The CNIO Cancer Conference on MYC and the Transcriptional Control of Proliferation and Oncogenesis took place between 11 and 13 June 2007, in Madrid, Spain, and was organized by M. Eilers, R.N. Eisenman and J. León.
Introduction
The MYC genes and proteins have attracted broad and sustained interest over the past 25 years owing to the bewildering range of biological processes controlled by MYC. At a recent CNIO Cancer Conference on MYC and the Transcriptional Control of Proliferation and Oncogenesis, MYCologists discussed new aspects of the molecular function and regulation of MYC, and its consequences on cell physiology and tumour formation. This meeting provided an enormous amount of new and interesting information, making it difficult to cover all the topics that were addressed. Here, we concentrate on the discussion of the most relevant questions in the MYC field and highlight some of the most important new findings. A more detailed understanding of the function of MYC, particularly in disease-related processes, will hopefully allow the development of new therapeutic strategies targeting either MYC itself or MYC-regulated pathways. For background reading, we suggest the excellent recent reviews by Adhikary & Eilers (2005), Grandori et al (2000) and Oster et al (2002).
R. Eisenman (Seattle, WA, USA) opened the meeting by presenting the questions he considered to be of crucial importance for clarifying the role of MYC in the regulation of cell physiology. We summarize and expand these questions, which will be important motivators of future research, and define the aspects of MYC that we need to understand to be able to manipulate this protein in disease.
What are the molecular functions of MYC?
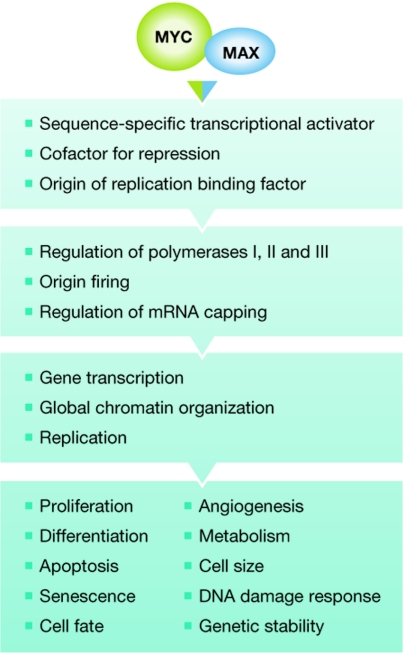
It is well established that MYC operates as a multifunctional transcriptional regulator, but it is possible that it also has other activities beyond the regulation of gene transcription (Fig 1). One such additional function is the role of MYC in replication. D. Dominguez-Sola (New York, NY, USA) and C. Grandori (Seattle, WA, USA) reported that MYC can stimulate replication in the absence of gene transcription by enhancing the activity of origins of replication. This comes at the price of elevated replication-associated DNA damage and the subsequent checkpoint activation, which ultimately results in the inhibition of replication (Dominguez-Sola et al, 2007), indicating that the fine-tuning of MYC function in replication is important for efficient S-phase progression.
Figure 1.
Summary of the functions of MYC–MAX complexes with increasing complexity from top to bottom.
MYC might also have activities that are independent of its heterodimeric partner MYC-associated factor X (MAX). Because the interaction of MYC with MAX is mediated by helices 1 and 2, and the leucine zipper of their respective bHLHZip domains, we would expect that in the absence of MAX the hydrophobic surface of the MYC dimerization domain would interact with another protein or bind to another domain of MYC. Work by M. Cole's laboratory (Lebanon, NH, USA) suggests that MYC can affect gene expression in the absence of its bHLHZip domain by enhancing mRNA cap methylation, resulting in increased protein synthesis (Cowling & Cole, 2007). This activity occurs independently of DNA binding; however, whether the full-length protein can perform this function in the absence of MAX remains to be seen. Furthermore, J. León (Santander, Spain) reported that Myc represses the c-Jun promoter in PC12 neuronal cells lacking Max. P. Gallant (Zürich, Switzerland) reported the characterization of Drosophila melanogaster mutants affecting Myc, Max and Mnt, which are the only members of the Myc/Max/Mad (Mad for Max dimerization protein) family found in flies. The analysis of different mutants and combinations of mutants suggests that Myc might have Max-independent functions. In particular, fly development is more severely disturbed in ΔMyc/ΔMnt than in ΔMax animals. It will be interesting to see whether the postulated Max-independent functions can now be defined genetically.
To which DNA sequence elements does MYC bind?
The MYC–MAX complex binds to E-box sequences with the consensus 5′-CACGTG-3′ or variations thereof (Luscher & Larsson, 1999). Assuming an even distribution of potential binding sites, the MYC–MAX complex should interact with DNA at least every 4 kb, adding up to a total of roughly one million binding sites in the human genome. Although such calculations might be too simplistic, it nevertheless reveals an obvious problem: how do MYC–MAX complexes—and, for that matter, any other DNA-specific transcriptional regulators—recognize true binding sites? This phenomenon remains poorly understood, although it is worth noting that the binding of MYC to chromatin is seen preferentially in regions with open chromatin defined by specific histone marks (Guccione et al, 2006). C. Dang (Baltimore, MD, USA) reported that in P493-6 B cells—a well-used model cell line with tetracycline-regulatable MYC—MYC-specific chromatin immunoprecipitation, in combination with cloning and sequencing of the bound DNA, revealed approximately 4,000 MYC-binding sites (Zeller et al, 2006). Thus, accessibility seems to be important for binding-site selection in addition to other parameters (see below).
Tumour cells generally show high expression levels of MYC compared with their non-transformed counterparts. Although normal proliferating cells have between several hundred and one or two thousand MYC molecules, tumour cells can have up to tens of thousands. In addition, depending on the status of the cell cycle, substantial differences might also be apparent in normal cells, for example during the transition from a resting G0 state into early G1 of the cell cycle. Despite the substantial number of MYC molecules in some cells, it is unlikely that enough MYC–MAX complexes are available to occupy simultaneously all potential sites. Therefore, what are the consequences of changes in the levels of MYC? Is the extra MYC used to elevate binding stoichiometry or are additional new sites used? In addition to a topological selection, other criteria must be considered, including the affinity to response elements, factors that modulate recruitment to DNA, factors that compete for E-box elements and kinetic properties. It is possible that additional transcriptional regulators, which communicate with MYC or specific chromatin marks—for example, core histone modifications—augment the local concentration of MYC at specific chromosomal sites. This might, in turn, be a prerequisite to the efficient loading of MYC–MAX complexes onto binding sites. Indeed, it has been reported previously that in the presence of elevated MYC levels, low affinity sites become increasingly occupied (Fernandez et al, 2003). Depending on where the extra MYC binds, we might expect that either more profound, dominant regulation of specific target genes is achieved or, alternatively, new target genes might become regulated. Although this issue has not been clarified, findings by S. Cory (Melbourne, Australia) suggest that it is important, as differences in Myc levels in transgenic mice affect the type of haematopoietic malignancy observed. Solving this question might define potential therapeutic targets downstream of MYC.
How does MYC function on a DNA response element?
MYC interacts with many proteins that are involved in the control of gene transcription, including the histone acetyl transferases GCN5, TIP60 and CBP/p300, the P-TEFb kinase that targets RNA polymerase II, and the SCFSKP2 E3 ligase complex, which is also involved in ubiquitin-dependent MYC degradation. Recent results suggest that MYC also associates with enzymes that are involved in controlling histone methylation. R. Eisenman described its interaction with and inhibition of the activity of JARID1—a JmjC-dependent trimethyl histone 3 lysine 4 (H3K4) demethylase (Secombe et al, 2007)—and B. Lüscher (Aachen, Germany) reported that MYC recruits an MLL histone H3K4 methyl transferase complex, suggesting that it can control H3K4 methylation—a mark associated with active promoters. Furthermore, the cyclin E–CDK2 (CDK for cyclin-dependent kinase) complex can be recruited to promoters by MYC and this affects MYC phosphorylation at Ser 62 (see below) and its transcriptional activity, as described by L.-G. Larsson (Stockholm, Sweden). S. Hann (Nashville, TN, USA) reported that, among non-enzymatic cofactors, MYC associates with the nucleolar phosphoprotein nucleophosmin. This interaction seems to promote MYC-driven proliferation, but blocks the apoptotic function of MYC. Together, these data suggest that MYC can recruit a considerable number of proteins to promoters, several of them with enzymatic activities. MYC box II—a functionally important element within the amino-terminally located transactivation domain of MYC—is important for the recruitment of some, but not all, of these factors, offering the possibility that more than one cofactor might bind to one MYC molecule at the same time. It will be interesting to understand the dynamics and the combinatories of MYC–cofactor interaction at promoters.
In addition to the gene-specific effects, MYC also seems to have a global effect on chromatin; overall histone acetylation decreases and methylation is altered in neuronal cells in response to disruption of MYCN (Knoepfler et al, 2006). D. Felsher (Stanford, CA, USA) showed that the shutdown of MYC in MYC-driven tumour cells results in a decrease of global histone H4—but not histone H3—acetylation and of global histone H3K4 trimethylation (Wu et al, 2007). These changes relate to the cellular senescence induced in the absence of MYC and suggest that this is an important mechanism in tumour regression. In addition, these findings further support the association of MYC with trimethylation of H3K4.
Which signals control MYC expression and function?
MYC is overexpressed in most human tumours and its expression is regulated in response to many physiological and pathophysiological cues. Despite many reports of individual signal transduction pathways and transcriptional regulators that impinge on MYC gene expression, we still lack a comprehensive concept that integrates the available information into a model of MYC expression (Liu & Levens, 2006). MYC gene transcription is highly complex, in particular with respect to the many signals that need to be integrated at the promoter. Among other signals, MYC expression is known to be regulated by the WNT signalling pathway and O. Sansom (Glasgow, UK) showed that Myc is an essential downstream target of WNT during the development of colorectal cancer (Sansom et al, 2007). Cole reported that MYC negatively regulates DKK1 and SFRP1—two inhibitors of the WNT pathway—providing evidence for a positive feedback loop (Cowling et al, 2007). In summary, these findings strengthen the idea that MYC interacts with the WNT pathway and suggest that such an interaction is important for both normal development and malignant cell proliferation.
For the past 20 years, we have been aware that MYC is modified after translation; however, we are still far from understanding in detail how this protein is controlled by signal transduction pathways (Vervoorts et al, 2006). Crucial regulation occurs at MYC box 1—which contains phosphorylation sites at Thr 58 and Ser 62. Phosphorylation at Thr 58 by glycogen synthase kinase (GSK) 3 is dependent on phosphorylation at Ser 62, and after Thr 58 is phosphorylated, the phosphatase PP2A and the F-box protein FBW7 are recruited and initiate poly-ubiquitination and subsequent proteasomal degradation. New results from M. Eilers (Marburg, Germany) show that the ubiquitin-specific protease USP28 interacts with MYC. This interaction is mediated by the α-isoform of FBW7 and therefore USP28 stabilizes MYC in the nucleus, whereas the nucleolar MYC is unaffected because it is degraded by the FBW7γ isoform (Popov et al, 2007). J. Westermarck (Tampere, Finland) reported that cancerous inhibitor of PP2A (CIP2A) interacts with MYC and prevents PP2A-mediated dephosphorylation of Ser 62, thereby stabilizing MYC (Junttila et al, 2007). CIP2A is overexpressed in some human tumours, which supports its possible tumorigenic function. The regulatory role of Ser 62 was further supported by Larsson, who showed that interferon-γ-signalling inhibits Ser 62 phosphorylation by cyclin E–CDK2 through p27Kip1, thereby inducing MYC degradation. Furthermore, R. Sears (Portland, OR, USA) found that axin, which is known to promote β-catenin degradation, seems to act as a scaffold that brings together Ser 62-phosphorylated Myc with a complex consisting of GSK3, Pin1, PP2A and Fbw7, thereby promoting its proteasomal degradation. Together, these studies show that phosphorylation and ubiquitination fine-tune MYC function. Individual MYC modifications and combinations thereof will probably create distinct chemical surfaces that are specifically recognized by proteins and, therefore, the various isoforms of MYC might interact differentially with the many MYC-binding proteins.
Target genes and processes relevant for MYC function
The role of MYC in promoting proliferation, cell cycle progression and apoptosis is well established, whereas the function of MYC as a regulator of cell growth (cell size), which involves processes such as ribosome biogenesis, protein translation and energy metabolism, has become clearer only recently (Adhikary & Eilers, 2005; Oskarsson & Trumpp, 2005). D. Eick (Munich, Germany) reported on the importance of the MYC target gene products pescadillo homologue 1 (Pes1), block of proliferation 1 (Bop1), WD repeat domain 12 (WDR12)—the PeBoW complex—and UTP18 (for human small subunit processome component 18) in ribosomal RNA processing, and R. White (Glasgow, UK) described the mechanisms of regulation of RNA Pol III-transcribed genes by MYC. Together with the talks by Cole and Hann mentioned previously, this underlines the role of MYC in nucleolar function and mRNA translation. Increased glycolysis and decreased respiration are characteristic of many cancer cells. Dang reported that hypoxia-inducible factor 1 interferes with MYC function, at least in part by inducing the MYC antagonist MAX interactor 1 (MXI1), resulting in the negative regulation of mitochondrial biogenesis and oxygen consumption. MYC stimulates the gene encoding the transcriptional coactivator PGC-1b (for polar granule component 1b)—one of the many genes involved in mitochondrial biogenesis—the loss of which is associated with reduced respiration (Zhang et al, 2007). D. Ayer (Salt Lake City, UT, USA) provided evidence that Mondo A—a member of a parallel MYC/MAX/MAD-associated network—functions as an energy sensor that shuttles between the mitochondrial membrane and the nucleus, where it regulates transcription. Together, these studies further validate the role of MYC in the control of basic cellular processes.
The role of MYC in DNA damage response and senescence was highlighted in several talks. MYC can activate the DNA damage response by causing replication stress, either directly by controlling origin firing as mentioned previously (Dominguez-Sola and Grandori) or indirectly by regulating specific target genes (Felsher). Eilers reported that HectH9—which poly-ubiquitinates MYC through Lys 63 linkages—targets the DNA damage checkpoint regulators TopBP1 (DNA topoisomerase II binding protein 1) and ATR (ataxia telangiectasia and Rad3 related) for destruction by conjugating Lys 48-linked poly-ubiquitin chains. Furthermore, the Myc interaction partner Msx-interacting zinc finger 1 (Miz1) inhibits HectH9 ubiquitination of TopBP1 and ATR, resulting in their stabilization and in activation of the ATR checkpoint in response to ultraviolet light. When MYC is overexpressed, it interferes with this pathway by binding to Miz1, which results in abrogation of the DNA damage checkpoint and thereby in an escape from senescence or apoptosis. Thus, it seems that MYC induces DNA damage but also has ways to interfere with the consequences.
Many studies have addressed the question of how MYC transforms cells. Complex animal experiments reveal that MYC cooperates with several other factors to achieve this. B. Amati (Milan, Italy) reported that in an Eμ-Myc tumour model, Tip60—one of the HAT cofactors recruited by MYC to chromatin—functions as a haplo-insufficent tumour suppressor. Furthermore, in several human tumours, the TIP60 gene suffers mono-allelic loss, which strengthens the idea that it has tumour suppressor activity (Gorrini et al, 2007). How does Tip60 suppress lymphomagenesis? Amati and colleagues showed that the Myc-induced DNA damage response—which acts as an anti-cancer barrier—is primarily dependent on Tip60. C. Schmitt (Berlin, Germany) described how the inactivation of ataxia telangiectasia mutated (ATM) leads to accelerated development of lymphomas in a similar Eμ-MYC model, again suggesting that MYC-induced DNA damage response inhibits tumorigenesis (Reimann et al, 2007).
Several studies have shown that the cyclin E–CDK2 kinase is an important downstream effector of MYC. Amati presented evidence of Cdk2 requirement for efficient transformation. As cyclin E–CDK2 phosphorylates MYC (Larsson)—thereby affecting its activity and stability—these molecules interact in a positive feedback loop that might efficiently prevent exit from the cell cycle. MYC is also known to promote ubiquitin/proteasome-mediated degradation of the CDK-inhibitor p27, resulting in abnormally reduced levels of p27, which is a characteristic of many tumours. As reported by J. Cleveland (Jupiter, FL, USA) and León, one possible explanation is that the transcription of Skp2 and Cks1—two subunits of the SCFSkp2 E3 ligase that targets p27—is activated by Myc. Cleveland further showed that Myc-driven lymphomagenesis is impaired and p27 levels restored in a Cks1−/− background. Thus, p27 has an important role in the functional interaction between cyclin E–CDK2 and MYC.
Induction of Arf is a well-known proapoptotic pathway promoted by Myc. Cleveland reported that overexpressed B-cell lymphoma 2 (Bcl2)—which is normally repressed by Myc—blocks Arf induction by Myc and thereby contributes to lymphoma development in the Eμ-myc model. Furthermore, Schmitt showed that induction of Arf by Myc occurs through FoxO3.
Felsher used several ‘Myc on/off' tumour models, in which he could show that switching off Myc led to tumour regression by senescence (Wu et al, 2007). This was not ATM or ATR dependent, but required functional p53, Rb and p16. p53 was apparently required for Tsp1 induction and repression of angiogenesis. G. Evan (San Francisco, CA, USA) used conditional MycER β-cell tumour model systems to show that active Myc potently induced vascularization of the tumour owing to VEGF release. He showed that this effect was triggered by Myc-induced IL-1β production (Shchors et al, 2006). Myc-induced chemokines recruited mast cells to the tumour site, which had an important role in promoting angiogenesis (Soucek et al, 2007). Furthermore, Evan reported that inactivation of Myc using the dominant-negative omoMyc system in an intestinal mouse tumour model led to complete collapse of the normal epithelium as well as the Myc-driven tumours. Interestingly, the normal cells recovered after reactivation of Myc, whereas the tumour cells did not. This suggests that targeting MYC function might be of therapeutical benefit for certain tumours. M. Henriksson (Stockholm, Sweden) reported the identification of several lead compounds that target Myc activity (Mo & Henriksson, 2006). Several reports suggested the possibility of a MYC ‘de-stabilizing therapy' by interfering with proteins regulating phosphorylation and ubiquitination of MYC such as Usp28 (Eilers), CIP2A (Westermarck), CDK2 (Larsson), axin (Sears) and statins (Felsher) or inhibiting downstream targets of MYC such as ODC (Cleveland). These results, together with some recently published findings, suggest that there might be ways to pharmacologically target MYC function after all.
What do we learn by studying MYC in development?
It is evident that the past 15 years of Myc knockout and over-expression studies in model organisms have revealed new and important insights (de la Cova & Johnston, 2006; Pirity et al, 2006). The Drosophila model has provided new information on Myc target genes and chromatin regulation and has also provided the basis of the concepts of Myc regulation of cell size, Max-independent functions (Gallant), and programmed cell competition, as discussed by L. Johnston (New York, NY, USA) and E. Moreno (Madrid, Spain). Moreno discussed the seemingly important role that programmed cell death has in stem-cell homoeostasis, which is relevant both for insects and vertebrates, and also for cancer stem cells in mammals. The field of normal and cancer stem cells has recently gained much attention, and many presentations highlighted the role of MYC in stem-cell regulation, including self-renewal of stem cells, and the proliferation and differentiation of progenitor cells.
Although the common perception has been that MYC blocks or delays cell differentiation—as shown for myeloid leukaemia cells (León) and for B cells (Cleveland)—it is becoming increasingly clear that in many situations MYC can also promote differentiation. By using a conditional Myc knockout model, I. Moreno de Alborán (Madrid, Spain) showed that MYC is required for all stages of B-cell development because it controls genes such as Pax5, which is essential for B-cell differentiation. P. Hurlin (Portland, OR, USA) reported that MycN—but not Myc—is required for the generation of Fgf-responsive, undifferentiated mesenchymal cells that are important for cartilage formation in limb buds (Ota et al, 2007), thereby showing different developmental functions for different Myc family members. S. Aznar Benitah (Barcelona, Spain) described the regulation of Myc in the stem-cell compartment of adult mouse skin, where Myc DNA binding and dimerization with Max is negatively regulated by phosphorylation of the Myc carboxyl terminus. This process is controlled by integrin signalling in the stem-cell niche through Rac1 and the p21-activated kinase 2 (Pak2). Regulation of Rac1 signalling results in low levels of Myc activity in stem cells, high levels in proliferating and differentiating progenitor cells, and low levels in terminally differentiated cells.A. Trumpp (Lausanne, Switzerland) reported similar findings in haematopoietic bone marrow stem cells, where Myc seems to control the self-renewal process (Wilson & Trumpp, 2006). Loss of Myc results in severe anaemia owing to the low proliferation of progenitors and the expansion of the—mostly resting—stem-cell compartment, whereas Myc overexpression leads to a loss of the self-renewal and depletion of stem cells owing to excess differentiation. Surprisingly (and perhaps disappointingly?) for many MYCologists, Trumpp revealed that the early embryonic lethality found in earlier studies of Myc knockout mice was due to placental failure. Embryos with Myc deletion in embryonic stem cells but with intact Myc in the placenta survived longer, although they eventually died owing to mal-development of the haematopoietic system. Unexpectedly, no obvious defects were observed in other tissues in the absence of Myc.
Conclusion
This year's meeting revealed many new and surprising findings about the function and regulation of MYC. Although we do not have full answers to the burning questions in the field, the new information that became available at the meeting strengthens the framework of knowledge in which we can expect to see the development of more complete answers.
Bernhard Lüscher (left) and Lars-Gunnar Larsson
Acknowledgments
We thank all MYCologists for providing the wealth of information that makes the MYC field so vibrant. We apologize to colleagues whose work could not be mentioned specifically owing to space limitations. We thank J. Vervoorts and H. Schuchlautz for critical reading of the manuscript. Work in our laboratories was funded by the Deutsche Forschungsgemeinschaft, the Interdisciplinary Centre for Clinical Research ‘BIOMAT' and the START program of the Medical School of the RWTH Aachen University (to B.L.) and the Swedish Cancer Foundation, Swedish Childhood Cancer Foundation, Swedish Research Council and Human Frontiers Science Program (to L.-G.L.).
References
- Adhikary S, Eilers M (2005) Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol 6: 635–645 [DOI] [PubMed] [Google Scholar]
- Cowling VH, Cole MD (2007) The Myc transactivation domain promotes global phosphorylation of the RNA polymerase II carboxy-terminal domain independently of direct DNA binding. Mol Cell Biol 27: 2059–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowling VH, D'Cruz CM, Chodosh LA, Cole MD (2007) c-Myc transforms human mammary epithelial cells through repression of the Wnt inhibitors DKK1 and SFRP1. Mol Cell Biol 27: 5135–5146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cova C, Johnston LA (2006) Myc in model organisms: a view from the flyroom. Semin Cancer Biol 16: 303–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, Galloway DA, Gu W, Gautier J, Dalla-Favera R (2007) Non-transcriptional control of DNA replication by c-Myc. Nature 448: 445–451 [DOI] [PubMed] [Google Scholar]
- Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, Cocito A, Amati B (2003) Genomic targets of the human c-Myc protein. Genes Dev 17: 1115–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorrini C et al. (2007) Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature 448: 1063–1067 [DOI] [PubMed] [Google Scholar]
- Grandori C, Cowley SM, James LP, Eisenman RN (2000) The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol 16: 653–699 [DOI] [PubMed] [Google Scholar]
- Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall' Olio V, Zardo G, Nervi C, Bernard L, Amati B (2006) Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol 8: 764–770 [DOI] [PubMed] [Google Scholar]
- Junttila MR et al. (2007) CIP2A inhibits PP2A in human malignancies. Cell 130: 51–62 [DOI] [PubMed] [Google Scholar]
- Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN (2006) Myc influences global chromatin structure. EMBO J 25: 2723–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Levens D (2006) Making myc. Curr Top Microbiol Immunol 302: 1–32 [DOI] [PubMed] [Google Scholar]
- Luscher B, Larsson LG (1999) The basic region/helix-loop-helix/leucine zipper domain of Myc proto-oncoproteins: function and regulation. Oncogene 18: 2955–2966 [DOI] [PubMed] [Google Scholar]
- Mo H, Henriksson M (2006) Identification of small molecules that induce apoptosis in a Myc-dependent manner and inhibit Myc-driven transformation. Proc Natl Acad Sci USA 103: 6344–6349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oskarsson T, Trumpp A (2005) The Myc trilogy: lord of RNA polymerases. Nat Cell Biol 7: 215–217 [DOI] [PubMed] [Google Scholar]
- Oster SK, Ho CS, Soucie EL, Penn LZ (2002) The myc oncogene: MarvelouslY Complex. Adv Cancer Res 84: 81–154 [DOI] [PubMed] [Google Scholar]
- Ota S, Zhou ZQ, Keene DR, Knoepfler P, Hurlin PJ (2007) Activities of N-Myc in the developing limb link control of skeletal size with digit separation. Development 134: 1583–1592 [DOI] [PubMed] [Google Scholar]
- Pirity M, Blanck JK, Schreiber-Agus N (2006) Lessons learned from Myc/Max/Mad knockout mice. Curr Top Microbiol Immunol 302: 205–234 [DOI] [PubMed] [Google Scholar]
- Popov N, Wanzel M, Madiredjo M, Zhang D, Beijersbergen R, Bernards R, Moll R, Elledge SJ, Eilers M (2007) The ubiquitin-specific protease USP28 is required for MYC stability. Nat Cell Biol 9: 765–774 [DOI] [PubMed] [Google Scholar]
- Reimann M, Loddenkemper C, Rudolph C, Schildhauer I, Teichmann B, Stein H, Schlegelberger B, Dorken B, Schmitt CA (2007) The Myc-evoked DNA damage response accounts for treatment resistance in primary lymphomas in vivo. Blood 110: 2996–3004 [DOI] [PubMed] [Google Scholar]
- Sansom OJ, Meniel VS, Muncan V, Phesse TJ, Wilkins JA, Reed KR, Vass JK, Athineos D, Clevers H, Clarke AR (2007) Myc deletion rescues Apc deficiency in the small intestine. Nature 446: 676–679 [DOI] [PubMed] [Google Scholar]
- Secombe J, Li L, Carlos L, Eisenman RN (2007) The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Genes Dev 21: 537–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shchors K, Shchors E, Rostker F, Lawlor ER, Brown-Swigart L, Evan GI (2006) The Myc-dependent angiogenic switch in tumors is mediated by interleukin 1β. Genes Dev 20: 2527–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek L, Lawlor ER, Soto D, Shchors K, Brown Swigart L, Evan GI (2007) Mast cells are required for angiogenesis and macroscopic expansion of Myc-induced pancreatic islet tumors. Nat Med 13: 1211–1218 [DOI] [PubMed] [Google Scholar]
- Vervoorts J, Luscher-Firzlaff J, Luscher B (2006) The ins and outs of MYC regulation by posttranslational mechanisms. J Biol Chem 281: 34725–34729 [DOI] [PubMed] [Google Scholar]
- Wilson A, Trumpp A (2006) Bone-marrow haematopoietic-stem-cell niches. Nat Rev Immunol 6: 93–106 [DOI] [PubMed] [Google Scholar]
- Wu CH, van Riggelen J, Yetil A, Fan AC, Bachireddy P, Felsher DW (2007) Cellular senescence is an important mechanism of tumor regression upon c-Myc inactivation. Proc Natl Acad Sci USA 104: 13028–13033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller KI et al. (2006) Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci USA 103: 17834–17839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Gao P, Fukuda R, Kumar G, Krishnamachary B, Zeller KI, Dang CV, Semenza GL (2007) HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell 11: 407–420 [DOI] [PubMed] [Google Scholar]