Abstract
Understanding the molecular pathways mediating neuronal function in retinas can be greatly facilitated by the identification of genes regulated in the retinas of different mutants under various light conditions. We attempted to conduct a gene chip analysis study on the genes regulated during rhodopsin kinase (Rhok−/−) and arrestin (Sag−/−) knockout and double knockouts in mice retina. Hence, mice were exposed to constant illumination of 450 lux or 6,000 lux on dilated pupils for indicated periods. The retinas were removed after the exposure and processed for microarray analysis. Double knockout was associated with immense changes in gene expression regulating a number of apoptosis inducing transcription factors. Subsequently, network analysis revealed that during early exposure the transcription factors, p53, c-MYC, c-FOS, JUN, and, in late phase, NFκB, appeared to be essential for the initiation of light-induced retinal rod loss, and some other classical pro- and antipoptotic genes appeared to be significantly important as well.
1. INTRODUCTION
The molecular analysis of knockouts provides us with a plenty of knowledge on the functions of genes in mammals. Thus, the characterization of knockouts in mouse retinas is of great importance in our understanding of the mechanisms of signaling networks in the visual system. Rods and cones in vertebrate retina transform visual information into neuronal signals. In mouse rod photoreceptors, light activates rhodopsin, a G-protein-coupled receptor, which is then phosphorylated by rhodopsin kinase [1–4]. Visual arrestin terminates the light response by selectively binding to phosphorylated rhodopsin [5, 6]. Upon illumination and transducin, a G-protein specific to rod photoreceptor cells turns on and calcium influx occurs [7].
Alternatively in mammals, exposure to light can induce photoreceptor cell death and retinal degeneration. The retina of transgenic mice with a null mutation in the gene encoding rhodopsin kinase [8] or arrestin [9] had been sensitized to light damage [10] and revealed prolonged rhodopsin signaling. Furthermore, mouse rod photoreceptor cells lacking the α-subunit of transducin revealed that light-activated rhodopsin and phototransduction signaling were no longer connected [11]. In addition, under certain conditions, the absence of c-FOS [12] or the absence [13] or modification [14] of Rpe65 prevented light-induced degeneration. In previous studies, two different pathways of photoreceptor-cell apoptosis induced by light, transducin-dependent (low light), and AP-1 dependent (bright light), were suggested [15]. Excessive levels of light induced caspase-independent photoreceptor apoptosis have also been proposed during retinal development [16]. However, the molecular signaling networks that initiate the retinal degeneration cascade are not fully understood [17, 18].
The rationale of the study was to delineate the signal transduction networks by taking account of the gene expression changes at different time points and light intensities. In this study, two key gene knockouts in phototransduction, such as rhodopsin kinase (Rhok−/−), arrestin (Sag−/−), and rhodopsin kinase/arrestin (Rhok−/−/Sag−/−), were tested by measuring the expression levels of thousands of genes for their roles in phototransduction signaling in light-induced retinal degeneration.
2. MATERIALS AND METHODS
2.1. Animals
All procedures concerning animals were performed in accordance with the Association for Research in Vision and Ophthalmology (ARVO, MD, USA) Statement on the use of animals in ophthalmic and vision research. Rhodopsin kinase (Rhok−/−) and arrestin (Sag−/−) knockout mice were generated [8, 9]. These mice were crossed to each other to obtain the double-deficient mice, rhodopsin kinase arrestin (Rhok−/−/Sag−/−). All mice including wild-type (WT) were reared in dark until the given experiments were performed. Wild-type mice were derived from an initial cross of 129Sv and C57BL/6. The mice used in this study ranged from 6 to 8 weeks of age.
2.2. Light illumination
The mice reared in dark were placed in aluminum foil-wrapped polycarbonate cages that were covered with stainless steel wire tops to protect them from uncontrolled light exposure. Fluorescent lamps gave off light from an opening at the top of the cage. They were supplied with food and water at the bottom of the cage. Constant illumination of 450 lux on dilated pupils (1% Cyclogyl, Alcon; 5% Phenylephrine, Ciba Vision), or 6% on dilated pupils for indicated periods (1 hour for 450 and 80 minutes for 6 000 lux) was generated by diffuse, cool, white florescent lamps. The temperature was kept at 25°C during irradiation. After light exposure, the mice retinas were either analyzed immediately or after a given period in darkness. Retinas were removed rapidly through a slit in the cornea and frozen in liquid nitrogen until total RNA was extracted by the Trizol method (Invitrogen Life Technologies). The retinas from three to four mice were pooled to make the corresponding sample.
2.3. Microarray analysis
With 3 μg of total RNA from retinas as starting material, first strand cDNA was synthesized using T7-oligo dT primer and SuperScript II (Invitrogen Life Technologies). Second strand cDNA was synthesized with second strand buffer (Invitrogen Life Technologies), DNA polymerase I (New England Biolabs), DNA ligase (New England Biolabs), and RNase H (Invitrogen Life Technologies). cDNA was extracted using phenol:chloroform:isoamyl alcohol, precipitated with ethanol, washed with 80% and 100% cold ethanol, and air dried. The dried pellet was then dissolved in 22μL of nuclease-free water and stored at −20°C. In vitro transcription was performed using the RNA Transcript Labeling Kit (Enzo Diagnostics) to produce hybridizable biotin-labeled RNA targets. The cDNA was used as a template in the presence of a mixture of unlabeled NTPs and biotinylated CTP and UTP. After in vitro transcription, cRNA was purified using RNeasy Mini Kit (Qiagen Inc.). The fragmented cRNA, generated by incubating at 94°C for 35 minutes, was applied to the Affymetrix GeneChip U74Av2 array (total 12,488 probe sets) and hybridized at 40°C for 16 hours. After hybridization, the array was washed several times and stained with streptavidine-conjugated phycoerythrin in the GeneChip Fluidics Station 400 (Affymetrix, Inc.). The arrays were scanned by the Agilent Scanner (Agilent Technologies) and analyzed with GeneChip Analysis Suite 5.0 (Affymetrix, Inc.).
2.4. Network analysis
For each array, genes that were regulated more than or equal to 0.5 and less than or equal to −0.5 in log2 ratio were loaded onto the Ingenuity Pathways Analysis program(http://analysis.ingenuity.com) to identify possible gene networks or pathways.
3. RESULTS AND DISCUSSION
3.1. The general patterns of regulation
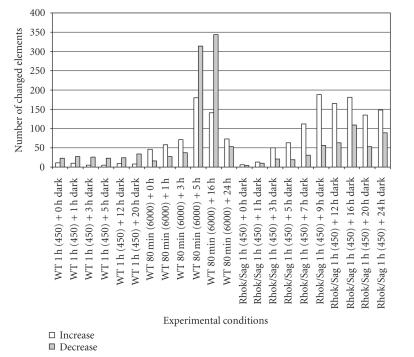
To identify the genes regulated by different knockout conditions, we performed DNA microarrays. The number of changed genes (≥2 folds) in each condition was measured (see Figure 1). The initial screening was done in the c-Fos−/− knockout as well as the transducin (Gnat−/−) knockout to understand the nature of disturbance in signaling mechanisms (manuscript in preparation). However, herewith we will be discussing on the initial changes in gene expression with special focus on Rhok−/−/Sag−/− knockout mice. The experiments were also extended to include a low light condition (450 lux) and a high light condition (6 000 lux) for a series of dark adaptation periods up to 24 hours in wildtype and Rhok−/−/Sag−/− mice, which might provide a visional molecular progress on the mechanisms of light-induced apoptosis condition.
Figure 1.
Number of upregulated and downregulated genes in wild-type and rhodopsin kinase/arrestin knock-out (Rhok−/−/Sag−/−) after dark adaptation for indicated periods.
Wildtype and knockout (Rhok−/−/Sag−/−) mice were exposed to low light (450 lux) with dilated pupils for 1 hour and again placed back into a dark room for up to 20 hours (see Figure 1). A subgroup of wildtype mice were also exposed to bright light (6 000 lux) with dilated pupils for 80 minutes and placed back into a dark room for an indicated period of time (up to 24 hours). There were only a few elements (tens out of total 12 488 probe sets) regulated in wildtype under low light (450 lux) condition during the test up to 20 hours. However, mutants (Rhok−/−/Sag−/−) placed under low light (450 lux) for 1 hour and wildtype mice placed under bright light (6 000 lux) for 80 minutes had considerably more changed genes with the lapse of time. One notion is that only mutant (Rhok−/−/Sag−/−) can cause apoptosis in this condition and there were significantly different molecular changes noted in mutants, which were different from wildtype.
3.2. Significant early phase gene expression in Rhok−/−/Sag−/− mutant
As shown in Table 1, many transcription factors were highly upregulated at the early stage of dark adaptation in the Rhok−/−/Sag−/− mutant exposed to low light (450 lux with dilation) for 1 hour. For example, FBJ osteosarcoma oncogene (c-FOS), CCAAT/enhancer binding protein delta (C/EBP delta), early growth response 1 (EGR1), brain derived neurotrophic factor (BDNF), activating transcription factor 4 (ATF4), fos-like antigen 1 (FRA1), activating transcription factor 3 (ATF3), and growth arrest and DNA-damage-inducible 45 beta (GADD45 beta) were highly expressed. The induction of these transcription factors could potentially trigger the production of their downstream target genes. These genes may have been regulated by different mechanisms, yet coordinately expressed at an early stage in the light-exposed mutant mice (Rhok−/−/Sag−/−) when retinal degeneration occurs and, therefore, may have roles to cooperatively play in light-induced apoptotic cellular signaling networks.
Table 1.
Significant or specific regulations in early stage of Rhok−/−/Sag−/− mutant. L0: low light for 0 hour in wild-type; L1: low light for 1 hour in wild-type; and so on. H0, high light for 0 hour in wild-type; H1: high light for 1 hour in wild-type; and so on. M0: low light for 0 hour in Rhok−/−/Sag−/− mutant; M1: low light for 1 hour in Rhok−/−/Sag−/− mutant; and so on. The numbers are in log2 ratio. Refer to the text for details.
| Gene Name | Accession | L0 | L1 | L3 | L5 | L12 | L20 | H0 | H1 | H3 | H5 | H16 | H24 | M0 | M1 | M3 | M5 | M7 | M9 | M12 | M16 | M20 | M24 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FBJ osteosarcoma oncogene | V00727 | 2.8 | 1 | 0 | 1.6 | 1.5 | 0 | 2 | 1.5 | 3 | 3.4 | 2.2 | 0.7 | 1.5 | 1.4 | 2.5 | 1.9 | 2.5 | 2.6 | 2.1 | 2.1 | 1.7 | 1.1 |
| connective tissue growth factor | M70642 | 0.9 | 0 | 0.8 | 0 | 0 | 0 | 0 | 0 | 0 | 1.4 | 0 | −1.3 | 1.1 | 1.8 | 1.1 | 1 | 1.9 | 1.7 | 1.6 | 1.9 | 1.3 | 1.3 |
| CCAAT/enhancer binding protein (C/EBP), delta | X61800 | 0 | 0 | 0 | 0 | −0.6 | −0.5 | 0 | 0 | 2.6 | 3.6 | 3 | 2.9 | 1.1 | 0 | 2.5 | 2.9 | 3.6 | 4.2 | 4 | 4.1 | 4 | 4.3 |
| early growth response 1 | M28845 | 2.5 | 0 | 1.1 | 1.8 | 1.5 | 0 | 0 | 2.3 | 3 | 3.2 | 1.6 | 0 | 1 | 1.4 | 2.8 | 1.5 | 2.4 | 2.9 | 2.4 | 2.4 | 1.4 | 0.6 |
| prostaglandin D2 synthase (21 kDa, brain) | AB006361 | −0.7 | −0.8 | 0 | −0.8 | −0.5 | −0.3 | 0 | 1 | 1.2 | 0.9 | 0 | −0.3 | 0.9 | 1 | 1.2 | 1 | 0.9 | 0.5 | 0.9 | 1.1 | 0.8 | 0.6 |
| brain derived neurotrophic factor | X55573 | 0.6 | 0 | 0 | 0 | 0 | 0 | 1 | 0.8 | 0 | 1.3 | 1.4 | 0.6 | 0.9 | 0 | 0 | 0 | 1 | 1.2 | 0.8 | 1.2 | 0.6 | 0.9 |
| nuclear receptor subfamily 4, group A, member 1 | X16995 | 2.1 | 0.7 | 0 | 0.4 | 0.7 | 0 | 1.3 | 0 | 0 | 1.2 | 0 | 0 | 0.9 | 0 | 0 | 0 | 0 | 0.4 | 0.5 | 0.6 | 0.9 | 0 |
| prostaglandin D2 synthase (21 kDa, brain) | AB006361 | −0.5 | −1 | 0.4 | −0.8 | −0.8 | −0.4 | 0.6 | 1 | 1.3 | 1.1 | 0 | 0 | 0.8 | 1.2 | 1.2 | 0.5 | 0.7 | 0.6 | 1 | 1.1 | 0.9 | 0.8 |
| activating transcription factor 4 | M94087 | 1 | 1 | 0.5 | 0 | 0 | 0 | 0.7 | 0.7 | 0 | 0 | 0 | 0 | 0.7 | 1.2 | 1.3 | 1.2 | 1.1 | 0.6 | 0.6 | 0.5 | 0 | 0 |
| retinol binding protein 1, cellular | X60367 | 0 | −0.4 | 0 | −0.4 | 0 | 0 | 0.6 | 0.9 | 0.6 | 0.9 | 1 | 0.6 | 0.7 | 0.6 | 0.6 | 0 | 0.9 | 1.3 | 1.8 | 2.2 | 1.9 | 2.4 |
| metallothionein 2 | K02236 | 0 | 0 | 0.9 | 0 | 0 | 0 | 0 | 1.4 | 2 | 3.7 | 0 | 1.2 | 0.6 | 1.2 | 2.5 | 2.6 | 4 | 3.8 | 4.1 | 3.7 | 3.1 | 2.7 |
| metallothionein 1 | V00835 | 0 | 0.6 | 1 | 0.4 | −0.4 | 0 | 0 | 0.9 | 1.7 | 3 | 1 | 0.8 | 0 | 1.1 | 2.1 | 2.5 | 3.7 | 3.8 | 3.7 | 4 | 2.6 | 2.6 |
| Bcl2-associated athanogene 3 | AV373612 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.7 | 2.8 | 0 | 0 | 0 | 0 | 5.2 | 0 | 6.4 | 6.6 | 6 | 6.8 | 6.1 | 6.2 |
| fos-like antigen 1 | AF017128 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.2 | 0 | 0 | 0 | 0 | 3.7 | 0 | 4.6 | 4.8 | 4.9 | 4 | 3.7 | 0 |
| growth arrest and DNA-damage-inducible 45 beta | AV138783 | 1.2 | 0.9 | 0 | 0 | 0 | 0 | 1.3 | 1.1 | 2.2 | 3.3 | 3 | 2 | 0 | 0 | 3.1 | 2 | 3.6 | 4.4 | 4.5 | 4.2 | 3.7 | 3.5 |
| activating transcription factor 3 | U19118 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 2.9 | 2.5 | 2.3 | 2.6 | 2.7 | 2.9 | 2.7 |
| cytokine inducible SH2-containing protein 3 | AV374868 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.8 | 3.1 | 2.9 | 2.9 | 0 | 0 | 2.6 | 2.4 | 3.3 | 3.2 | 3.7 | 3.3 | 3.3 | 3.3 |
| myocyte enhancer factor 2C | AI426400 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | −0.6 | −1 | −0.9 | 0 | 0 | −0.7 | −1.2 | 0 | −0.5 | −0.7 | −0.7 | −1.3 | −1.3 | −1.7 |
| Similar to rhodopsin (opsin 2, rod pigment) | M36699 | 0 | −0.2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | −0.3 | 0 | −1.2 | −1.4 | −1.6 | −1.2 | −1.4 | −1.5 | −0.9 | −1.5 |
| hexokinase 2 | Y11666 | 0 | 0 | 0 | 0 | 0 | 0 | −0.5 | −0.7 | 0 | 0 | 0 | −0.7 | −0.4 | −1 | −1.3 | −1.2 | −1.3 | −1 | −1.2 | −1 | −0.6 | −1.2 |
| expressed sequence AA960287 | AW061237 | −0.4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | −0.6 | 0 | 0 | −0.6 | 0 | −0.9 | −1 | −1.4 | −0.8 | −1.1 | −1.3 | −0.8 | −1.2 |
| rod outer segment membrane protein 1 | AV356715 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | −0.4 | 0 | 0 | 0 | −0.3 | −0.6 | −0.6 | −1 | −0.9 | −1.5 | −1 | −1 | −1 | −0.6 | −0.9 |
| WNT1 inducible signaling pathway protein 1 | AF100777 | −0.4 | 0 | 0 | 0 | 0 | 0 | −0.3 | −0.5 | −0.6 | 0 | 0 | 0 | −0.6 | −0.9 | −0.8 | 0 | −0.9 | −1.1 | −1.4 | −1.4 | −1 | −1.5 |
| high mobility group box 2 | X67668 | 0 | 0 | −0.8 | 0 | 0 | 0 | 0 | −0.7 | −1.2 | 0 | 0 | 0 | −0.6 | −0.9 | −1.3 | −1 | 0 | 0 | −1 | −1 | −0.8 | 0 |
| ESTs | C78037 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | −0.8 | 0 | 0 | 0 | −1.2 | 0 | −0.9 | −1.1 | −0.7 | −1.3 |
| kallikrein 9 | M17979 | 0.6 | 0 | 0 | −0.4 | 0 | −1.4 | 0 | −0.8 | −1.7 | −3.9 | 0 | −3.8 | −0.8 | 0 | −0.9 | −0.8 | −0.7 | 0 | −0.9 | 0 | −1.4 | −0.9 |
| ISL1 transcription factor, LIM/homeodomain, (islet-1) | AJ132765 | −0.2 | −0.5 | 0 | 0 | −0.3 | 0 | −0.5 | −0.7 | 0 | 0 | 0 | 0 | −0.8 | −1.5 | −0.6 | −0.5 | −0.7 | 0 | 0 | 0 | −0.7 | 0 |
| RIKEN cDNA 1110013B16 gene | AW123271 | 0 | 0 | −0.5 | 0 | 0 | −0.3 | 0 | 0 | 0 | 0 | −1.6 | 0 | −1.3 | −0.9 | −1.1 | −1.3 | −1.4 | −0.6 | −1.2 | −1.3 | −1.4 | −1.1 |
3.3. The transcriptional regulations in the apoptosis pathway surveyed by GenMAPP
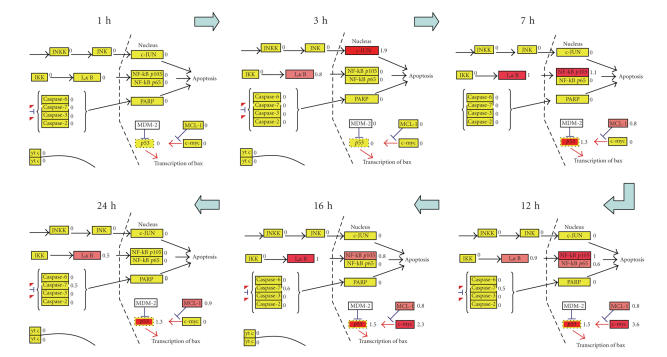
Studying the signal transduction pathways and transcriptional regulation using DNA microarray can be a tremendous challenge to biologists. Obviously, novel bioinformatic tools are required to gain biological insights out of microarray data. Here, we surveyed the transcriptional regulation of the apoptosis pathway focusing on Rhok−/−/Sag−/− mice using the GenMAPP program (see Figure 2) [19]. In wildtypes, including both low and bright light conditions, none of the genes in the hypothetical apoptosis pathway provided by the GenMAPP were differentially expressed. However, we observed a serial regulation of genes involved in apoptosis in Rhok−/−/Sag−/− mutants. After 1 hour of light exposure (450 lux with dilation) followed by 3 hours of dark adaptation, some AP1 components including c-JUN were highly upregulated. This upregulation was observed till 5 hours of dark adaptation, disappeared at 7 hours, and never surfaced again till 24 hours, which was the whole duration of the experiment. Instead, IκB and NF-κB p105 subunit were upregulated and these high expression levels were maintained for a specific period of time. IκB expression was maintained for 24 hours while NF-κB p105 expression went back to normal at 20 hours of dark adaptation. AP1 was reported to be an essential component in light-induced apoptosis [12, 15], which seems to be authentic in the Rhok−/−/Sag−/− mouse model system. The expression level of another AP1 component, c-FOS, also turned out to be highly induced after 1 hour of light exposure and maintained for at least 24 hours in Rhok−/−/Sag−/− mice (see Table 1). This c-FOS was also induced in wildtype mice under the both low and high light conditions and further studies in our lab are delineating this process.
Figure 2.
Apoptosis map. Expression data of Rhok−/−/Sag−/− mice was overlaid onto a hypothetical apoptosis map using the GenMAPP (http://www.genmapp.org) program. Each rectangle represents a gene in the pathway. The number on the right of the rectangle corresponds to the log 2 ratio. Genes highlighted in yellow are unchanged. Pink and red denote upregulated genes while dark and light green correspond to downregulated genes. Rhok−/−/Sag−/− mice were exposed to 1 hour of low light (450 lux) and adapted for the indicated time period of darkness.
3.4. The molecular functions of regulated genes in light-induced apoptosis
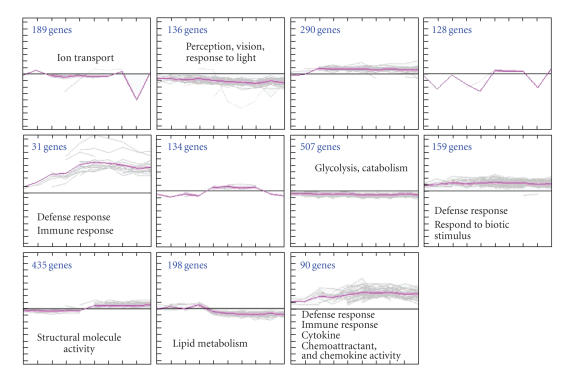
Cluster Assignment for Biological Inference (CLASSIFI) analysis [20] was performed on wild-type (450 lux) and 6 000 lux) and mutant (Rhok−/−/Sag−/−) time point data. There were not any significant GO clusters for the wild-type mice but the case is opposite in the mutant data with many significant GO coclustering in Figure 3. There were many defense/immune response-related gene clusters (highly upregulated genes). There is also one vision-related cluster with mostly downregulated genes, indicating photoreceptor cell damages. The significant cutoff use for this CLASSIFI analysis was 2.17×10-5, estimated using the Bonnferoni correction with an alpha of 0.05.
Figure 3.
CLASSIFI analysis was performed on mutant (Rhok−/−/Sag−/−). The significant cutoff use for this CLASSIFI analysis was 2.17×10-5, estimated using the Bonnferoni correction with an alpha of 0.05.
3.5. The expression of other classical pro- and antiapoptotic genes
Surprisingly, we have found the expressions of other classical pro- and antiapoptotic genes, which were different from what we might have expected. For example, the expression levels of proapoptotic genes, caspase 1, 2, 3, 6, 7, 8, 9, 11, 12, and 14 did not change at all in Rhok−/−/Sag−/− mice when retina cells degeneration occurred after light exposure. The expression levels of these caspases did not change in wild-type under both low (450 lux) and bright (6 000 lux) light conditions. Previously, it has been reported that the expression of caspase 3 was distinctly upregulated in blue light-induced apoptosis in photoreceptor cells [21].
There were other elements such as Bcl2 families which were distinctly working on cell death. Our data indicate that Bcl2 family including BAX, BCL2L10, BAD, BAK1, BAG3, BOK, BAL2L, BCL2L11, BAG1, BCL2L2, and BAD were found to be unchanged, suggesting that the light-induced enzymatic apoptosis may not be regulated at the level of transcription but rather by the activities of proteins that were already present normally. On the other hand, there was also the generation of new transcripts of active molecules which then induced the apoptotic death cascade. p53 and c-MYC were upregulated at 7 or 9 hours after dark adaptation and were maintained at higher levels up to later time points (20 to 24 hours). The transcription factor AP1, c-FOS, and JUN family seemed to be essential for the initiation of light-induced retinal rod loss, while other classical pro- and antiapoptotic genes appeared to be also important in our model system (see Figure 2 and Table 1).
The current study describes about the light-induced gene regulation and transcriptional responses in mouse retinas after genes knockout. As shown in Table 1, many transcription factors were highly upregulated at the early stage of dark adaptation in the Rhok−/−/Sag−/− mutant exposed to low light (450 lux with dilation). The induction of transcription factors, such as c-FOS, C/EBP delta, EGR1, BDNF, ATF4, FRA1, ATF3, and GADD45 beta could potentially trigger the production of their downstream target genes. When they were coordinately expressed at early stages in light-exposed mutant mice (Rhok−/−/Sag−/−), it is likely that they function cooperatively in the cellular signaling networks to induce retinal degeneration. Inferring signal transduction pathways and transcription regulation using DNA microarray data and legendary literature-based interaction information can be a tremendous challenge. While the transcription regulation in the apoptosis pathway derived from the GenMAPP program (see Figure 2) is useful to understand the mechanism, there are many possible networks which we do not know yet. The initial trigger for condition-specific transcription in complex animals often comes from several groups of regulatory transcription factors. According to the dictates of what best contributes to organism survival and selection, individual genes have binding sites to accommodate an assortment of different types of transcription factors. There is no surprise, when complicated biological events are affected, that individual genes are involved in multiple pathways. This signal flow leads to the expression of genes responsible for transcriptional regulation, transport, defense response, immune response, signal transduction, and vision in retinas of Sag−/− mice (see Figure 3).
4. CONCLUSION
Mild or excessive light accelerates the cell death process in certain knockout mice. In this study, we found out the signal transduction networks by analyzing the gene expression changes during different time points of key phototransduction gene knockout in mice. Herewith, we revealed many gene transcripts essential for the initiation of light-induced rod degeneration and proposed important networks fabricated in pro- and antiapoptotic signaling.
ACKNOWLEDGMENT
This work was supported by the Korea Research Foundation Grant funded by the Korean Government (MOEHRD) (KRF-2006-311-C00482) and the Korea Science and Engineering Foundation (KOSEF) Grant, funded by the Korea government (MOST) (R01-2007-000-20533-0) to “S. Choi.” J. Krishnan was supported by BK21 program.
References
- 1.Arshavsky VY. Rhodopsin phosphorylation: from terminating single photon responses to photoreceptor dark adaptation. Trends in Neurosciences. 2002;25(3):124–126. doi: 10.1016/s0166-2236(00)02094-4. [DOI] [PubMed] [Google Scholar]
- 2.Maeda T, Imanishi Y, Palczewski K. Rhodopsin phosphorylation: 30 years later. Progress in Retinal and Eye Research. 2003;22(4):417–434. doi: 10.1016/s1350-9462(03)00017-x. [DOI] [PubMed] [Google Scholar]
- 3.Senin II, Koch K-W, Akhtar M, Philippov PP. Ca2+-dependent control of rhodopsin phosphorylation: recoverin and rhodopsin kinase. Advances in Experimental Medicine and Biology. 2002;514:69–99. doi: 10.1007/978-1-4615-0121-3_5. [DOI] [PubMed] [Google Scholar]
- 4.Ridge KD, Abdulaev NG, Sousa M, Palczewski K. Phototransduction: crystal clear. Trends in Biochemical Sciences. 2003;28(9):479–487. doi: 10.1016/S0968-0004(03)00172-5. [DOI] [PubMed] [Google Scholar]
- 5.Dolph PJ. Arrestin: roles in the life and death of retinal neurons. Neuroscientist. 2002;8(4):347–355. doi: 10.1177/107385840200800410. [DOI] [PubMed] [Google Scholar]
- 6.Miller WE, Lefkowitz RJ. Arrestins as signaling molecules involved in apoptotic pathways: a real eye opener. Science's STKE: Signal Transduction Knowledge Environment. 2001;2001(69):pe1. doi: 10.1126/stke.2001.69.pe1. [DOI] [PubMed] [Google Scholar]
- 7.Makino CL, Wen X-H, Lem J. Piecing together the timetable for visual transduction with transgenic animals. Current Opinion in Neurobiology. 2003;13(4):404–412. doi: 10.1016/s0959-4388(03)00091-6. [DOI] [PubMed] [Google Scholar]
- 8.Chen C-K, Burns ME, Spencer M, et al. Abnormal photoresponses and light-induced apoptosis in rods lacking rhodopsin kinase. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(7):3718–3722. doi: 10.1073/pnas.96.7.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J, Simon MI, Matthes MT, Yasumura D, LaVail MM. Increased susceptibility to light damage in an arrestin knockout mouse model of Oguchi disease (stationary night blindness) Investigative Ophthalmology and Visual Science. 1999;40(12):2978–2982. [PubMed] [Google Scholar]
- 10.Choi S, Hao W, Chen C-K, Simon MI. Gene expression profiles of light-induced apoptosis in arrestin/rhodopsin kinase-deficient mouse retinas. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(23):13096–13101. doi: 10.1073/pnas.201417498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calvert PD, Krasnoperova NV, Lyubarsky AL, et al. Phototransduction in transgenic mice after targeted deletion of the rod transducin α-subunit. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(25):13913–13918. doi: 10.1073/pnas.250478897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hafezi F, Steinbach JP, Marti A, et al. The absence of c−fos prevents light-induced apoptotic cell death of photoreceptors in retinal degeneration in vivo . Nature Medicine. 1997;3(3):346–349. doi: 10.1038/nm0397-346. [DOI] [PubMed] [Google Scholar]
- 13.Grimm C, Wenzel A, Hafezi F, Yu S, Redmond TM, Remé CE. Protection of Rpe65-deficient mice identifies rhodopsin as a mediator of light-induced retinal degeneration. Nature Genetics. 2000;25(1):63–66. doi: 10.1038/75614. [DOI] [PubMed] [Google Scholar]
- 14.Samardzija M, Wenzel A, Naash M, Remé CE, Grimm C. Rpe65 as a modifier gene for inherited retinal degeneration. European Journal of Neuroscience. 2006;23(4):1028–1034. doi: 10.1111/j.1460-9568.2006.04639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hao W, Wenzel A, Obin MS, et al. Evidence for two apoptotic pathways in light-induced retinal degeneration. Nature Genetics. 2002;32(2):254–260. doi: 10.1038/ng984. [DOI] [PubMed] [Google Scholar]
- 16.Donovan M, Cotter TG. Caspase-independent photoreceptor apoptosis in vivo and diffrential expression of apoptotic protease activating factor-1 and caspase-3 during retinal development. Cell Death & Differentiation. 2002;9(11):1220–1231. doi: 10.1038/sj.cdd.4401105. [DOI] [PubMed] [Google Scholar]
- 17.Wenzel A, Grimm C, Samardzija M, Remé CE. Molecular mechanisms of light-induced photoreceptor apoptosis and neuroprotection for retinal degeneration. Progress in Retinal and Eye Research. 2005;24(2):275–306. doi: 10.1016/j.preteyeres.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 18.Roca A, Shin K-J, Liu X, Simon MI, Chen J. Comparative analysis of transcriptional profiles between two apoptotic pathways of light-induced retinal degeneration. Neuroscience. 2004;129(3):779–790. doi: 10.1016/j.neuroscience.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 19.Dahlquist KD, Salomonis N, Vranizan K, Lawlor SC, Conklin BR. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nature Genetics. 2002;31(1):19–20. doi: 10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- 20.Lee JA, Sinkovits RS, Mock D, et al. Components of the antigen processing and presentation pathway revealed by gene expression microarray analysis following B cell antigen receptor (BCR) stimulation. BMC Bioinformatics. 2006;7:237. doi: 10.1186/1471-2105-7-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu J, Gorman A, Zhou X, Sandra C, Chen E. Involvement of caspase-3 in photoreceptor cell apoptosis induced by in vivo blue light exposure. Investigative Ophthalmology & Visual Science. 2002;43(10):3349–3354. [PubMed] [Google Scholar]