Abstract
The remarkably high specificity of the coagulation proteases towards macromolecular substrates is provided by numerous interactions involving the catalytic groove and remote exosites. For FVIIa [activated FVII (Factor VII)], the principal initiator of coagulation via the extrinsic pathway, several exosites have been identified, whereas only little is known about the specificity dictated by the active-site architecture. In the present study, we have profiled the primary P4–P1 substrate specificity of FVIIa using positional scanning substrate combinatorial libraries and evaluated the role of the selective active site in defining specificity. Being a trypsin-like serine protease, FVIIa had P1 specificity exclusively towards arginine and lysine residues. In the S2 pocket, threonine, leucine, phenylalanine and valine residues were the most preferred amino acids. Both S3 and S4 appeared to be rather promiscuous, however, with some preference for aromatic amino acids at both positions. Interestingly, a significant degree of interdependence between the S3 and S4 was observed and, as a consequence, the optimal substrate for FVIIa could not be derived directly from a subsite-directed specificity screen. To evaluate the role of the active-site residues in defining specificity, a series of mutants of FVIIa were prepared at position 239 (position 99 in chymotrypsin), which is considered to be one of the most important residues for determining P2 specificity of the trypsin family members. This was confirmed for FVIIa by marked changes in primary substrate specificity and decreased rates of antithrombin III inhibition. Interestingly, these changes do not necessarily coincide with an altered ability to activate Factor X, demonstrating that inhibitor and macromolecular substrate selectivity may be engineered separately.
Keywords: activated Factor VIIa (FVIIa), antithrombin III (AT III), coagulation, protease, substrate specificity, serine protease inhibitor (serpin)
Abbreviations: ACC, 7-amino-4-carbamoylmethylcoumarin; APC, activated protein C; Arg152 (c15) etc., specific amino acid residues with chymotrypsin numbering in parentheses; ATIII, antithrombin III; BHK cell, baby hamster kidney cell; cmk, chloromethyl ketone; FVII, Factor FVII; FVIIa, activated FVII; FIX, Factor IX; FIXa, activated FIX; Fmoc, fluoren-9-ylmethoxycarbonyl; FX, Factor X; FXa, activated FX; Gla, γ-carboxyglutamic acid; ΔΔGT‡, change in transition state stabilization energy; LMW heparin, low-molecular-mass heparin; MUGB, 4-methylumbelliferyl p-guanidinobenzoate; PEG, poly(ethylene glycol); rFVIIa, recombinant FVIIa; PS-SCL, positional scanning substrate combinatorial library; serpin, serine protease inhibitor; t1/2, half-life; TF, tissue factor; sTF, soluble TF; TFPI, TF pathway inhibitor
INTRODUCTION
Coagulation FVIIa [activated FVII (Factor VII)] is a trypsin-like serine protease responsible for triggering blood coagulation when it associates with its cofactor TF (tissue factor), which is exposed upon vascular injury [1,2]. TF-bound FVIIa is an efficient activator of both FIX (Factor IX) and FX (Factor X) which, on the platelet surface, ultimately results in a burst of thrombin, fibrin deposition and the formation of a haemostatic plug [1]. Thus initiation of blood coagulation represents a powerful and highly amplified system, and, for obvious reasons, it must be tightly controlled in order to restrict thrombosis at the site of vessel injury and to prevent complete occlusion of the damaged vessel. The enzymatic activity of FVIIa is regulated by two physiological inhibitors. The FVIIa–TF complex is efficiently inhibited by the Kunitz inhibitor TFPI (TF pathway inhibitor) in the presence of FXa (activated FX) by forming an inhibitory TFPI–FXa–FVIIa–TF quaternary complex [3]. FVIIa can also be inhibited by the serpin (serine protease inhibitor) ATIII (antithrombin III), and the inhibition is accelerated many folds by sulfated glycosaminoglycans lining the vascular wall [4]. Interactions taking place at regions remote from the active-site cleft, at so-called exosites, also influence the rate of TFPI inhibition of the FVIIa–TF complex and FXa [5], whereas the direct interaction between ATIII and its target proteases is mediated primarily through active-site interactions and few exosite interactions [6].
From a molecular perspective, the interaction between FVIIa and TF is characterized by a large interface between their extracellular domains [7]. The activation of FIX and FX by this complex is mediated by extensive interactions involving both the Gla-EGF1 (γ-carboxyglutamic acid domain and the first epidermal growth factor-like domain) region and the protease domains of FIX and FX. Thus the activation is largely considered to be controlled by exosite interactions remote from the active site [8,9] and, thus, ground state stabilization (Km) for the activation reaction may be largely independent of the active site. Consequently, we hypothesized that the inhibitory profile and the ability to act as an initiator of coagulation may constitute two independent paradigms in terms of substrate recognition. Thus, when altering substrate specificity, there are a number of different strategies available, among which the most obvious are: (i) blocking sites where large residues are preferred, and (ii) generating new cavities [10,11]. Numerous attempts at engineering specific enzyme–substrate interactions have been described in the literature; the general conclusion reached from these studies has been that major changes to substrate specificity often require global changes to the protein structure [12,13]. In contrast with the challenge associated with generating new substrate specificities via the introduction of specific interactions, the generation of FVIIa variants that do not interact with ATIII, but maintain the ability to activate FX, appears to be of an entirely simpler nature.
In order to identify the most optimal subsite for altering FVIIa specificity, we first evaluated the substrate specificity of FVIIa using PS-SCLs (positional scanning substrate combinatorial libraries). One aspect that such libraries do not always fully address is the potential interdependence between the different subsites. Thus, in order to get a more detailed insight into the primary substrate selectivity of FVIIa, we generated and analysed a number of purified peptide substrates, as well as a three-dimensional structure of FVIIa–sTF (soluble TF) in complex with a cmk (chloromethyl ketone) derivative mimicking an optimal substrate based on screening of the peptide substrates. On the basis of the findings from these studies, a number of variants were generated by substituting residues within the active-site cleft that form direct interactions with the side chains of the substrate [14,15]. However, for reasons of simplicity, only one position will be evaluated in the present study. From other trypsin family members, such as tPA (tissue-type plasminogen activator), FXa, APC (activated protein C) and thrombin [15–17], position 99 (chymotrypsin numbering) has proven important for the S2 specificity of this family. Consequently, in order to reduce the rate of inhibition of FVIIa by circulating inhibitors while retaining proteolytic activity towards FX, we initially focused on this position in our attempts to alter the primary specificity of FVIIa.
MATERIALS AND METHODS
Materials
rFVIIa (recombinant FVIIa) and sTF (residues 1–219) were from Novo Nordisk. Plasma-derived FX and FXa were from Enzyme Research Laboratories. Plasma-derived ATIII was from American Diagnostica. LMW heparin (low-molecular-mass heparin) from porcine intestinal mucosa (molecular mass ≈ 5000 Da) was from Calbiochem. The chromogenic substrates S-2288 (D-Ile-Pro-Arg-p-nitroanilide) and S-2765 (benzyloxycarbonyl-D-Arg-Gly-Arg-p-nitroanilide) were from Chromogenix. The inhibitor Trp-Tyr-Thr-Arg-cmk was synthesized by SynPep. All oligonucleotides were from MWG-Biotech. All restriction endonucleases were from New England Biolabs. SDS/PAGE was performed on 4–12% (w/v) NuPAGE Novex Bis-Tris gels from Invitrogen. All other reagents were of analytical grade from Sigma–Aldrich.
Mutagenesis, protein expression and purification
The wild-type FVII expression plasmid pLN174 [18] was used as template for site-directed mutagenesis using the QuikChange® II XL Site-Directed Mutagenesis Kit (Stratagene). Plasmids were prepared using a QIAprep spin mini-prep kit (Qiagen), and the mutations were verified by DNA sequencing. BHK (baby hamster kidney) cells were transfected with the FVII expression plasmids using the FuGENE6™ transfection reagent (Roche), and the FVII variants were purified as described previously [19]. After auto-activation, the concentration of the FVIIa variants was determined by ELISA. All FVIIa variants were active-site titrated with the ester substrate MUGB (4-methylumbelliferyl p-guanidinobenzoate) essentially as described previously [20].
PS-SCLs
ACC (7-amino-4-carbamoylmethylcoumarin)-based PS-SCLs were synthesized at Novo Nordisk by solid phase synthesis on Fmoc (fluoren-9-ylmethoxycarbonyl)–ACC resins essentially as described previously [19]. The four synthesized PS-SCLs consisted of 18 sublibraries, with each sublibrary containing a mixture of tetrapeptide substrates incorporating one of the 18 proteinogenic amino acids (methionine and cysteine excluded) at P1, P2, P3 or P4 [21]. In the P2, P3 and P4 libraries, arginine was fixed at P1 (preferred by FVIIa), and the last two positions were randomized. In the P1 library, the P1 residue was varied and the P2, P3 and P4 residues were all randomized. The concentration of the libraries was determined by amino acid analysis and absorbance measurements at 325 nm. The optimal emission and excitation parameters for assaying the PS-SCLs were determined by cleaving one of the P4 sublibraries with trypsin. Each sublibrary was assayed at final concentrations of 3, 6, 9 and 12 μM in black 96-well fluorescence plates in a total reaction volume of 200 μl. The assays were performed at room temperature (25 °C) in a buffer containing 50 mM Hepes (pH 7.4), 100 mM NaCl, 5 mM CaCl2 and 0.01% Tween 80. The hydrolysis reactions were initiated by addition of a final concentration of 100 nM FVIIa variant and 150 nM sTF. Kinetics were monitored with a SpectraMax Gemini EM fluorescence microplate reader with excitation at 380 nm, emission at 455 nm and cut-off at 420 nm. The tetrapeptide substrates in each sublibrary were assayed at a concentration far below the expected Km and, thus, linear fitting of the initial rates yielded a set of apparent second-order rate constants for hydrolysis of each sublibrary, which were normalized to the rate of the sublibrary with the highest activity.
Assay of ACC-linked tetrapeptides
A total of 23 ACC-linked tetrapeptides were synthesized on Fmoc–ACC resins essentially as described previously [22]. The concentration of the peptides was determined by amino acid analysis and reversed phase-HPLC. Each peptide was assayed in duplicate at final concentrations ranging from 0.78–50 μM in black 96-well fluorescence plates in a total reaction volume of 200 μl. The assays were performed at room temperature in a buffer containing 50 mM Hepes (pH 7.4), 100 mM NaCl, 5 mM CaCl2 and 0.01% Tween 80. The hydrolysis reactions were initiated by adding a final concentration of 10 nM rFVIIa and 100 nM sTF, and kinetics were monitored using a SpectraMax Gemini EM fluorescence microplate reader with excitation at 380 nm, emission at 455 nm and cut-off at 420 nm. kcat/Km values were determined by linear fitting of the data as [S]0 (substrate concentration at time 0) ≪Km.
FVIIa activity assays
All proteins were diluted in 50 mM Hepes (pH 7.4), 100 mM NaCl, 5 mM CaCl2 and 1 mg/ml BSA. The amidolytic activity was assayed with the chromogenic substrate S-2288 (final concentration, 0.2–10 mM) in a total reaction volume of 200 μl in 96-well plates. The reactions were initiated by addition of 10 nM of each FVIIa variant in the presence of 100 nM sTF. The absorbance increase was measured continuously at 405 nm in a SpectraMax 190 microplate reader. kcat and Km values were determined by fitting the data to the Michaelis–Menten equation by non-linear regression. When Km >5 mM, only kcat/Km was determined by linear regression assuming [S]0≪Km.
The kinetic parameters of FX activation were determined by incubating 10 nM of each FVIIa variant with 100 nM sTF and 0.1–6.4 μM FX (all final concentrations) for 20 min at room temperature in a total reaction volume of 100 μl in 96-well plates. A total of 50 μl of stop buffer [50 mM Hepes (pH 7.4), 100 mM NaCl and 20 mM EDTA] was added, followed by the addition of 50 μl of 2 mM S-2765, and the absorbance increase was measured continuously at 405 nm in a SpectraMax 190 microplate reader. The kcat/Km values were determined, and the amount of FXa generated was estimated from a standard curve generated with final concentrations of 0.5–10 nM active-site-titrated FXa.
Kinetics of ATIII inhibition
The apparent second-order rate constant of inhibition, kinh, was determined by the progress-curve method monitoring competing hydrolytic and inhibition reactions [23]. All reagents were equilibrated at room temperature, and the assay was performed in a buffer containing 50 mM Hepes (pH 7.4), 100 mM NaCl, 5 mM CaCl2, 1 mg/ml BSA and 0.1% PEG [poly(ethylene glycol)] 8000 in a total reaction volume of 200 μl in 96-well plates. To maintain pseudo-first order conditions during the assay, a minimum 10-fold excess of ATIII over FVIIa was used. In a 96-well plate, 3 μM LMW heparin was mixed with 50–750 nM ATIII. The reactions were started by adding 20 μl of 2 mM S-2288 ensuring [S]0 <Km, followed by addition of FVIIa and sTF to final concentrations of 5 and 50 nM respectively. The absorbance increase was measured in a SpectraMax 190 microplate reader at 405 nm for 30 min to obtain progress curves at each inhibitor concentration. kobs values were determined by fitting the progress curves to eqn (1):
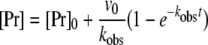 |
(1) |
where [Pr]0 and [Pr] are the accumulation of product at assay start (t=0) and at any time respectively, visualized by a change in absorbance after addition of a chromogenic substrate. kLim and KD values were obtained by fitting the kobs values plotted against the inhibitor concentrations to a hyperbolic function, which accounts for substrate binding [eqn (2)]:
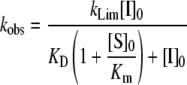 |
(2) |
where KD is the dissociation constant for binding of FVIIa to the ATIII–heparin complex to form the non-covalent Michaelis complex, and kLim is the observed rate constant at saturation for formation of the covalent protease–inhibitor complex. The apparent second-order rate constant of inhibition, kinh, is given by kinh=kLim/KD.
FVIIa–ATIII complex breakdown
Covalent complex breakdown was measured by making ATIII–FVIIa complexes at high concentrations of FVIIa and with ATIII in excess to ensure fast and complete inhibition. The reactions were then extensively diluted into a substrate-containing solution with [S]0≫Km and the restoration of protease activity was monitored continuously. The assay was performed in a buffer containing 50 mM Hepes (pH 7.4), 100 mM NaCl, 5 mM CaCl2, 1 mg/ml BSA and 0.1% PEG 8000. rFVIIa (0.5 μM) was incubated with 2 μM sTF and 1.25 or 2.5 μM ATIII. A 4-fold excess of LMW heparin over ATIII was used. The reactions were incubated at room temperature for 1 h. Each reaction was diluted 400, 450 and 500 times into reaction mixtures (200 μl) in a 96-well plate with a final concentration of 4 mM S-2288 and 100 μg/ml polybrene. The absorbance increase at 405 nm was measured for 12 h in a SpectraMax 190 microplate reader. The first-order rate constant of complex breakdown, kbrkdn, was determined by fitting the absorbance curves to a single exponential function. The observed first-order rate constant was converted into a t1/2 (half-life), according to the relationship t1/2=ln2/kbrkdn.
Data analysis
KaleidaGraph 3.6 from Synergy software or GraphPad Prism 4.0 was used to fit the data to the appropriate equations.
Crystallization conditions
The complex between Trp-Tyr-Thr-Arg-cmk–FVIIa and sTF1-209 was purified using a Superdex75 16/60 prep-grade column (Amersham Biosciences) and concentrated to approx. 10 mg/ml using a Centriprep centrifugation device with a molecular mass cut-off of 50 kDa. A buffer composed of 10 mM Tris/HCl (pH 7.5), 100 mM NaCl and 2 mM CaCl2 was used. Crystal plates were obtained using the hanging-drop vapour diffusion method using 0.1 M sodium citrate, 16.5–15.5% (w/v) PEG 4000 and 12% (v/v) propan-1-ol (pH 5.6) as the precipitating agent. A final protein concentration in the drop of approx. 3 mg/ml was used. Seeding was performed using crystal needles obtained as described previously [7].
Crystal data collecting and processing
Crystallographic data collection was performed on the Swiss Light Source synchrotron beam line PXI. Data reductions were performed with the XDS software package [24]. The structure was solved by the molecular-replacement method using the MR facility in the CCP4 software package and the PDB (Protein Data Bank) entry 1DAN as a search model [25]. Model building was performed using Coot software [26], and iterative refinement (initially performed as a rigid body refinement) was performed using RefMac5 in the CCP4 software package [25].
RESULTS
Specificity profiling of rFVIIa
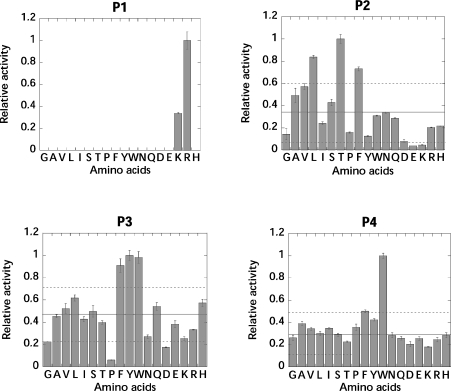
PS-SCLs were used to elucidate the substrate preference in the S1-, S2-, S3- and S4-binding pockets of rFVIIa. The libraries constitute a powerful tool for determination of protease specificity, since a specificity profile for the individual subsites can be rapidly obtained by monitoring the enzymatic liberation of fluorogenic ACC. The PS-SCLs used in the present study were verified by amino acid analysis (results not shown), and the P1 library has been validated previously [27] with three different proteases (trypsin, caspase 3 and subtilisin Carlsberg), showing the expected primary specificities. Specificity profiling of rFVIIa in the presence of sTF showed, as expected, that rFVIIa only accepted arginine and lysine residues in the S1 pocket, with arginine being favoured three-fold more than lysine. In the S2 pocket, β-branched or hydrophobic amino acids were allowed, with threonine, leucine and phenylalanine being the preferred residues (see Figure 1). In the S3 position, the large aromatic residues phenylalanine, tyrosine and tryptophan were preferred, whereas the S4 subsite appeared to be highly promiscuous, although a slight preference for tryptophan was detected. An identical specificity profile of rFVIIa was obtained in the absence of sTF, indicating that sTF did not alter the substrate specificity of rFVIIa (results not shown).
Figure 1. Specificity profile of rFVIIa in the presence of sTF.
The specificity profile was measured in a buffer composed of 50 mM Hepes (pH 7.4), 100 mM NaCl, 5 mM CaCl2 and 0.01% Tween 80. In each library, the apparent second-order rate constants of substrate hydrolysis were normalized to the sublibrary with the highest activity. Solid line, mean value of all of the columns; dotted line, 1 S.D. of all of the columns. Single-letter amino-acid notation is used.
Assay of proteases with the PS-SCLs provides a complete specificity profile in the non-prime sites of the active-site cleft. However, only one substrate-binding pocket is profiled at a time and, therefore, no steric considerations or interdependency between the sites are investigated. To elucidate any interdependency between the S1–S4-binding pockets, rFVIIa was assayed with a range of single ACC-linked tetrapeptides having arginine in P1, threonine or leucine in P2, tryptophan, tyrosine, leucine or alanine in P3, and tryptophan, phenylalanine or asparagine in P4. The substrates were selected partly to address different levels of activity, based on the specificity profiles, and partly to explore the space around the natural FX cleavage site Asn-Leu-Thr-Arg↓Ile-Val-Gly-Gly. For technical reasons, the Asn-Leu-Thr-Arg-ACC peptide was omitted and, thus, rFVIIa was assayed with a total of 23 different ACC-linked tetrapeptides in the presence of sTF.
The kcat/Km values obtained for cleavage of the ACC-linked tetrapeptides by rFVIIa confirmed the preference for aromatic residues in both P3 and P4 (Table 1). The highest kcat/Km values were obtained for the Trp-Ala-Thr-Arg-ACC and Trp-Tyr-Thr-Arg-ACC peptides. The best peptide, Trp-Ala-Thr-Arg-ACC, was cleaved 3.6-fold more efficiently than the Asn-Leu-Thr-Arg-ACC peptide, which represented the P4–P1 cleavage site in the activation peptide of FX. Peptides with P4 asparagine residues were cleaved slowly compared with peptides with P4 tryptophan or P4 phenylalanine residues, except with the Asn-Trp-Thr-Arg-ACC peptide, which was cleaved with a rate similar to the Trp-Trp-Thr-Arg-ACC and Phe-Trp-Thr-Arg-ACC peptides, probably due to the favourable P3 tryptophan residue. For all peptides, rFVIIa was more active towards peptides with P2 threonine than P2 leucine by a factor of approx. 2.5. Thus the clear P2 threonine specificity was independent of the nature of the P3 and P4 residues of the peptides.
Table 1. kcat/Km values for cleavage of ACC-linked tetrapeptides, in the format P4-P3-P2-Arg-ACC, by rFVIIa in the presence of sTF.
Values are means±S.D. The assay was carried out in 50 mM Hepes (pH 7.4), 100 mM NaCl, 5 mM CaCl2 and 0.01% Tween 80. n.d., not determined.
| kcat/Km (mM−1·s−1) | |||||
|---|---|---|---|---|---|
| P2 residue | P3 residue… | Alanine | Leucine | Tyrosine | Tryptophan |
| Threonine | |||||
| P4 residue | |||||
| Asparagine | 2.6±0.04 | 13.8±0.1 | n.d. | 19.6±0.4 | |
| Phenylalanine | 19.4±0.5 | 12.2±0.5 | 24.5±0.2 | 24.6±0.3 | |
| Tryptophan | 49.8±0.6 | 25.8±0.3 | 44.0±0.4 | 23.7±0.2 | |
| Leucine | |||||
| P4 residue | |||||
| Asparagine | 1.1±0.005 | 5.7±0.07 | 9.8±0.04 | 11.2±0.1 | |
| Phenylalanine | 9.7±0.1 | 7.8±0.2 | 7.4±0.1 | 7.2±0.2 | |
| Tryptophan | 19.2±0.3 | 11.7±0.08 | 18.0±0.08 | 6.1±0.04 | |
The discrimination between two substrates was determined by the relative binding of their transition states to the enzyme. ΔΔGT‡ (changes in transition state stabilization energy) of the peptides was calculated using the relative difference in kcat/Km values [28], where ΔΔGT‡ is negative if the X-X-X-Arg peptide is a better substrate than the Y-Y-Y-Arg peptide (where X and Y are any amino acid):
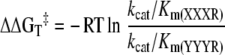 |
(3) |
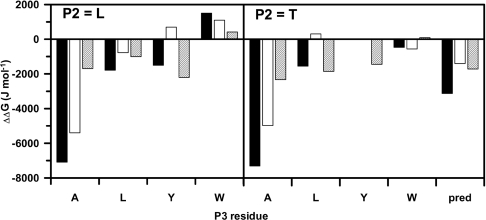
Using this equation, it was possible to evaluate the impact of a single substitution on the overall transition state stabilization energy as well as to assess whether there was interdependence between the different subsites. Interestingly, even with the relatively small number of substrates tested, there appeared to be quite a significant degree of interdependence, as is evident from Figure 2. Thus the effect on ΔΔGT‡ of a single amino-acid substitution was highly context-dependent, for example, in the extreme case, the ΔΔGT‡ for the P4 asparagine to tryptophan substitution in the Asn-Ala-Thr-Arg-ACC peptide was −7.3 kJ·mol−1, whereas the same substitution in the Asn-Trp-Thr-Ala-ACC peptide was only −0.4 kJ·mol−1. For reference, in a system with no subsite interdependence, a ΔΔGT‡ of −3.2 kJ·mol−1 would have been expected based on the subsite profiling.
Figure 2. Interdependence of FVIIa subsites for the cleavage of ACC-linked tetrapeptides.
The interdependence of FVIIa subsites is illustrated by ΔΔGT‡ values for cleavage of ACC-linked tetrapeptides (P4-P3-P2-Arg-ACC) for the substitution of P4 asparagine to tryptophan (closed bar), asparagine to phenylalanine (open bar) and phenylalanine to tryptophan (hatched bar) as a function of the P3 residues (alanine, leucine, tyrosine and tryptophan) with P2 as either leucine (left-hand panel) or threonine (right-hand panel). Single-letter amino-acid notation is used. The pred column reflects the ΔΔGT‡ value anticipated based on the subsite profiling. It is evident that there is a significant impact of P3 on the substitution of P4 residues, but only minor influence of P2.
Three-dimensional structure of Trp-Tyr-Thr-Arg-cmk–FVIIa in complex with sTF1-209
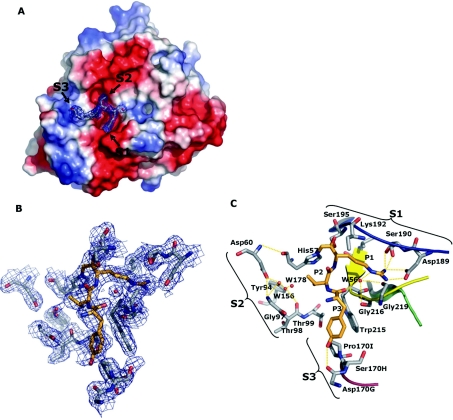
To gain an increased insight into the manner in which an optimal substrate, as determined by the PS-SCL technique, binds to rFVIIa, the three-dimensional structure of rFVIIa–sTF in complex with Trp-Tyr-Thr-Arg-cmk was determined (Figure 3). The structure of the complex resolved to 2.05 Å (where 1 Å=0.1 nm) based on 52511 unique reflections observed giving 100% completeness for the complete bin range 2.05–39.16 Å. Overall, the structure is highly similar to the structure of FVIIa–TF inhibited with Phe-Phe-Arg-cmk reported by Banner et al. [7]. The Cα rmsd (root mean square deviation) between the protease domain of the two structures was 0.3 Å. The covalently bound inhibitor was identified in the active site in the initial electron density map and built by excluding it during initial refinements. Interestingly, only the three first residues were clearly visible in the electron density map, i.e. arginine, threonine and tyrosine (see Figure 3). Thus, to ensure that the inhibitor was intact, the complex was analysed by MALDI–TOF-MS (matrix-assisted laser-desorption ionization–time-of-flight MS), which demonstrated that the inhibitor had the expected sequence (results not shown).
Figure 3. Crystal structure of Trp-Tyr-Thr-Arg-cmk–FVIIa–sTF1-209 obtained at a 2.05 Å resolution.
(A) The structure is shown in the standard orientation. The orientation of (B) and (C) is changed slightly to give an optimal view. (A) View of the protease domain in an electro-potential surface plot with the inhibitor shown with a 2Fo-Fc electron-density map with 2σ cut-off. Colour-coding: negative charges are in red, positive charges of in blue, carbon is in grey, oxygen is in red and nitrogen is in blue. The S1, S2 and S3 pockets are indicated with arrows. Note the striking difference between S1 and S2 compared with S3 in which the former are distinct cavities with defined boundaries and high degree of electro-negativity, whereas the S3 pocket is flat with an almost neutral electro-potential surface. No obvious S4 pocket was identified surrounding the active site in accordance with the lack of electron density for the P4 residue. (B) Close-up of selected residues within a 5 Å sphere of the covalently bound inhibitor shown with a 2Fo-Fc electron-density map with 2σ cut-off. As indicated, the electron density traced the active-site residues well. Carbons of the inhibitor are coloured orange to distinguish them from protein carbons. (C) Specific interactions between the selected active-site residues and covalently bound inhibitor are highlighted [the view is slightly tilted compared with (B)]. The S1, S2 and S3 composition is shown with brackets, and residue identifications are with chymotrypsin numbering. Hydrogen-bonding networks are shown in orange dotted lines. The following secondary elements are coloured accordingly: Lys336–Gly346 is in blue, His373–Tyr377 is in green, Val362–Ala369 is in yellow and Gln313–Asn322 is in red.
The arginine residue in the P1 position was strongly defined and had a similar binding motif as observed in the structure reported by Banner et al. [7]. The P2 threonine residue was positioned adjunct to the catalytic triad with a distance of 3.4 Å to the catalytic His193 (c57), which was also the closest contact to any residue in the S2 pocket. The residue was within hydrogen-bonding distance with the two water molecules W156 and W178 and could be part of a hydrogen-bonding network of the S2 pocket involving the para-positioned hydroxy group of Tyr234 (c94), the carboxylate group of Asp196 (c60), the carbonyl of Thr238 (c98) and amides of both Gly237 (c97) and Thr238 (c98). No other contacts were identified for the P2 residue.
The P3 tyrosine residue interacted with the Gln313–Pro321 (c170–c170I) loop and the para-positioned hydroxy group of the side chain of the tyrosine residue hydrogen bonds with the carbonyl group of Asp319 (c170G), whereas the benzyl moiety interacts via hydrophobic contacts with Pro321 (c170I) and Trp364 (c215). The carbonyl group of the tyrosine residue hydrogen bonds with the amide group of Gly367 (c216) as well as with water molecule W56, as described earlier. Thus the observation that the preferred residue is tyrosine, even compared with phenylalanine and tryptophan, is reflected by the extra hydrogen-bond donor of the side chain, which then leads to the more stabilized pocket as observed for the Gln313–Pro321 loop (c170-c170I).
Expression and characterization of FVIIa variants
A number of FVIIa variants modified at position 239 (c99) were generated to explore the effect of different side-chain sizes. The variants were expressed in BHK cells and purified to homogeneity by a combination of anion-exchange chromatography and immuno-affinity chromatography. After complete auto-activation, the FVIIa variants appeared as two bands on SDS/PAGE under reducing conditions identical with wild-type FVIIa, without visible signs of degradation (results not shown). All FVIIa variants were active-site titrated with MUGB. The active-site concentrations determined by the MUGB titrations were >75% relative to the concentrations determined by ELISA for all FVIIa variants except T239G.
Enzymatic activity of FVIIa variants
The enzymatic activity of the FVIIa variants towards small peptidyl substrates was analysed with the chromogenic peptide S-2288 and the fluorogenic peptide Trp-Ala-Thr-Arg-ACC (Table 2). The amidolytic activity towards S-2288 was decreased greatly for variants T239A, T239G and T239Y (4.8-, 10.9- and 10.8-fold respectively, relative to wild-type rFVIIa). For variant T239I, the kcat/Km values for cleavage of S-2288 were only decreased slightly compared with wild-type FVIIa. The amidolytic activity of all of the FVIIa variants was improved when measured with the Trp-Ala-Thr-Arg-ACC peptide, but the kcat/Km values were still lower compared with wild-type rFVIIa.
Table 2. kcat/Km values for cleavage of S-2288 and Trp-Ala-Thr-Arg-ACC, as well as activation of FX, by rFVIIa and the FVIIa variants in the presence of sTF.
Values are means±S.D. The assay was carried out in 50 mM Hepes (pH 7.4), 100 mM NaCl, 5 mM CaCl2 and 1 mg/ml BSA.
| kcat/Km (mM−1·s−1) | |||
|---|---|---|---|
| FVIIa variant | S-2288 | Trp-Ala-Thr-Arg-ACC | FX |
| rFVIIa | 26.12±2.77 | 44.66±0.11 | 550±30 |
| T239A | 5.45±0.15 | 28.59±0.08 | 69±5 |
| T239G | 2.40±0.036 | 17.59±0.10 | 23±0.6 |
| T239I | 19.71±2.26 | 29.01±0.07 | 461±12 |
| T239Y | 2.47±0.18 | 6.08±0.02 | 74±5 |
The FVIIa variants were analysed for their ability to activate FX in the presence of sTF (Table 2). The proteolytic activity was shown to be decreased 8-, 24- and 7-fold for variants T239A, T239G and T239Y respectively, relative to wild-type rFVIIa, whereas the kcat/Km value for activation of FX by variant T239I was comparable with that of wild-type rFVIIa.
The influence of residue 239 (c99) on substrate specificity
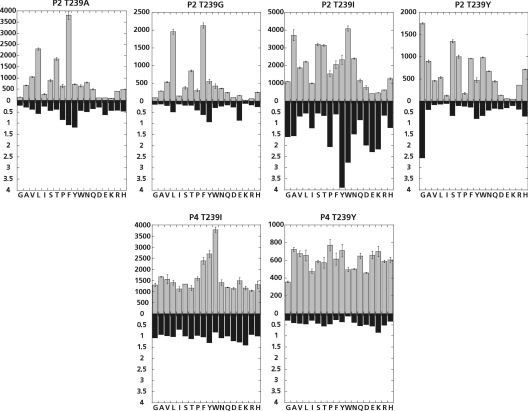
The effect of the introduced mutations on the substrate specificity was monitored with the PS-SCLs (Figure 4). Because of the limited amounts of purified FVIIa variants, T239A and T239G were only profiled with the P2 library, whereas T239I and T239Y were profiled with both the P2 and P4 libraries. Phenylalanine, threonine and leucine were favourable P2 residues when Thr239 was substituted with either alanine or glycine but, relative to wild-type rFVIIa, the majority of the P2 sublibraries were not favoured. When isoleucine or tyrosine residues were introduced at position 239 (c99), large changes in specificity were observed. T239I had a 2.5- and 4-fold increased preference towards tryptophan and tyrosine in P2 respectively, compared with wild-type rFVIIa. No changes in P4 specificity were observed, indicating that the introduced isoleucine residue was pointing into the S2 pocket and was not influencing the specificity in the S4 pocket. T239Y had a 2.5-fold increased preference towards P2 glycine relative to wild-type rFVIIa. The P4 specificity was also affected by this mutation, with all amino acids being nearly equally well tolerated as the P4 residue, and no clear preference for tryptophan was observed as seen in the P4 profile for wild-type rFVIIa. However, all of the P4 sublibraries were disfavoured for T239Y relative to wild-type rFVIIa.
Figure 4. Specificity profiles of FVIIa variants modified at position 239 measured in the presence of sTF.
The specificity profile was measured in a buffer composed of 50 mM Hepes (pH 7.4), 100 mM NaCl, 5 mM CaCl2 and 0.01% Tween 80. The columns in grey show the apparent second-order rate constants of substrate hydrolysis (M−1·s−1) of each sublibrary. The closed bars show the apparent second-order rate constants of substrate hydrolysis relative to wild-type rFVIIa for each sublibrary. Single-letter amino-acid notation is used.
Kinetic parameters for ATIII inhibition of FVIIa variants
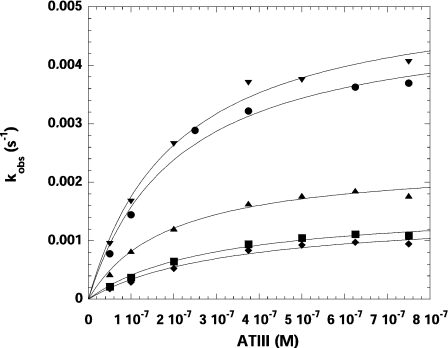
The stoichiometry of inhibition for inhibition of wild-type rFVIIa and the FVIIa variants by ATIII in the presence of LMW heparin could not be determined accurately as the titration curves moved to higher [I]0/[E]0 ratios as a function of time, indicating breakdown of the ATIII–rFVIIa complexes (results not shown). The apparent second-order rate constant of inhibition (kinh) of wild-type rFVIIa and the FVIIa variants by ATIII was determined by the progress-curve method (Figure 5) with the results shown in Table 3. The wild-type rFVIIa–sTF complex was inhibited by ATIII in the presence of LMW heparin with an apparent second-order rate constant of inhibition of 26917 M−1·s−1. Compared with wild-type rFVIIa, variants T239A and T239G were inhibited very slowly by ATIII with kinh reduced 4.4- and 5.7-fold respectively, reflecting their low enzymatic activities. In contrast, T239Y was inhibited slightly faster than wild-type rFVIIa. Interestingly, T239I had a 40% decrease in kinh relative to wild-type rFVIIa, but maintained almost full activity against both S-2288 and FX.
Figure 5. Rate of inhibition of rFVIIa and the FVIIa variants by ATIII.
Rate of inhibition of rFVIIa and the FVIIa variants by ATIII was determined by the progress-curve method under pseudo-first order conditions measured in the presence of sTF and LMW heparin in 50 mM Hepes (pH 7.4), 100 mM NaCl, 5 mM CaCl2, 1 mg/ml BSA and 0.1% PEG 8000. kLim and KD values were determined by plotting the kobs values against the concentration of ATIII and fitting the data to eqn (2) (see Materials and methods section). The apparent second-order rate constant of inhibition, kinh, was determined as the ratio kLim/KD. ●, rFVIIa; ■, T239A; ◆, T239G; ▲, T239I; ▼, T239Y.
Table 3. The apparent second-order rate constants for inhibition, kinh, of the FVIIa variants by ATIII determined as the ratio kLim/KD measured in the presence of sTF and LMW heparin.
Values are means±S.D. The first-order rate constants of complex breakdown, kbrkdn, and t1/2 determined as the average of six independent measurements. t1/2 was calculated by the relationship t1/2=ln2/kbrkdn. The assay was carried out in 50 mM Hepes (pH 7.4), 100 mM NaCl, 5 mM CaCl2, 1 mg/ml BSA and 0.1% PEG 8000.
| FVIIa variant | kLim (10−3·s−1) | KD (10−7 M) | kinh (M−1·s−1) | kbrkdn (10−5·s−1) | t1/2 (h) |
|---|---|---|---|---|---|
| rFVIIa | 4.9±0.32 | 1.81±0.33 | 26917±5237 | 3.43 | 5.6 |
| T239A | 1.6±0.10 | 2.60±0.41 | 6160±1058 | 2.85 | 6.8 |
| T239G | 1.5±0.14 | 1.14±0.71 | 4688±1155 | 1.61 | 12.0 |
| T239I | 2.4±0.12 | 1.45±0.22 | 16338±2564 | 10.20 | 1.9 |
| T239Y | 5.3±0.29 | 1.74±0.26 | 30591±4906 | 2.08 | 9.3 |
FVIIa–ATIII complex breakdown
The first-order rate constant of covalent complex breakdown, kbrkdn, was determined for wild-type rFVIIa and the FVIIa variants by measuring the restoration of amidolytic activity over 12 h of pre-formed serpin–protease complexes. Similar values of kbrkdn were obtained when the covalent complexes were made with 2.5- and 5-fold excess of ATIII relative to FVIIa, confirming complete inhibition prior to measuring the complex breakdown. The obtained t1/2 for the complex of ATIII and wild-type rFVIIa was 5.6 h (Table 3). The covalent complexes between ATIII and variants T239A, T239G and T239Y were more stable than the wild-type complex, with t1/2 of 6.8, 12 and 9.3 h respectively. In contrast, T239I formed an unstable complex with ATIII having a t1/2 of 1.9 h.
DISCUSSION
The serine proteases of the haemostatic system are hallmarked by a marked and distinctive specificity towards endogenous substrates. This is required to tightly regulate the specific proteolytic events that convert inactive precursors in blood into active enzymes in the reactions leading to blood coagulation and fibrinolysis. FVIIa plays a crucial role in the haemostatic system by initiating blood coagulation upon vascular injury. In the present study, the substrate and inhibitor specificity of FVIIa has been evaluated using small peptidyl substrates, physiological macromolecular substrates as well as site-directed mutagenesis of key residues in the active-site cleft of FVIIa. As expected, rFVIIa only accepted positively charged amino acids in the S1 pocket, with a 3-fold higher preference towards arginine relative to lysine. In the P2 position, hydrophobic or β-branched amino acids were favoured (threonine> leucine> phenylalanine>valine) and aromatic amino acids were preferred in the S3 pocket. Thus the specificity profile clearly demonstrates that rFVIIa is not a very discriminative protease with a low degree of active-site specificity. The selectivity index defined by the ratio between the apparent second-order rate constant of the best and worst sublibrary is 26 in the P2 specificity profile of rFVIIa. Thus the P2 selectivity for rFVIIa appears to be more like trypsin, which has a selectivity index of 12, than FXa or thrombin, which are characterized by selectivity indexes of 290 and 19000 respectively [29]. Recently, the P2 and P3 specificity of FVIIa has been profiled using fluorogenic peptide microarrays [30]. In that study, threonine and valine were also found to be favourable P2 residues; however, in P3, the investigators found that glutamine, arginine, asparagine and proline residues were preferred, in contrast with the preference towards aromatic amino acids found in the present study. The basis for this difference remains unclear; however, it may be due to a possible interdependence between S3 and S4, as the study by Gosalia et al. [30] utilizes alanine in P4 and the present study uses a degenerated pool of amino acids.
In the P4 position, there was a pronounced preference towards tryptophan, which is in agreement with a study searching for extensions to the A-183 FVIIa inhibitor peptide [31], where it was found that the best sequence extending into the active site of rFVIIa also had a tryptophan residue in P4. From modelling studies, the P4 tryptophan was expected to make favourable π − π interactions with Trp364 (c215) in the bottom of the putative S4 pocket. However, the structure of the Trp-Tyr-Thr-Arg-cmk–FVIIa–sTF complex had no electron density corresponding to the P4 tryptophan (see Figure 3), indicating that there is not a well-defined S4 pocket in FVIIa, despite the clear preference observed within the libraries. This could be explained by suboptimal interactions between the P4 residue and the S4 pocket, which would result in a poorly defined electron density, because of high temperature factors, and suggest that FVIIa may not have a well-defined S4 subsite. This notion is to some extent supported by the structure as well as the fact that the natural substrate for FVIIa is the Asn*-Leu-Thr-Arg-Ile-Val-Gly-Gly site of FX, which harbours a glycosylated asparagine (Asn*) residue in the P4 position.
A review of a large number of proteases in the PDB database has revealed that proteases generally bind their substrates in an extended binding conformation, which is to a large degree defined by β-strand-type backbone interactions [32,33]. The PS-SCLs profile the primary selectivity in the S1–S4-binding pockets of a protease independently, without accounting for cross-talk between subsites and without the requirements for optimal alignment of the peptide backbone. To address this issue, interdependency between the S1–S4-binding sites of rFVIIa was investigated by determining the ΔΔGT‡ upon a single substitution in the substrate, for example, Trp-Ala-Thr-Arg-ACC to Phe-Ala-Thr-Arg-ACC etc. As demonstrated by the derived results shown in Figure 2, there is a significant degree of interdependence between the different subsites, as indicated by the relatively large differences associated with identical amino acid substitutions in different scaffolds, i.e. the ΔΔGT‡ for a P4 substitution of asparagine to tryptophan or phenylalanine is very different for substrates with different P3 residues, whereas only minor differences are observed between similar substrates with leucine or threonine in P2. Thus S3 selectivity is probably driven or influenced by P4 occupancy, and this could potentially explain the difference between the two rather similar approaches used to describe FVIIa specificity.
Interestingly, the effects of changing from an asparagine residue in P4 of Asn-Ala-Thr-Arg-ACC to phenylalanine or tryptophan far exceeds the predicted effects from the subsite profiling, as ΔΔGT‡ values of −1.4 and −3.1 kJ·mol−1 respectively, were expected rather than the observed −5 and −7 kJ·mol−1 respectively. This is in contrast with the changes observed for the phenylalanine to tryptophan substitution, which are very close to what should be anticipated from the profiling. One explanation for this observation is that optimal subsite occupancy does not translate to one optimal substrate. As a result, if P3 contains tryptophan or phenylalanine the influence of P4 is ablated, in agreement with the observation that no electron density is visible for the P4 tryptophan in the X-ray structure of the Trp-Tyr-Thr-Arg-cmk–FVIIa–sTF complex. Consequently, the effect of substituting asparagine for a large aromatic amino acid in P4 will be underestimated in the PS-SCLs, due to the requirement for a suboptimal P3 substituent for efficient catalysis.
Since the predominating specificity of FVIIa appears to be located to S1 and S2, we examined the S2-binding site for residues that may alter substrate recognition. Thr239 (c99) is localized at the interface between the S2- and S4-binding pocket in the active-site cleft and could potentially influence the substrate specificity in these binding pockets (see Figure 3). The effect of substituting Thr239 with alanine, glycine, isoleucine or tyrosine was evaluated. All variants had very low amidolytic and proteolytic activity relative to wild-type rFVIIa, except variant T239I, which had an activity comparable with wild-type rFVIIa. Thus, just like FIXa [34], FVIIa is highly sensitive to mutations in this position, indicating that it is important in the substrate and inhibitor recognition by FVIIa.
Substitution of Thr239 with tyrosine caused a 2.5-fold increase in preference towards P2 glycine and a broader P4 specificity profile compared with wild-type rFVIIa. Interestingly, despite the very low catalytic activity of T239Y, this variant was inhibited by ATIII at a rate comparable with wild-type rFVIIa. This is presumably due to the observed P2 glycine specificity of T239Y, as ATIII has a glycine residue in the P2 position of the reactive centre loop. This is in accordance with FXa which has a tyrosine residue in this position, a P2 preference towards glycine and rapidly inhibited by ATIII, which indicated that the T239Y mutation in FVIIa transmitted a FXa-like P2 specificity. The increased propensity of P2 glycine-specific proteases to inhibition by ATIII has also been demonstrated with Gla-domainless APC T254Y (c99), showing an increased preference towards chromogenic substrates with P2 glycine residues, and a 181-fold more efficient inhibition by ATIII compared with Gla-domainless APC [15]. Substitution of Thr239 in FVIIa with isoleucine resulted in a 40% decrease in the apparent second-order rate constant of inhibition by ATIII relative to wild-type rFVIIa, indicating an important role of residue 239 in the interaction with ATIII. Interestingly, the T239I mutation did not affect the proteolytic activity towards FX markedly. The general importance of residue c99 on the rate of inhibition by ATIII has been demonstrated with the thrombin variants L281Y, L281T and L281G (c99), where the apparent second-order rate constant was decreased 2-, 42- and 65-fold respectively, indicating that the larger hydrophilic S2 pockets of FVIIa and APC are not optimal for interacting with ATIII [15]. T239I favoured large aromatic P2 residues revealed by a 2.5- and 4-fold increased preference towards P2 tryptophan and P2 tyrosine respectively, relative to wild-type rFVIIa. However, T239I also had a slightly increased P2 glycine preference (1.5-fold), but was inhibited slower by ATIII compared with T239Y and wild-type rFVIIa, thus indicating that the inhibition of FVIIa by ATIII might be more complex. These findings adequately support the notion derived from the structure, which indicates that the preference for threonine is not associated with specific residue interactions.
Studies of FXa have demonstrated that the P2 glycine preference does not solely determine the specificity towards ATIII indicated by the findings that interactions at secondary sites outside the P6–P3′ region also influence the inhibition of FXa by ATIII [35]. In addition, an exosite within strand three of β-sheet C of ATIII has been identified and has been shown to be important in the reaction of ATIII with FIXa and FXa [36].
The importance of residues within the putative S2 pocket of proteases to the inhibition by serpins has been confirmed by a mutagenesis study of APC [37]. Substitution of residues within the active-site cleft of APC resulted in variants exhibiting resistance to inhibition by protein C inhibitor and α1-antitrypsin, yet maintained anticoagulant activity. When Thr254 (c99) was substituted with a serine residue, the t1/2 in human plasma was prolonged 2-fold due to reduced rates of inactivation by circulating serpins found in plasma. The effect was more pronounced when the T254S (c99) mutation was combined with mutations of residue Leu194 (c40), yielding up to 11-fold prolongation of the plasma t1/2 [37]. Thus it appears that residue c99 in homologous enzymes such as FVIIa and APC influences the inhibitor specificity towards different serpins.
The present study of FVIIa and the one on APC variants described by Berg et al. [37] shows how it is possible to engineer the primary specificity of serine proteases without compromising their biological function in the propagation and regulation of the coagulation cascade. These interesting findings reinforce the notion that the in vivo specificity of proteases towards their macromolecular substrates is, to a large degree, dictated by exosite interactions. As a result, it is clearly possible to separately engineer the recognition of macromolecular substrates and inhibitors. Thus, by substituting key residues in the active site cleft of FVIIa, the specificity towards peptidyl substrates as well as towards physiological inhibitors, e.g. ATIII, can be altered, thereby influencing the rate of inactivation without significantly altering the ability to activate FX.
References
- 1.Davie E. W., Fujikawa K., Kisiel W. The coagulation cascade: initiation, maintenance, and regulation. Biochemistry. 1991;30:10363–10370. doi: 10.1021/bi00107a001. [DOI] [PubMed] [Google Scholar]
- 2.Perona J. J., Craik C. S. Structural basis of substrate specificity in the serine proteases. Protein Sci. 1995;4:337–360. doi: 10.1002/pro.5560040301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broze G. J., Jr, Warren L. A., Novotny W. F., Higuchi D. A., Girard J. J., Miletich J. P. The lipoprotein-associated coagulation inhibitor that inhibits the factor VII-tissue factor complex also inhibits factor Xa: insight into its possible mechanism of action. Blood. 1988;71:335–343. [PubMed] [Google Scholar]
- 4.Olson S. T., Bjork I., Sheffer R., Craig P. A., Shore J. D., Choay J. Role of the antithrombin-binding pentasaccharide in heparin acceleration of antithrombinproteinase reactions. Resolution of the antithrombin conformational change contribution to heparin rate enhancement. J. Biol. Chem. 1992;267:12528–12538. [PubMed] [Google Scholar]
- 5.Petersen L. C., Bjorn S. E., Olsen O. H., Nordfang O., Norris F., Norris K. Inhibitory properties of separate recombinant Kunitz-type-protease-inhibitor domains from tissue-factor-pathway inhibitor. Eur. J. Biochem. 1996;235:310–316. doi: 10.1111/j.1432-1033.1996.0310f.x. [DOI] [PubMed] [Google Scholar]
- 6.Chuang Y. J., Swanson R., Raja S. M., Bock S. C., Olson S. T. The antithrombin P1 residue is important for target proteinase specificity but not for heparin activation of the serpin. Characterization of P1 antithrombin variants with altered proteinase specificity but normal heparin activation. Biochemistry. 2001;40:6670–6679. doi: 10.1021/bi002933d. [DOI] [PubMed] [Google Scholar]
- 7.Banner D. W., D'Arcy A., Chene C., Winkler F. K., Guha A., Konigsberg W. H., Nemerson Y., Kirchhofer D. The crystal structure of the complex of blood coagulation factor VIIa with soluble tissue factor. Nature. 1996;380:41–46. doi: 10.1038/380041a0. [DOI] [PubMed] [Google Scholar]
- 8.Shobe J., Dickinson C. D., Edgington T. S., Ruf W. Macromolecular substrate affinity for the tissue factor–factor VIIa complex is independent of scissile bond docking. J. Biol. Chem. 1999;274:24171–24175. doi: 10.1074/jbc.274.34.24171. [DOI] [PubMed] [Google Scholar]
- 9.Baugh R. J., Dickinson C. D., Ruf W., Krishnaswamy S. Exosite interactions determine the affinity of factor X for the extrinsic Xase complex. J. Biol. Chem. 2000;275:28826–28833. doi: 10.1074/jbc.M005266200. [DOI] [PubMed] [Google Scholar]
- 10.Bone R., Silen J. L., Agard D. A. Structural plasticity broadens the specificity of an engineered protease. Nature. 1989;339:191–195. doi: 10.1038/339191a0. [DOI] [PubMed] [Google Scholar]
- 11.Bone R., Sampson N. S., Bartlett P. A., Agard D. A. Crystal structures of α-lytic protease complexes with irreversibly bound phosphonate esters. Biochemistry. 1991;30:2263–2272. doi: 10.1021/bi00222a032. [DOI] [PubMed] [Google Scholar]
- 12.Hedstrom L. Serine protease mechanism and specificity. Chem. Rev. 2002;102:4501–4524. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- 13.Hedstrom L., Szilagyi L., Rutter W. J. Converting trypsin to chymotrypsin: the role of surface loops. Science. 1992;255:1249–1253. doi: 10.1126/science.1546324. [DOI] [PubMed] [Google Scholar]
- 14.Chang Y. J., Hamaguchi N., Chang S. C., Ruf W., Shen M. C., Lin S. W. Engineered recombinant factor VII Q217 variants with altered inhibitor specificities. Biochemistry. 1999;38:10940–10948. doi: 10.1021/bi990055h. [DOI] [PubMed] [Google Scholar]
- 15.Rezaie A. R. Role of residue 99 at the S2 subsite of factor Xa and activated protein C in enzyme specificity. J. Biol. Chem. 1996;271:23807–23814. doi: 10.1074/jbc.271.39.23807. [DOI] [PubMed] [Google Scholar]
- 16.Vindigni A., Winfield M., Ayala Y., Di Cera E. Role of residue Y99 in tissue plasminogen activator. Protein Sci. 2000;9:619–622. doi: 10.1110/ps.9.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rezaie A. R. Role of Leu99 of thrombin in determining the P2 specificity of serpins. Biochemistry. 1997;36:7437–7446. doi: 10.1021/bi962965u. [DOI] [PubMed] [Google Scholar]
- 18.Persson E., Nielsen L. S. Site-directed mutagenesis but not γ-carboxylation of Glu-35 in factor VIIa affects the association with tissue factor. FEBS Lett. 1996;385:241–243. doi: 10.1016/0014-5793(96)00400-0. [DOI] [PubMed] [Google Scholar]
- 19.Persson E., Nielsen L. S., Olsen O. H. Substitution of aspartic acid for methionine-306 in factor VIIa abolishes the allosteric linkage between the active site and the binding interface with tissue factor. Biochemistry. 2001;40:3251–3256. doi: 10.1021/bi001612z. [DOI] [PubMed] [Google Scholar]
- 20.Payne M. A., Neuenschwander P. F., Johnson A. E., Morrissey J. H. Effect of soluble tissue factor on the kinetic mechanism of factor VIIa: enhancement of p-guanidinobenzoate substrate hydrolysis. Biochemistry. 1996;35:7100–7106. doi: 10.1021/bi9529770. [DOI] [PubMed] [Google Scholar]
- 21.Schechter I., Berger A. On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 22.Maly D. J., Leonetti F., Backes B. J., Dauber D. S., Harris J. L., Craik C. S., Ellman J. A. Expedient solid-phase synthesis of fluorogenic protease substrates using the 7-amino-4-carbamoylmethylcoumarin (ACC) fluorophore. J. Org. Chem. 2002;67:910–915. doi: 10.1021/jo016140o. [DOI] [PubMed] [Google Scholar]
- 23.Schechter N. M., Plotnick M. I. Measurement of the kinetic parameters mediating protease-serpin inhibition. Methods. 2004;32:159–168. doi: 10.1016/s1046-2023(03)00207-x. [DOI] [PubMed] [Google Scholar]
- 24.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 1993;26:795–800. [Google Scholar]
- 25.Potterton E., Briggs P., Turkenburg M., Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 26.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta. Crystallogr. D. Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 27.Snipas S. J., Wildfang E., Nazif T., Christensen L., Boatright K. M., Bogyo M., Stennicke H. R., Salvesen G. S. Characteristics of the caspase-like catalytic domain of human paracaspase. Biol. Chem. 2004;385:1093–1098. doi: 10.1515/BC.2004.142. [DOI] [PubMed] [Google Scholar]
- 28.Gron H., Meldal M., Breddam K. Extensive comparison of the substrate preferences of two subtilisins as determined with peptide substrates which are based on the principle of intramolecular quenching. Biochemistry. 1992;31:6011–6018. doi: 10.1021/bi00141a008. [DOI] [PubMed] [Google Scholar]
- 29.Bianchini E. P., Louvain V. B., Marque P. E., Juliano M. A., Juliano L., Le Bonniec B. F. Mapping of the catalytic groove preferences of factor Xa reveals an inadequate selectivity for its macromolecule substrates. J. Biol. Chem. 2002;277:20527–20534. doi: 10.1074/jbc.M201139200. [DOI] [PubMed] [Google Scholar]
- 30.Gosalia D. N., Salisbury C. M., Ellman J. A., Diamond S. L. High throughput substrate specificity profiling of serine and cysteine proteases using solution-phase fluorogenic peptide microarrays. Mol. Cell. Proteomics. 2005;4:626–636. doi: 10.1074/mcp.M500004-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Maun H. R., Eigenbrot C., Lazarus R. A. Engineering exosite peptides for complete inhibition of factor VIIa using a protease switch with substrate phage. J. Biol. Chem. 2003;278:21823–21830. doi: 10.1074/jbc.M300951200. [DOI] [PubMed] [Google Scholar]
- 32.Tyndall J. D., Fairlie D. P. Conformational homogeneity in molecular recognition by proteolytic enzymes. J. Mol. Recognit. 1999;12:363–370. doi: 10.1002/(SICI)1099-1352(199911/12)12:6<363::AID-JMR478>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 33.Tyndall J. D., Nall T., Fairlie D. P. Proteases universally recognize β strands in their active sites. Chem. Rev. 2005;105:973–999. doi: 10.1021/cr040669e. [DOI] [PubMed] [Google Scholar]
- 34.Sichler K., Kopetzki E., Huber R., Bode W., Hopfner K. P., Brandstetter H. Physiological fIXa activation involves a cooperative conformational rearrangement of the 99-loop. J. Biol. Chem. 2003;278:4121–4126. doi: 10.1074/jbc.M210722200. [DOI] [PubMed] [Google Scholar]
- 35.Chuang Y. J., Swanson R., Raja S. M., Olson S. T. Heparin enhances the specificity of antithrombin for thrombin and factor Xa independent of the reactive center loop sequence. Evidence for an exosite determinant of factor Xa specificity in heparin-activated antithrombin. J. Biol. Chem. 2001;276:14961–14971. doi: 10.1074/jbc.M011550200. [DOI] [PubMed] [Google Scholar]
- 36.Izaguirre G., Zhang W., Swanson R., Bedsted T., Olson S. T. Localization of an antithrombin exosite that promotes rapid inhibition of factors Xa and IXa dependent on heparin activation of the serpin. J. Biol. Chem. 2003;278:51433–51440. doi: 10.1074/jbc.M309266200. [DOI] [PubMed] [Google Scholar]
- 37.Berg D. T., Gerlitz B., Shang J., Smith T., Santa P., Richardson M. A., Kurz K. D., Grinnell B. W., Mace K., Jones B. E. Engineering the proteolytic specificity of activated protein C improves its pharmacological properties. Proc. Natl. Acad. Sci. U.S.A. 2003;100:4423–4428. doi: 10.1073/pnas.0736918100. [DOI] [PMC free article] [PubMed] [Google Scholar]