Figure 2.
Separation of filaments. (A) Fractions after a sucrose gradient, SDS–PAGE and immunoblotting. (B, C) AFM images of (B) soluble NM in fraction 3 and (C) filaments in fraction 5. Scale bars, 200 nm. AFM, atomic force microscopy; M, highly charged middle region; N, asparagine- and glutamine-rich region; SDS–PAGE, SDS–polyacrylamide gel electrophoresis.
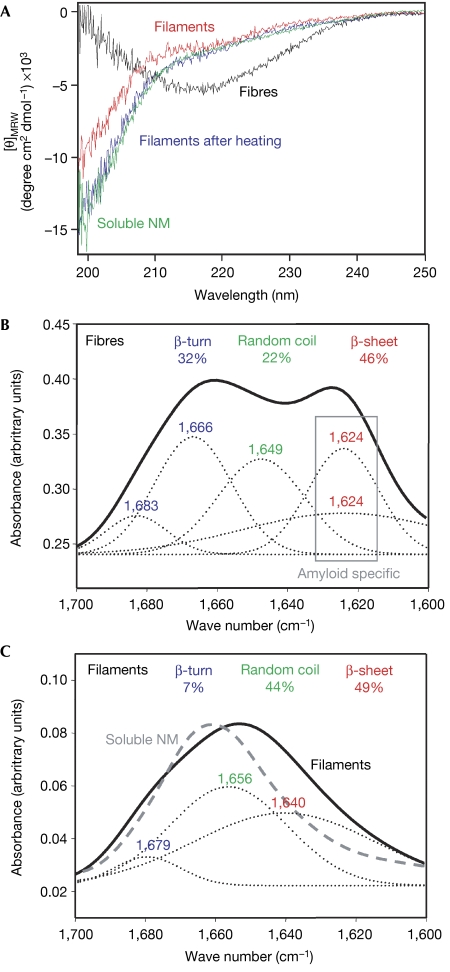
In contrast to intrinsically unstructured soluble NM, amyloid-like fibres resemble β-sheet-rich structures (Scheibel & Lindquist, 2001). As determined by far-UV circular dichroism (CD) spectroscopy, filaments showed a structure dominated by random coil with only little β-sheet content (Fig 3A). The secondary structure content of fibres and soluble NM was thermally stable, whereas the filaments lost their β-sheet content on heating, yielding CD spectra that were indistinguishable from that of soluble NM (Fig 3A). No remaining filaments were detected by AFM analysis after heating.
Figure 3.
Structural characterization of filaments. (A) Comparison of soluble (green), filamentous (red), fibrous NM (black) and filaments after heat treatment at 98°C (blue) by CD spectroscopy. (B, C) Comparison of (B) fibres and (C) filaments by FTIR spectroscopy using the amid I band (solid lines) between 1,600 and 1,700 cm−1. The secondary structure content was determined by deconvoluting the spectra using gaussian fits (dotted lines). Structure-content and -corresponding peaks are marked in identical colours: β-turn in blue, random coil in green and β-sheet in red. The FTIR spectrum of soluble NM is shown as a dashed grey line (C). CD, circular dichroism; FTIR, Fourier transformed infrared; [θ]MRW, mean residue weight ellipticities; M, highly charged middle region; N, asparagine- and glutamine-rich region.
Next, we used Fourier transformed infrared (FTIR) spectroscopy for detailed structural analysis. The amyloid-like fibres showed a spectrum indicative of high β-sheet content (46%) with amyloid-specific signals below 1,630 cm−1, high β-turn content (32%) and some random coil structure (22%; Fig 3B). In comparison, the filament spectrum showed a similar amount of β-sheets (49%) but a much higher amount of random coil structure (44%; Fig 3C). The β-sheet signal was above 1,630 cm−1, indicative of a non-amyloidogenic structure (Zandomeneghi et al, 2004). Strikingly, the β-turn content in the filament sample was significantly lower (7%) than in the fibre sample. The FTIR spectrum of soluble NM was different from that of both filaments and fibres, with a high content of random coil, and little β-sheet and β-turn structure (Fig 3C).
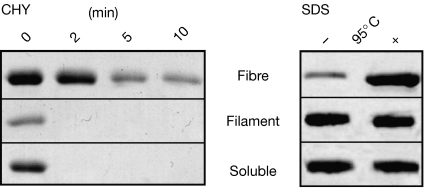
To investigate further the structural differences between filaments and fibres, we compared their protease sensitivity and SDS resistance. NM fibres show partial proteolytic resistance to chymotrypsin digestion, as the cleavage sites, which are restricted to the N-region, become inaccessible inside the rigidly packed β-sheet structure (Serio et al, 2000). Here, filaments showed protease sensitivity comparable with soluble NM (Fig 4, left panel). Furthermore, resistance to solubilization with SDS is often observed in β-sheet-rich proteins with compact structural packing (Manning & Colon, 2004), such as NM fibres (Serio et al, 2000). NM fibres could be only partly dissolved by SDS, whereas soluble and filamentous NM showed no resistance to SDS denaturation (Fig 4B, right panel).
Figure 4.
Stability of filaments, fibres and soluble NM analysed by Coomassie-stained SDS–PAGE. Left panel: protease digestion (1/50 w/w chymotrypsin) at 37°C for 0, 2, 5 and 10 min. Right panel: SDS stability (2% w/v SDS) before (−) and after (+) incubation at 95°C for 10 min. The protein bands indicate monomeric NM. CHY, chymotrypsin; M, highly charged middle region; N, asparagine- and glutamine-rich region; SDS–PAGE, SDS–polyacrylamide gel electrophoresis.
NM filaments are off-pathway for amyloid formation
To determine whether NM filaments are on-pathway intermediates, filaments were incubated for 4 weeks to investigate putative fibre assembly. Aliquots were continuously removed and analysed using CR binding (Fig 5), AFM and far-UV CD. No structural change was detected in the sample during the incubation period, indicating that either filaments are not on-pathway for fibre assembly or that they reflect very stable intermediates.
Figure 5.
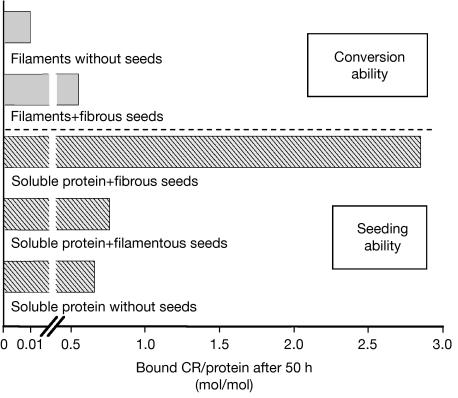
Amyloid formation and seeding ability of filaments. The amount of amyloid-like fibres was detected by CR binding after incubation for 50 h at 24 °C. Amyloid-forming ability of filaments: filaments (150 μg/ml each) were incubated with or without fibrous seeds (4% w/w). Seeding ability of filaments: soluble NM (150 μg/ml each) was incubated in the presence of fibrous seeds, filamentous seeds (4% w/w each) and without seeds (to exclude possible influences owing pipetting, 4% w/w soluble NM was added to simulate the addition of seeds). CR, congo red; M, highly charged middle region; N, asparagine- and glutamine-rich region.
Seeding was used to test whether filaments are stable intermediates. On addition of seeds to the end points of quiescent assembly reactions, a small amount of newly formed amyloid-like fibres was detected (Fig 5). In contrast to seeded reactions with identical concentrations of soluble NM, only approximately 20% of the sample was converted into amyloid-like fibres. It seems most likely that the protein that was converted in these reactions was the remaining soluble fraction of NM, which had not acquired a filamentous structure to begin with because the yield of fibres was stable over several days. Furthermore, when the end products of quiescent reactions were heated to denature the filaments, all of the protein was converted into fibres on the addition of seeds.
To investigate whether filaments themselves act as seeds, soluble NM was incubated in the presence of identical volumes and concentrations of further soluble NM, filaments or fibres. Fibrous NM seeds efficiently accelerated fibre assembly, but filaments and soluble NM were indistinguishably inactive, indicating that filaments do not represent seeds (Fig 5).
On the basis of our results, we conclude that filaments are an NM species that is structurally different from amyloid-like fibres that does not convert directly into fibres under the conditions used.
Oligopeptide repeats are critically involved in NM assembly
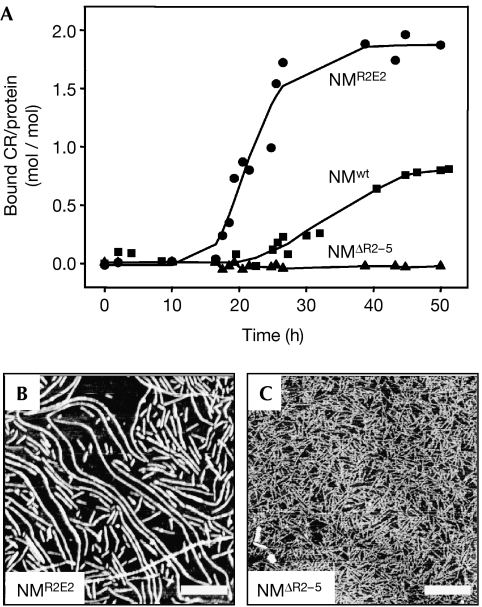
Finally, we tested whether the oligopeptide repeats of NM are involved in partitioning between the different assembly forms. Fibre assembly of wild-type NM (NMwt) was compared with that of the partial oligopeptide repeat deletion mutant NMΔR2−5 (lacking the last four repeats) and of the oligopeptide repeat expansion NMR2E2 (with two additional copies of the second repeat). In comparison with NMwt (Figs 1A, 6A), NMR2E2 showed a significantly shorter lag time (∼10 h) and a higher amount of bound CR at the end point of assembly (Fig 6A). Strikingly, NMΔR2−5 did not form detectable amounts of amyloid-like fibres even after 6 weeks of incubation (Fig 6A). AFM images showed a large number of fibres for NMR2E2 (Fig 6B), less for NMwt (Fig 1B) and none for NMΔR2−5 (Fig 6C). In the case of NMΔR2−5, mainly filaments were detected (Fig 6C).
Figure 6.
Influence of oligopeptide repeats. Unseeded fibre assembly reactions monitored by CR binding: (A) NMwt (squares), NMR2E2 (circles) and NMΔR2−5 (triangles); and 150 μg/ml each. After completion of the assembly reaction, samples were investigated by AFM: (B) NMR2E2 and (C) NMΔR2−5. Scale bars, 500 nm. AFM, atomic force microscopy; CR, congo red; M, highly charged middle region; N, asparagine- and glutamine-rich region; NMΔR2−5, partial oligopeptide repeat deletion mutant NM; NMR2E2, oligopeptide expansion mutant of NM; NMwt, wild-type NM.
Discussion
The new filamentous NM structure described here shows, beside its fibrous appearance, no common characteristics with amyloid fibres or protofibrils. NM filaments do not bind to the amyloid-specific dyes CR and ThT, are not stable against SDS treatment, and are not protease or heat resistant. Furthermore, filaments do not act as seeds for amyloid fibre assembly and do not self-assemble into fibres.
CD and FTIR analysis show a β-sheet-rich secondary structure for NM fibres, whereas the filaments are highly unstructured with little β-sheet content. Approximately 30% of the fibre structure reflects β-turns, which are almost absent in filaments. Furthermore, the FTIR maxima below 1,630 cm−1, which is indicative of a β-sheet structure characteristic for amyloids (Zandomeneghi et al, 2004), was only detected for fibres (at 1,624 cm−1) and not for filaments.
Therefore, we assume that filaments show a different structural arrangement from the amyloid-like fibres, having β-strands inside a β-sheet that are aligned perpendicular to the fibre axis (Geddes et al, 1968; Slotta et al, 2007). Filaments probably consist of β-strands connected by long unstructured loops, instead of β-turns as in the case of amyloid-like fibres. As shown for monomeric Aβ peptide (Mastrangelo et al, 2006) and for fibres of the yeast prion protein Ure2 (Chan et al, 2005), the height of two β-strands forming a β-sheet is in the low nanometre region, as determined by AFM. Taking into account that determining heights of biopolymers as measured by AFM depends on scanning parameters (including instrument and cantilevers), the height shown in our experiments is in the same region. Therefore, structurally the β-strands could be aligned in parallel to the filament axis. However, the exact arrangement of β-strands inside the filaments has yet to be determined.
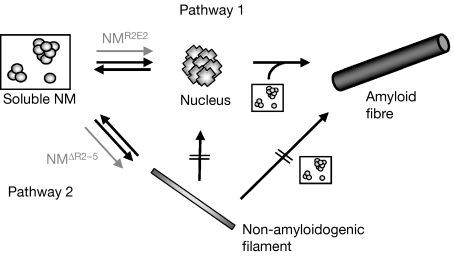
The presence of filaments indicates an alternative NM assembly pathway, which is off-pathway for amyloid formation (Fig 7). The formation of filaments is most probably a nucleation-independent reaction, as filament assembly was not accelerated on the addition of preformed filaments. Furthermore, filaments cannot be directly converted into amyloid-like fibres. The routing of NM into one of the assembly pathways is probably kinetically driven. The oligopeptide expansion mutant NMR2E2 forms nuclei faster than NMwt, leading to a higher amount of fibres and to no detectable amounts of filaments. In accordance with this hypothesis, the deletion mutant NMΔR2−5 assembles mainly into filaments. This hypothesis is supported further by the results of seeded fibre assembly reactions in which preformed fibres bypass the nucleation step and directly initiate fibre elongation; in seeded reactions, almost no filaments were detected.
Figure 7.
Model of Sup35–NM assembly pathways. Amyloid fibre formation follows the mechanism of nucleated conformational conversion (Pathway 1). Non-amyloidogenic filament assembly (Pathway 2) represents an alternative pathway: filaments are not directly assembling into amyloid-like fibres and do not act as seeds for soluble NM, indicating that they are not on-pathway for amyloid fibre assembly. Fibre assembly through the dissociation of filaments into soluble NM is not excluded. NMR2E2 is less prone to filament assembly than NMwt, whereas NMΔR2−5 has a higher tendency to form filaments (grey arrows). M, highly charged middle region; N, asparagine- and glutamine-rich region; NMΔR2−5, partial oligopeptide repeat deletion mutant; NMR2E2, oligopeptide expansion mutant of NM; NMwt, wild-type NM.
On- and off-pathway oligomers differing in size and shape have been reported for several amyloid-forming proteins (Collins et al, 2004; Chiti & Dobson, 2006; Kodali & Wetzel, 2007). Regardless of biologically relevant isoforms, the ability of amyloidogenic proteins to form distinct abnormal conformers reflects the complexity of the energetic landscape of folding as well as high conformational plasticity. NM filaments represent oligomers off pathway for amyloid formation with different characteristics to that of oligomers known from other amyloidogenic proteins. Our finding raises the question of whether the formation of off-pathway oligomers is a generic characteristic of amyloidogenic proteins and whether such oligomers have biological importance.
Methods
Protein purification. NMwt, NMR2E2 and NMΔR2−5 were produced and purified as described previously (Scheibel & Lindquist, 2001).
Analysis of fibre assembly. Purified NM was denatured in 8 M GdmCl overnight. Fibre assembly was initiated on dilution into 5 mM potassium phosphate (pH 6.8), 150 mM NaCl to a final concentration of 150 μg/ml protein and 80 mM GdmCl. The assembly reactions were carried out at 24 °C and monitored by the CR spectral-shift assay as described previously (Scheibel & Lindquist, 2001). In seeded reactions, fibre assembly was initiated by the addition of sonicated preformed amyloid-like fibres. ThT binding (excitation at 450 nm) was carried out at 24 °C with a final concentration of 1.25 μM NM and 3.75 μM ThT using a Spex FluoroMax-3 spectrophotometer (Jobin, Edison, NJ, USA).
Atomic force microscopy. Samples (10 μl) were placed on freshly cleaved mica. After 3 min of adsorption, samples were rinsed three times with Millipore-filtered distilled water and air dried. Tapping mode imaging was carried out on a Digital Instruments multimode scanning probe microscope (Veeco, Santa Barbara, CA, USA) using n+-silicon cantilevers (NCH-50, Nanosensors, Neuchatel, Switzerland).
Filament separation. Samples were sedimented in an Optima Max-E ultracentrifuge (Beckman, Fullerton, CA, USA) using a TLA-45 rotor at 130,000g for 30 min at 20°C. The supernatant was filtered twice through 0.22 μm filters (Whatman, Maidstone, UK). Separation of NM filaments and soluble protein by size-exclusion chromatography or filtration through a 100 kDa cut-off membrane was not feasible owing to unspecific interactions of NM with SEC matrices and membranes. To qualitatively separate them, a 20–60% continuous sucrose gradient (2.5 ml total volume) was prepared as described previously (Liebman et al, 2006). A 300 μl volume of NM (150 μg/ml) was applied to the sucrose gradient and centrifuged at 20°C and 100,000g overnight in an Optima Max-E ultracentrifuge. After centrifugation, fractions of 200 μl were successively removed from the top to the bottom of the tube, and analysed by SDS–PAGE and immunoblotting for NM, as well as by AFM. As it was not possible to separate filaments from the remaining soluble protein in a preparative scale, structural analysis was carried out with the mixture.
Structural analysis. Far-UV CD spectra were recorded using a Jasco J-715 spectropolarimeter (Jasco, Gross-Umstadt, Germany) at a protein concentration of 150 μg/ml in 5 mM potassium phosphate (pH 6.8), 150 mM NaCl and 80 mM GdmCl at 24 °C in a 0.1 cm path-length quartz cuvette (Hellma, Müllheim, Germany). All spectra were buffer corrected.
For FTIR spectroscopy, samples were prepared on CaF2 disks after a dialysis step to remove the remaining GdmCl (which disturbs the amid I signal), air dried and investigated by using an infrared microscope (Irscope, Bruker, Bremen, Germany) coupled to an immunofluorescence staining 66/s spectrometer (Bruker, Germany). Polarized absorbance spectra were recorded in the spectral range between 700 cm−1 and 6,000 cm−1 with a resolution of 4 cm−1. The secondary structure content was determined by deconvoluting the amid I band (1,600–1,700 cm−1) using gaussian fits. All measurements were carried out at 24 °C.
Stability analysis. SDS–PAGE sample buffer containing 2% (w/v) SDS was added to fibrous, filamentous and soluble NM (150 μg/ml each). The samples were analysed directly or after incubation for 10 min at 95°C by SDS–PAGE.
Soluble, filamentous and fibrous NM (150 μg/ml each) were incubated with 1/50 (w/w) bovine α-chymotrypsin (Sigma, St Louis, MO, USA) for various time points at 37°C and analysed by SDS–PAGE.
Acknowledgments
We thank S. Rammensee and G. Sawicki for experimental help, and C. Vendrely, L. Römer, V. Grimminger and T. Franzmann for critical comments on the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft (SFB 596, B14).
References
- Chan JCC, Oyler NA, Yau WM, Tycko R (2005) Parallel β-sheets and polar zippers in amyloid fibrils formed by residues 10–39 of the yeast prion protein Ure2p. Biochemistry 44: 10669–10680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiti F, Dobson CM (2006) Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem 75: 333–366 [DOI] [PubMed] [Google Scholar]
- Collins SR, Douglas A, Vale RD, Weissman JS (2004) Mechanism of prion propagation: amyloid growth occurs by monomer addition. PLoS Biol 2: e321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePace AH, Weissman JS (2002) Origins and kinetic consequences of diversity in Sup35 yeast prion fibers. Nat Struct Biol 9: 389–396 [DOI] [PubMed] [Google Scholar]
- Geddes AJ, Parker KD, Atkins ED, Beighton E (1968) ‘Cross-β' conformation in proteins. J Mol Biol 32: 343–358 [DOI] [PubMed] [Google Scholar]
- Glover JR, Kowal AS, Schirmer EC, Patino MM, Liu JJ, Lindquist S (1997) Self-seeded fibers formed by Sup35, the protein determinant of (PSI+), a heritable prion-like factor of S. cerevisiae. Cell 89: 811–819 [DOI] [PubMed] [Google Scholar]
- Harper JD, Wong SS, Lieber CM, Lansbury PT (1997) Observation of metastable Aβ amyloid protofibrils by atomic force microscopy. Chem Biol 4: 119–125 [DOI] [PubMed] [Google Scholar]
- Kelly JW (1998) The alternative conformations of amyloidogenic proteins and their multi-step assembly pathways. Curr Opin Struct Biol 8: 101–106 [DOI] [PubMed] [Google Scholar]
- Kodali R, Wetzel R (2007) Polymorphism in the intermediates and products of amyloid assembly. Curr Opin Struct Biol 17: 1–10 [DOI] [PubMed] [Google Scholar]
- Krishnan R, Lindquist SL (2005) Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature 435: 765–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebman SW, Bagriantsev SN, Derkatch IL (2006) Biochemical and genetic methods for characterization of (PIN+) prions in yeast. Methods 39: 23–34 [DOI] [PubMed] [Google Scholar]
- Liu JJ, Lindquist S (1999) Oligopeptide-repeat expansions modulate ‘protein-only' inheritance in yeast. Nature 400: 573–576 [DOI] [PubMed] [Google Scholar]
- Manning M, Colon W (2004) Structural basis of protein kinetic stability: resistance to dodecyl sulfate suggests a central role for rigidity and a bias toward β-sheet structure. Biochemistry 43: 11248–11254 [DOI] [PubMed] [Google Scholar]
- Mastrangelo IA, Ahmed M, Sato T, Liu W, Wang C, Hough P, Smith SO (2006) High-resolution atomic force microscopy of soluble Aβ42 oligomers. J Mol Biol 358: 106–119 [DOI] [PubMed] [Google Scholar]
- Parham SN, Resende CG, Tuite MF (2001) Oligopeptide repeats in the yeast protein Sup35p stabilize intermolecular prion interactions. EMBO J 20: 2111–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibel T, Buchner J (2006) Protein aggregation as a cause for disease. In Molecular Chaperones in Health and Disease; Handbook of Experimental Pharmacology, M Gaestel (ed) 172, pp 199–219. Berlin/Heidelberg, Germany: Springer [DOI] [PubMed] [Google Scholar]
- Scheibel T, Lindquist SL (2001) The role of conformational flexibility in prion propagation and maintenance for Sup35p. Nat Struct Biol 8: 958–962 [DOI] [PubMed] [Google Scholar]
- Scheibel T, Bloom J, Lindquist SL (2004) The elongation of yeast prion fibers involves separable steps of association and conversion. Proc Natl Acad Sci USA 101: 2287–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serio TR, Cashikar AG, Kowal AS, Sawicki GJ, Moslehi JJ, Serpell L, Arnsdorf MF, Lindquist SL (2000) Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science 289: 1317–1321 [DOI] [PubMed] [Google Scholar]
- Serpell LC (2000) Alzheimer's amyloid fibrils: structure and assembly. Biochim Biophys Acta 1502: 16–30 [DOI] [PubMed] [Google Scholar]
- Shorter J, Lindquist S (2005) Prions as adaptive conduits of memory and inheritance. Nat Rev 6: 435–450 [DOI] [PubMed] [Google Scholar]
- Slotta U, Hess S, Spieß K, Stromer T, Serpell L, Scheibel T (2007) Spider silk and amyloid fibrils: a structural comparison. Macromol Biosci 7: 183–188 [DOI] [PubMed] [Google Scholar]
- True HL, Lindquist SL (2000) A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature 407: 477–483 [DOI] [PubMed] [Google Scholar]
- Walsh DM, Lomakin A, Benedek GB, Condron MM, Teplow DB (1997) Amyloid β-protein fibrillogenesis. Detection of a protofibrillar intermediate. J Biol Chem 272: 22364–22372 [DOI] [PubMed] [Google Scholar]
- Wickner RB (1994) (URE3) as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science 264: 566–569 [DOI] [PubMed] [Google Scholar]
- Zandomeneghi G, Krebs MRH, McCammon MG, Fändrich M (2004) FTIR reveals structural differences between native β-sheet proteins and amyloid fibrils. Protein Sci 13: 3314–3321 [DOI] [PMC free article] [PubMed] [Google Scholar]