Abstract
Woronin body, a specialized peroxisome, is a unique organelle involved in septal pore sealing and protecting filamentous fungus from excessive cytoplasmic bleeding. We recently characterized the Aohex1 gene encoding the major protein of the Woronin body in the fungus Aspergillus oryzae. Although three-dimensional microscopy revealed plugging of the septal pore by Woronin body, the mechanism of its formation remains unknown. We report here a reduction in the oligomeric forms (dimeric and tetrameric) of AoHex1 upon λ-phosphatase treatment, which indicated that AoHex1 phosphorylation in vivo facilitates its oligomerization. Concomitant with the presence of a highly conserved predicted PKC (protein kinase C)-phosphorylatable site (Ser151), the recombinant AoHex1 was phosphorylated by PKC in vitro and the administration of the PKC inhibitors, bisindolylmaleimide I and chelerythrine, resulted in the reduction of the oligomeric forms of AoHex1 in vivo. While spherical dot-like Woronin bodies were visualized by expressing the dsred2–Aohex1 and egfp (enhanced green fluorescent protein)–Aohex1 constructs in A. oryzae, treatment with the PKC inhibitors caused an abnormal localization to ring-like structures. In addition to the reduced phosphorylation of the mutagenized recombinant AoHex1[S151A] (Ser151 to alanine substitution) by PKC in vitro, the overexpression of Aohex1[S151A] as dsred2 fusion against the wild-type background also showed reduction of the oligomeric forms of the endogenous AoHex1 and its perturbed localization to ring-like structures in vivo. In conclusion, the present study implicates the relevance of PKC-dependent phosphorylation of the Woronin body protein, AoHex1, for its multimerization and proper localization.
Keywords: Aspergillus oryzae, bisindolylmaleimide I, chelerythrine, phosphorylation, protein kinase C (PKC), Woronin body
Abbreviations: CD, Czapeks–Dox (not circular dichroism in this paper); DPY, dextrin polypeptone yeast extract; GFP, green fluorescent protein; EGFP, enhanced GFP; HEX1, hexagonal 1; PKA, protein kinase A; PKC, protein kinase C; PTS1, peroxisome-targeting signal 1
INTRODUCTION
Filamentous ascomycete and deuteromycete fungi possess the ‘Woronin body’, a unique organelle that seals the septal pore in the event of hyphal damage [1]. Septal pore plugging by this dense proteinaceous organelle prevents hyphal bleeding and excessive loss of cytoplasmic constituents. While the Woronin bodies have been identified in several fungi [1–3], recent studies in Neurospora crassa have provided insights into the gene encoding the protein, HEX1 (hexagonal 1), which constitutes the major portion of the Woronin body [4–6]. Following these studies, genes encoding the HEX1 protein have been characterized from other fungi [7–10]. The presence of the PTS1 (peroxisome-targeting signal 1) at the C-terminal end of HEX1 provides a ‘specialized peroxisome’ status for the Woronin body. N. crassa HEX1 self-assembles into a ‘hexagonal’ crystal lattice, resulting in the formation of a solid hexagonal core of the Woronin body [4]. The self-assembly of HEX1 has been explicitly investigated, which led to the identification of key amino acid residues involved in crystal contact [11]. Although such a critical analysis has provided insight into the formation of the Woronin body core in N. crassa, there are questions that remain to be addressed especially with regard to other molecular signals/factors contributing to Woronin body formation. Tenney et al. [5] have proposed that phosphorylation of HEX1 could lead to its multimerization.
Our recent microscopic analysis of hyphal lysis induced by hypo-osmotic shock has provided a three-dimensional view of septal plugging by Woronin body in Aspergillus oryzae [9]. In addition to obtaining high-resolution images of the Woronin body occluded at the septal pore, we also noted that the A. oryzae Woronin bodies did not exist as hexagonal solid cores but as spherical structures capable of squeezing themselves into the septal pore. In the present study, we found that AoHex1 was phosphorylated by PKC (protein kinase C), and treatment with PKC inhibitors, bisindolylmaleimide I or chelerythrine, caused mislocalization of AoHex1. Furthermore, mutational analysis of the highly conserved serine residue (Ser151) predicted as PKC-phosphorylatable site revealed a perturbed localization of AoHex1 to ring-like structures in vivo. These findings for the first time provide evidence implicating a role for PKC in the assembly and localization of AoHex1.
MATERIALS AND METHODS
Strains and culture conditions
Wild-type A. oryzae RIB40 and niaD300 (niaD−) strains served as the DNA donor and recipient for transformation experiments respectively. Escherichia coli DH5α and BL21 (DE3) cells were used for DNA manipulations and recombinant protein expression respectively. E. coli and A. oryzae were transformed as described earlier [12,13]. A. oryzae strains were cultured either in DPY (dextrin polypeptone yeast extract) complete medium (20 g of dextrin, 10 g of polypeptone, 5 g of yeast extract, 5 g of KH2PO4 and 0.5 g of MgSO4·7H2O per litre; pH 5.5) or in CD (Czapeks–Dox) minimal medium (3 g of NaNO3, 2 g of KCl, 1 g of KH2PO4, 0.5 g of MgSO4·7H2O, 0.02 g of FeSO4·7H2O and 20 g of glucose per litre; pH 5.5) [9,14]. CD medium was used as the selection medium for the A. oryzae niaD+ transformants that were grown in either CD non-inducing [with 2% (w/v) glucose] or CD inducing [with 2% (w/v) dextrin] media. The list of strains used in the present study is given in Table 1.
Table 1. Strains used in the present study.
| Strain | Parent strain | Genotype |
|---|---|---|
| RIB40 | Wild-type | |
| niaD300 | RIB40 | niaD− |
| NGHs1 | niaD300 | niaD− (P-amyB::egfp-Aohex1::T-amyB::niaD) |
| NDRH6 | niaD300 | niaD− (P-amyB::dsred2-Aohex1::T-amyB::niaD) |
| NDRH2[S151A] | niaD300 | niaD− (P-amyB::dsred2-Aohex1[S151A]::T-amyB::niaD) |
Plasmids for the expression of recombinant AoHex1 and AoHex1[S151A] in E. coli
The plasmid, pGsAohex1 [8], was used for the expression of recombinant AoHex1 in E. coli. To introduce point mutation on Aohex1 by replacement of the adenine and guanine nucleotides at positions 451 and 452 with guanine and cytosine nucleotides (underlined) respectively, the primers HexA-SA-f (5′-GGACACGGCGCCGTCCGTGCTC-3′), HexA-SA-r (5′-GAGCACGGACGGCGCCGTGTCC-3′), HexA-F2 (5′-TTACCCGGGATGGGATACTACGACGAC-3′; with a SmaI site at the 5′-end) and HexA-R2 (5′-GCGATATCCTACAGACGGGAAGACTG-3; with an EcoRV site at the 5′-end) were utilized. The plasmid, pThexAs [9], served as the template. A 480 bp PCR fragment amplified using the primers, HexA-F2 and HexA-SA-r, and a 90 bp PCR fragment amplified using the primers HexA-SA-f and HexA-R2 were mixed and a second PCR was performed by using the mixture as a template with the primers HexA-F2 and HexA-R2. The 531 bp PCR-amplified fragment was digested with SmaI and EcoRV, and cloned into pBlueScript SK+ to obtain the plasmid, pBsAohex1[S151A]. The mutation was confirmed by nucleotide sequencing. The SmaI–EcoRV-digested Aohex1[S151A] fragment was subcloned into SmaI site on pGEX4T-3 (Amersham Biosciences, Uppsala, Sweden) to obtain the plasmid pGsAohex1[S151A]. The plasmids, pGsAohex1 and pGsAohex1[S151A], were separately transformed into E. coli (BL21) and expressed as GST (glutathione S-transferase) fusions. Purification of the recombinant proteins was performed using glutathione–Sepharose (Amersham Biosciences) according to the manufacturer's instructions.
Protein extraction and detection of AoHex1
The A. oryzae strains (RIB40, NDRH6 and NDRH2[S151A]) were grown for 24 h in either CD (glucose) or CD (dextrin) liquid nutrient media as shaking cultures for a period of 24 h at 30 °C. To detect the effect of PKC inhibition on the in vivo multimerization of AoHex1, the PKC inhibitors, bisindolylmaleimide I (GF109203X; 5 μM; Calbiochem, La Jolla, CA, U.S.A.) or chelerythrine (25 μM; Sigma, St. Louis, MO, U.S.A.), were included in the liquid growth media respectively. Cell extracts were prepared by homogenizing the mycelia using liquid nitrogen in buffer A (50 mM Tris/HCl, pH 7.5, 1 mM PMSF and 1:100 protease inhibitor cocktail). Total cell lysates were initially centrifuged at 500 g to eliminate the cell debris, and then the supernatants obtained were subjected to further centrifugation (10000 g, 10 min and 4 °C) to obtain the pellet fractions enriched with the Woronin body protein. The pellet fractions were collected and resuspended in buffer A. Protein contents were determined by Bradford's method.
Dephosphorylation and phosphorylation assays
The 10000 g pellet fraction (∼100 μg of protein) was subjected to dephosphorylation assay in a volume of 100 μl consisting of 50 mM Hepes buffer (pH 7.5), 0.1 mM EDTA, 2 mM MnCl2, 5 mM dithiothreitol and 500 units of λ-protein phosphatase (Upstate Cell Signaling Solutions, Lake Placid, NY, U.S.A.). After λ-protein phosphatase addition the sample was incubated at 30 °C for 1 h. The reaction was stopped with 5× SDS-sample buffer, boiled for 5 min and resolved on SDS/15% PAGE gel. Western-blot analysis using AoHex1 antibodies was performed as previously described [9].
PKC-dependent phosphorylation assays of recombinant AoHex1 or AoHex1[S151A] were performed in either the presence or absence of the PKC inhibitors, bisindolylmaleimide I (5 μM) and H7 (100 μM; Sigma), by using approx. 25 μg of protein in a volume of 100 μl consisting of 50 mM Hepes buffer (pH 7.4), 10 mM MgCl2, 1 mM CaCl2, 2 μg of phosphatidylserine (Sigma), 0.2 μg of diolein (Sigma) and 50 ng of PKC (Invitrogen, Molecular Probes, Eugene, OR, U.S.A.). The reactions were initiated by the addition of 100 μM ATP, incubated at 30 °C for 30 or 60 min, terminated by the addition of 5× SDS-sample buffer followed by boiling for 5 min and loaded on to SDS/15% PAGE gels. Gels were processed for phosphoprotein staining using the Pro-Q Diamond phosphoprotein detection kit (Invitrogen, Molecular Probes) according to the manufacturer's protocol. Phosphorylation was detected using the fluorescent mode on a LAS-1000plus luminescent image analyser (Fuji Photo Film, Tokyo, Japan).
Construction of DsRed2- or EGFP [enhanced GFP (green fluorescent protein)]-fused expression plasmids for Woronin body visualization
The expression plasmids, pNDRH and pNDRH[S151A], consisting of dsred2-Aohex1 and dsred2-Aohex1[S151A] respectively under the control of amyB promoter and A. oryzae niaD gene as a marker were constructed as follows. The 670 bp PCR-amplified dsred2 fragment using primers DsRed-F (5′-GCGATATCATGGCCTCCTCCGAGAAC-3′) and DsRed-R (5′-TTACCCGGGCAGGAACGAGTGGTGGCG-3′) and pDsRed2 (Clontech) as a template was subcloned at SmaI site downstream of the amyB promoter on pUNA (consisting of A. oryzae niaD gene, amyB promoter and amyB terminator), generating the plasmid pNDR1. Subsequently, the 531 bp Aohex1 fragment obtained by digestion of pBAohex1 with SmaI–EcoRV was subcloned at SmaI site downstream (3′-side) of dsred2 gene in-frame on pNDR1 to generate the plasmid pNDRH. In a similar manner, the 531 bp Aohex1[S151A] fragment obtained by digestion of pBAohex1[S151A] with SmaI–EcoRV was subcloned at SmaI site downstream of dsred2 in-frame on pNDR1 to generate the plasmid pNDRH[S151A]. Plasmids, pNDRH and pNDRH[S151A], were transformed separately into A. oryzae niaD300 strain. Two transformants, NDRH6 and NDRH2[S151A], were utilized for microscopy. To construct the EGFP–AoHex1 expression plasmid, the Aohex1 cDNA was amplified using the primers GFP–HexAN (5′-CTGTACATGGGATACTACGACGACGAC-3′) and GFP–HexAC (5′-TGTACACTACAGACGGGAAGACTGGAT-3′), added with Bsp1407I site and ligated at the relevant site of egfp gene in pBEGFP-F [9]. The fusion construct, egfp–Aohex1, was excised with EcoRV and XbaI and inserted between SmaI and XbaI sites located downstream of amyB promoter of pUNA. The resultant plasmid pUNAGHs was introduced into A. oryzae niaD300 strain and the transformant NGHs1 was used for microscopy.
Fluorescence microscopy
Conidia of strains NDRH6, NDRH2[S151A] and NGHs1 were inoculated in 100 μl of CD dextrin medium on coverslips and after cultivation for a period of 18–20 h at 30 °C were observed by fluorescence microscopy using an Olympus BX52 microscope (Olympus, Tokyo, Japan). Three-dimensional microscopy was performed using IX70 inverted fluorescence microscope (Olympus) equipped with a confocal scanning system CSU21 (Yokogawa Electronics), image-intensifier unit (Hamamatsu Photonics, Hamamatsu-City, Japan) and an AP imager camera (Hamamatsu Photonics). The images were processed using IPlab (Scanalytics) and Vox Blast (VayTek).
RESULTS
Phosphorylation of AoHex1 in vivo facilitates its multimerization
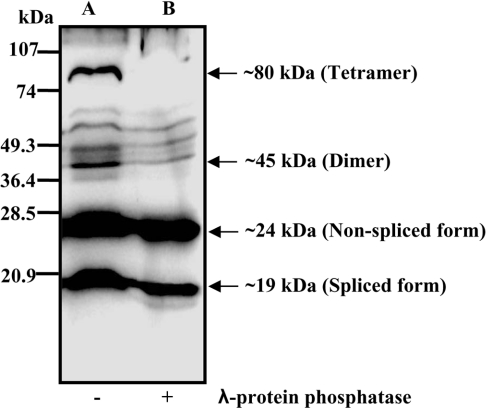
Our previous results on the in vitro detection of AoHex1 in A. oryzae cellular extracts demonstrated the presence of the non-spliced (∼24 kDa) and the spliced forms of AoHex1 (∼19 kDa) in compliance with two mRNA transcripts (0.68 and 0.53 kb) observed by RT (reverse transcriptase)–PCR [9]. These two forms of AoHex1 were enriched in the 10000 g pellet fraction [9]. In addition to the two monomeric forms of AoHex1, two more major bands of ∼45 and ∼80 kDa were also observed in Western blots (Figure 1, lane A). Considering the results of Tenney et al. [5] in N. crassa, these bands may correspond to the dimeric and tetrameric forms of AoHex1. Interestingly, these bands were highly resistant to solubilization in SDS but disappeared upon λ-protein phosphatase treatment (Figure 1, lane B). A similar observation by Tenney et al. [5] also suggested that the phosphorylation of the Woronin body protein in vivo might be necessary for the formation of a stable oligomeric structure.
Figure 1. Multimeric AoHex1 dissociates into monomeric forms by dephosphorylation assay.
Proteins extracted from A. oryzae (niaD300) grown in DPY liquid medium for 24 h were centrifuged at 10000 g, and the pellet fraction was treated with λ-protein phosphatase and probed with AoHex1 antibodies. Lanes A and B represent the control (absence of λ-protein phosphatase) and the λ-protein phosphatase-treated samples respectively. Arrows indicate the major forms of AoHex1 detected.
PKC phosphorylates recombinant AoHex1
To gain preliminary insights into the amino acid residues that could be the targets of various kinases, the AoHex1 sequence was scanned using the Phosphobase and NetPhosK databases [15]. As shown in Figure 2(A), predicted PKA (protein kinase A) phosphorylation sites at positions Ser25, Ser29, Ser59 and Ser174, and PKC phosphorylation sites at positions Ser25, Thr32, Thr105, Ser139, Ser151 and Ser173 were identifiable. An examination of the conservation of these residues among AoHex1 homologues revealed the conservation in only the Ser151 residue (Figure 2B). Interestingly, the amino acids flanking the Ser151 (GHGSVRAL) resembled the PKC substrate peptide sequence (RKGSVRQRKE) [16], indicating the higher possibility of its phosphorylation by PKC.
Figure 2. Predicted PKA and PKC phosphorylation sites on AoHex1.
(A) Predicted PKA and PKC phosphorylation sites are indicated by asterisks and underlined respectively. (B) The conservation in Ser151 residue among AoHex1 homologues and the flanking sequence showing homology to the PKC substrate peptide are shown. The arrowhead indicates the highly conserved Ser151 residue. Respective positions of amino acids in different fungal species are also indicated.
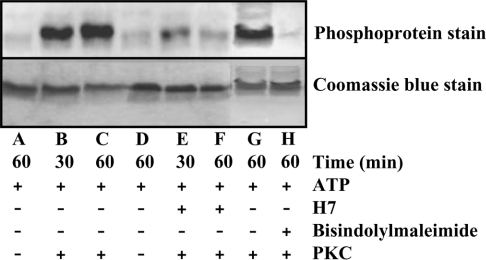
Considering the conservation between the PKC substrate peptide sequence and the sequence flanking the Ser151 residue, we sought to examine the PKC-dependent phosphorylation of AoHex1 in vitro. As shown in Figure 3 (phosphoprotein stain), the recombinant AoHex1 protein could be phosphorylated by PKC (lanes B and C). No significant variation in the phosphorylation was noted in the reactions performed for 30 or 60 min. Furthermore, PKC-dependent phosphorylation of AoHex1 was ascertained either in the absence of PKC (Figure 3, lanes A and D) or by inclusion of PKC inhibitors, H7 and bisindolylmaleimide I, in the phosphorylation reactions (lanes E, F and H). The observed reduction in the phosphorylation of AoHex1 in the presence of H7 and bisindolylmaleimide I clearly demonstrated that PKC phosphorylated AoHex1 in vitro.
Figure 3. PKC-dependent phosphorylation of AoHex1.
The recombinant AoHex1 protein (∼25 μg) was subjected to in vitro PKC phosphorylation assay as mentioned in the Materials and methods section. To assess PKC-dependent phosphorylation, the PKC inhibitors, H7 (100 μM; lanes E and F) and bisindolylmaleimide I (5 μM; lane H), were included in some reaction mixtures. After SDS/PAGE, the gel was directly processed for staining by incubating in fixation solution [50% (v/v) methanol and 10% (v/v) acetic acid] overnight, washed (three times) with deionized water, stained with Pro-Q Diamond phosphoprotein stain for 5 h and destained (1 M sodium acetate, pH 4.0, and 20% acetonitrile) for 1 h. For detection of phosphorylation the gel was scanned using the luminescent image analyser.
Inhibition of PKC results in the disappearance of the multimeric forms of AoHex1 and also causes abnormal localization of the Woronin body protein
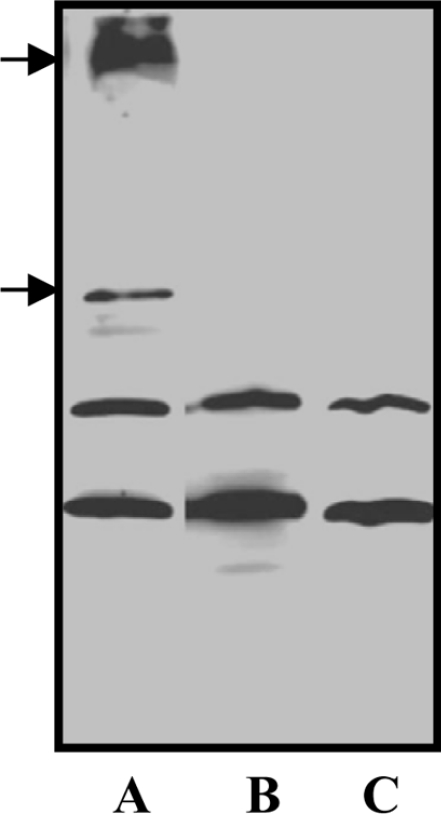
In order to verify if PKC-dependent phosphorylation of AoHex1 in vivo facilitates its aggregation into the multimeric forms, we next grew the A. oryzae wild strain (RIB40) in the presence of the PKC inhibitors, bisindolylmaleimide I and chelerythrine, with the notion that if indeed PKC phosphorylation brought about the assembly of AoHex1 then the treatment with a PKC inhibitor would abolish the formation of the multimeric forms. As shown in Figure 4, we confirmed that the pellet fractions (10000 g; enriched with AoHex1) obtained from the wild strain showed the presence of the dimeric and tetrameric forms of AoHex1 (Figure 4, lane A, indicated by arrows), but the bisindolylmaleimide I- and chelerythrine-treated culture did not contain the aggregated forms of AoHex1 (Figure 4, lanes B and C), confirming that phosphorylation of AoHex1 by PKC in vivo facilitates its multimerization.
Figure 4. PKC inhibitors affect the multimerization of AoHex1.
Proteins extracted from 24 h cultures of the RIB40 wild-type strain cultured in CD (dextrin) medium were centrifuged at 10000 g, and the respective pellet fractions were probed with AoHex1 antibodies. Lanes A, B and C represent samples from the strain cultured in the absence and in the presence of bisindolylmaleimide I or chelerythrine respectively.
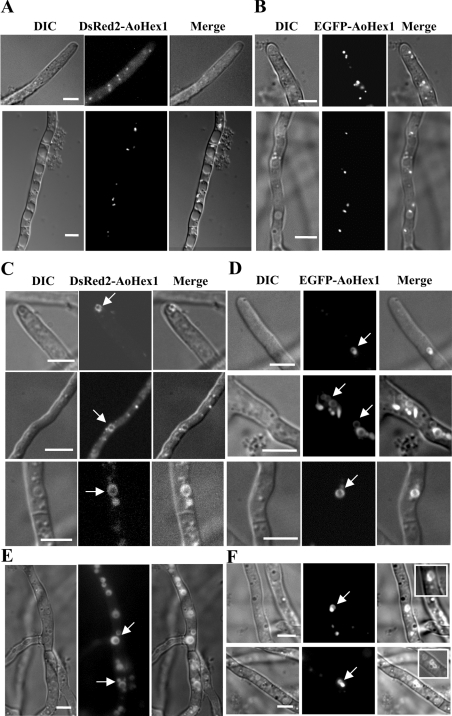
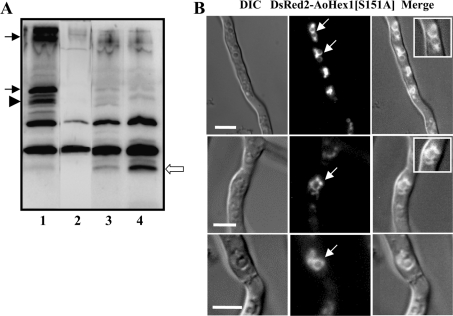
Since the administration of PKC inhibitors resulted in the inhibition of multimerization of AoHex1, we found it pertinent to investigate if PKC-dependent phosphorylation of AoHex1 also influenced the formation of the Woronin body in vivo. For this purpose, the A. oryzae NDRH6 and NGHs1 strains expressing DsRed2–AoHex1 and EGFP–AoHex1 fusion proteins respectively were treated with the PKC inhibitors. Using either bisindolylmaleimide I or chelerythrine at concentrations of 5 and 10 μM respectively did not have any growth-inhibitory effects. While under normal growth conditions (in the absence of PKC inhibitors) the DsRed2–AoHex1 and EGFP–AoHex1 were localized to small dot-like spherical Woronin bodies within the hyphal compartments (Figures 5A and 5B), the inhibitor-treated strains showed abnormal localization of DsRed2–AoHex1 and EGFP–AoHex1 to ring-like structures (Figures 5C–5F, indicated by arrows). Three-dimensional microscopy of these structures revealed that they are some tubular structures arranged in an abnormal ‘ring-like’ form (Figure 6). Administration of another PKC inhibitor, H7 (50 μM), also showed similar mislocalization pattern (results not shown). This result implicated that proper localization of AoHex1 into Woronin body depends on its state of phosphorylation and subsequent multimerization.
Figure 5. PKC inhibitors treatment affects the localization of the Woronin body protein in A. oryzae.
The NDRH6 and the NHGs1 strains expressing the DsRed2–AoHex1 and the EGFP–AoHex1 fusion proteins respectively were cultured in 100 μl of CD (dextrin) media either in the absence (A, B) or in the presence of the PKC inhibitor, bisindolylmaleimide I (5 μM; C, D) or chelerythrine (10 μM; E, F) for 20 h at 30 °C and observed by fluorescence microscopy. Arrows indicate the ring-like structures (C–F) observed in bisindolylmaleimide I- or chelerythrine-treated cultures. Scale bars, 5 μm. DIC, differential interference contrast.
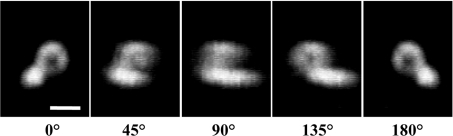
Figure 6. Three-dimensional image analysis of the ‘ring-like’ structure.
Confocal fluorescent image of a ‘ring-like’ structure is reconstructed three-dimensionally and shown from different angles. Scale bar, 2 μm.
The Ser151 to alanine mutation reduces the phosphorylation of AoHex1 in vitro
To verify if the highly conserved Ser151 residue was the site of PKC-dependent phosphorylation, site-directed mutagenesis was performed to alter the Ser151 residue to alanine, and the recombinant AoHex1[S151A] was examined for PKC-dependent phosphorylation in vitro. As shown in Figure 7 (phosphoprotein stain; lane 4), the recombinant AoHex1[S151A] showed a slight reduction in the phosphorylation by PKC, indicating that the Ser151 residue was the target of PKC-dependent phosphorylation. Since the recombinant AoHex1[S151A] was able to be phosphorylated, but to a lower extent in comparison with the wild-type AoHex1 (Figure 7, lane 1), we speculate that there may be other sites of phosphorylation by PKC on AoHex1. The PKC inhibitors, bisindolylmaleimide I and H7, diminished the phosphorylation in both the samples (Figure 7, lanes 2, 3, 5 and 6).
Figure 7. Mutation of Ser151 resulted in reduction of PKC-dependent phosphorylation of AoHex1.
Recombinant AoHex1 (lanes 1–3) and AoHex1[S151A] (lanes 4–6) were subjected to PKC phosphorylation assay either in the presence or in the absence of the PKC inhibitors, bisindolylmaleimide I (5 μM) or H7 (100 μM), as described in the Materials and methods section.
Overexpression of the mutated AoHex1[S151A] interferes with multimerization of the endogenous AoHex1, and causes abnormal localization of the Woronin body protein
To investigate the role of Ser151 phosphorylation in the multimerization and localization of AoHex1, we expressed the mutagenized form of AoHex1[S151A] to mimic a non-phosphorylated state. The cell lysates from the NDRH2[S151A] strain expressing the AoHex1[S151A] fused with DsRed2 in the wild-type background were examined by Western-blot analysis using AoHex1 antibodies, and the effect of overexpression of the AoHex1[S151A] on multimerization of the endogenous AoHex1 was analysed. While the multimeric forms (∼45 and ∼80 kDa) of the endogenous AoHex1 were not detected (Figure 8A, lane 4) in the NDRH2[S151A] strain, the NDRH6 strain expressing the wild-type AoHex1 as DsRed2 fusion (as a control) showed the presence of the multimeric forms (Figure 8A, lane 1, indicated by arrows). This indicated that the expression of the DsRed2–AoHex1 fusion (wild-type) did not influence multimerization of the endogenous AoHex1. In addition, the treatment of the NDRH6 strain with either bisindolylmaleimide I or chelerythrine abolished the oligomerization (Figure 8A, lanes 2 and 3). These results collectively indicated that the mutated AoHex1[S151A] interferes with the multimerization of the endogenous AoHex1.
Figure 8. Overexpression of the DsRed2–AoHex1[S151A] fusion protein inhibited the multimerization of the endogenous AoHex1.
(A) Proteins extracted from 24 h cultures of the NDRH6 and the NDRH2[S151A] strains were centrifuged at 10000 g, and the pellet fractions were electrophoresed and probed with AoHex1 antibodies. Lanes 1–4 represent the samples from the NDRH6 strain, the NDRH6 strain cultured in the presence of bisindolylmaleimide I or chelerythrine, and the NDRH2[S151A] strain respectively. Arrows indicate the multimeric forms (∼45 and ∼80 kDa) of AoHex1, and the arrowhead indicates the approx. 43 kDa band coinciding with the calculated molecular mass of the DsRed2–AoHex1 fusion protein. The approx. 17 kDa which migrated below the approx. 19 kDa spliced form of AoHex1 is indicated by an unfilled arrow. (B) Visualization of ring-like structures by the expression of DsRed2–AoHex1[S151A] fusion protein in A. oryzae. The NDRH2[S151A] strain was cultured in 100 μl of CD (dextrin) medium for 20 h at 30 °C and observed by fluorescence microscopy. Arrows indicate the ring-like structures observed. Scale bars, 5 μm.
While an approx. 43 kDa band coinciding with the calculated molecular mass of the DsRed2–AoHex1 fusion protein could also be detected clearly in the 10000 g pellet fraction of the NDRH6 strain (Figure 8A, lane 1), treatment with bisindolylmaleimide I or chelerythrine did not show the presence of this band (Figure 8A, lanes 2 and 3). Interestingly, the approx. 43 kDa band was detected in the supernatant fraction from the bisindolylmaleimide I- or chelerythrine-treated NDRH6 strain (results not shown). Similarly, the mutated AoHex1[S151A] fusion protein was also detected in the supernatant fraction but not in the pellet (Figure 8A, lane 4). These results indicated the inability of the fusion protein to assemble with the endogenous AoHex1 during chelerythrine treatment and the overexpression of the mutated AoHex1[S151A].
The NDRH2[S151A] strain apart from showing a reduction in the oligomeric forms (∼45 and ∼80 kDa) also showed an increase in a band of approx. 17 kDa which migrated below the approx. 19 kDa spliced form of AoHex1 (Figure 8A, lane 4, indicated by an unfilled arrow). Appearance of the approx. 17 kDa band seems significant considering the report by Tenney et al. [5], which suggested the possibilities of this polypeptide either not being enclosed within the Woronin bodies or passing through the Woronin body membrane. The observed multimerization defect of AoHex1 due to bisindolylmaleimide I or chelerythrine treatment and the overexpression of AoHex1[S151A] indicates that phosphorylation of AoHex1 may serve as an important step towards the multimerization of the AoHex1 protein.
Since results presented above implied that the point mutation (Ser151 to alanine) altered the multimeric status of AoHex1, and also reduced its ability to be phosphorylated by PKC in vitro, we next examined if the mutation also influenced the localization pattern of AoHex1. As shown above in Figure 5(A), in the NDRH6 strain expressing the wild-type AoHex1 fused with DsRed2 several small spherical Woronin bodies were noted all along a hyphal compartment and also close to the septa. However, a distinct localization pattern of DsRed2–AoHex1[S151A] to ring-like structures was observed in the NDRH2[S151A] strain (Figure 8B). These ring-like structures resembled the structures observed upon treatment of the NDRH6 strain with PKC inhibitors (Figures 5C–5F). DAPI (4′,6-diamidino-2-phenylindole) staining of mycelia to identify if the ring-like structures were nuclei revealed that these structures did not correspond to nuclei (results not shown) and ER Tracker Blue White staining only to a partial extent showed co-localization of these structures with the endoplasmic reticulum network (results not shown). Although we are yet to confirm the identity of these ring-like structures, we speculate that these could be some abnormal structures formed due to the improper formation of the Woronin body.
From the above analysis it was noted that both the PKC inhibitor treatment and the overexpression of AoHex1[S151A] showed similar effects on multimerization and localization of AoHex1. Therefore it is inferred that phosphorylation of Ser151 in AoHex1 by PKC promotes the assembly and the proper localization of AoHex1.
DISCUSSION
The formation of the dense core of the Woronin body is mainly thought to occur by the self-assembling property of the HEX1 protein in N. crassa [4,11]. Based on their study on the HEX1 crystal lattice, Yuan et al. [11] proposed three groups of intermolecular interactions that are involved in the formation of the hexagonal core of the Woronin body. Apart from the self-assembling property of HEX1, the first evidence that phosphorylation of HEX1 might also be another factor contributing to the formation of the multimeric core of the Woronin body comes from the report of Tenney et al. [5]. Hence, other factors that influence the assembly of the Woronin body protein or contribute to the formation of the Woronin body core are not clearly understood. The present study provides evidence on the relevance of PKC-dependent phosphorylation for the multimerization of the Woronin body protein by (i) employing in vitro dephosphorylation/phosphorylation assays, (ii) administration of PKC inhibitors and (iii) site-directed mutagenesis of the highly conserved predicted PKC-phosphorylable site (Ser151 to alanine) on AoHex1.
Dephosphorylation assay (Figure 1) indicated that phosphorylation of AoHex1 by an unknown kinase might facilitate its oligomerization in vivo. Based on the phosphorylation site prediction (Figure 2A) and conservation observed between the PKC substrate (RKGSVRQRKE) [16], and the residues (GHGSVR) flanking the Ser151 (Figure 2B), the phosphorylation of Ser151 of AoHex1 by PKC was focused in the present study. The affinity of a fungal PKC for its substrate peptide (RK#GSVR*QRKE) was demonstrated previously [16], and it was shown that a replacement of the lysine residue (indicated by #) with histidine residue did not alter the Km value. The fact that the sequence flanking the Ser151 residue on AoHex1 (GH#GSVR*) has a histidine residue in place of the lysine residue (indicated by #), and similarly has one basic residue (indicated by asterisk) coinciding with the PKC substrate peptide sequence indicated Ser151 to be the most probable phosphorylation site for PKC on AoHex1. Western-blot analysis on the bisindolylmaleimide I- or chelerythrine-treated strains and the NDRH2[S151A] strain showed reduction in the oligomeric forms of AoHex1 (Figures 3 and 8), indicating that phosphorylation of AoHex1 by PKC in vivo results in AoHex1 oligomerization. The reduction in the oligomeric forms of AoHex1 even in the NDRH2[S151A] may be attributed to the interference in multimerization of the endogenous AoHex1 by the AoHex1[S151A] mutated construct. Interestingly, the Ser151 residue is located outside of the three groups of the intermolecular interaction proposed for the formation of the HEX1 crystal lattice. It may thus be possible that phosphorylation of Ser151 by PKC influences the conformation of AoHex1 assembly.
We further investigated the role of PKC phosphorylation in localization of AoHex1 by the administration of the PKC inhibitors and overexpression of the AoHex1[S151A]. Interestingly, in contrast with the spherical Woronin bodies observed under normal conditions (Figures 5A and 5B), bisindolylmaleimide I and chelerythrine treatment altered the localization of both the DsRed2–AoHex1 and EGFP–AoHex1 fusion proteins to ring-like structures (Figures 5C–5F). Expression of Aohex1[S151A] as dsred2 fusion showed similar ring-like localization (Figure 8B). These ring-like structures seemed to be abnormal structures present in both the apical and basal compartments of the hyphae, indicating the requirement of PKC-dependent phosphorylation for the proper assembly of AoHex1 into the Woronin body.
Although the most conserved predicted PKC phosphorylatable site was point-mutated, the phosphorylation of AoHex1 still occurred but to a lower extent when compared with the wild-type AoHex1 (Figure 7). While the Ser151 is a highly conserved residue among the several AoHex1 homologues, it is possible that the phosphorylation of other sites by PKC modulates the extent of AoHex1 oligomerization. A recent analysis of the phosphorylated sites on human choline acetyltransferase sequence (345-QSSR*KL-350) has identified the PKC-dependent phosphorylation of the Ser346 residue (underlined) [17]. Incidentally, a similar sequence is also present at the C-terminus of AoHex1 (QSSR*L) that includes the PTS1 signal, SRL. While in the present study the results indicating that PKC-dependent phosphorylation of AoHex1 leads to its multimerization and proper localization are significant, further characterization of the phosphorylated sites on AoHex1 and their mutational analyses in future will reveal newer insights into the cellular signals that contribute to the regulation/assembly of the Woronin body protein in A. oryzae.
Acknowledgments
The present study was supported by funds from the Program for Promotion of Basic Research Activities for Innovative Biosciences (Japan).
References
- 1.Markham P., Collinge A. J. Woronin bodies of filamentous fungi. FEMS Microbiol. Rev. 1987;46:1–11. [Google Scholar]
- 2.Collinge A. J., Markham P. Ultrastructure of hyphal tip bursting in Penicillium chrysogenum. FEMS Microbiol. Lett. 1992;70:49–54. doi: 10.1016/0378-1097(92)90561-2. [DOI] [PubMed] [Google Scholar]
- 3.Markham P. Occlusions of septal pores in filamentous fungi. Mycol. Res. 1994;98:1089–1106. [Google Scholar]
- 4.Jedd G., Chua N. H. A new self-assembled peroxisomal vesicle required for efficient resealing of the plasma membrane. Nat. Cell Biol. 2000;2:226–231. doi: 10.1038/35008652. [DOI] [PubMed] [Google Scholar]
- 5.Tenney K., Hunt I., Sweigard J., Pounder J. I., McClain C., Bowman E. J., Bowman B. J. Hex-1, a gene unique to filamentous fungi, encodes the major protein of the Woronin body and functions as a plug for septal pores. Fungal Genet. Biol. 2000;31:205–217. doi: 10.1006/fgbi.2000.1230. [DOI] [PubMed] [Google Scholar]
- 6.Tey W. K., North A. J., Reyes J. L., Lu Y., Jedd G. Polarized gene expression determines Woronin body formation at the leading edge of the fungal colony. Mol. Biol. Cell. 2005;16:2651–2659. doi: 10.1091/mbc.E04-10-0937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curach N. C., Te'o V. S., Gibbs M. D., Bergquist P. L., Nevalainen K. M. Isolation, characterization and expression of the hex1 gene from Trichoderma reesei. Gene. 2004;331:133–140. doi: 10.1016/j.gene.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 8.Asiegbu F. O., Choi W., Jeong J. S., Dean R. A. Cloning, sequencing and functional analysis of Magnaporthe grisea MVP1 gene, a hex-1 homolog encoding a putative ‘Woronin body’ protein. FEMS Microbiol. Lett. 2004;230:85–90. doi: 10.1016/S0378-1097(03)00858-9. [DOI] [PubMed] [Google Scholar]
- 9.Maruyama J., Juvvadi P. R., Ishi K., Kitamoto K. Three-dimensional image analysis of plugging at the septal pore by Woronin body during hypotonic shock inducing hyphal tip bursting in the filamentous fungus Aspergillus oryzae. Biochem. Biophys. Res. Commun. 2005;331:1081–1088. doi: 10.1016/j.bbrc.2005.03.233. [DOI] [PubMed] [Google Scholar]
- 10.Soundararajan S., Jedd G., Li X., Ramos-Pamplona M., Chua N. H., Naqvi N. I. Woronin body function in Magnaporthe grisea is essential for efficient pathogenesis and for survival during nitrogen starvation stress. Plant Cell. 2004;16:1564–1574. doi: 10.1105/tpc.020677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan P., Jedd G., Kumaran D., Swaminathan S., Shio H., Hewitt D., Chua N. H., Swaminathan K. A HEX-1 crystal lattice required for Woronin body function in Neurospora crassa. Nat. Struct. Biol. 2003;10:264–270. doi: 10.1038/nsb910. [DOI] [PubMed] [Google Scholar]
- 12.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 13.Kitamoto K. Molecular biology of the Koji molds. Adv. Appl. Microbiol. 2002;51:129–153. doi: 10.1016/s0065-2164(02)51004-2. [DOI] [PubMed] [Google Scholar]
- 14.Maruyama J., Nakajima H., Kitamoto K. Visualization of nuclei in Aspergillus oryzae with EGFP and analysis of the number of nuclei in each conidium by FACS. Biosci. Biotechnol. Biochem. 2001;65:1504–1510. doi: 10.1271/bbb.65.1504. [DOI] [PubMed] [Google Scholar]
- 15.Blom N., Sicheritz-Ponten T., Gupta R., Gammeltoft S., Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- 16.Lendenfeld T., Kubicek C. P. Characterization and properties of protein kinase C from the filamentous fungus Trichoderma reesei. Biochem. J. 1998;330:689–694. doi: 10.1042/bj3300689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dobransky T., Doherty-Kirby A., Kim A. R., Brewer D., Lajoie G., Rylett R. J. Protein kinase C isoforms differentially phosphorylate human choline acetyltransferase regulating its catalytic activity. J. Biol. Chem. 2004;279:52059–52068. doi: 10.1074/jbc.M407085200. [DOI] [PubMed] [Google Scholar]