Abstract
A genetic locus suppressing DNA underreplication in intercalary heterochromatin (IH) and pericentric heterochromatin (PH) of the polytene chromosomes of Drosophila melanogaster salivary glands, has been described. Found in the In(1)scV2 strain, the mutation, designated as Su(UR)ES, was located on chromosome 3L at position 34.8 and cytologically mapped to region 68A3-B4. A cytological phenotype was observed in the salivary gland chromosomes of larvae homozygous and hemizygous for Su(UR)ES: (i) in the IH regions, that normally are incompletely polytenized and so they often break to form “weak points,” underreplication is suppressed, breaks and ectopic contacts disappear; (ii) the degree of polytenization in PH grows higher. That is why the regions in chromosome arm basements, normally β-heterochromatic, acquire a distinct banding pattern, i.e., become euchromatic by morphological criteria; (iii) an additional bulk of polytenized material arises between the arms of chromosome 3 to form a fragment with a typical banding pattern. Chromosome 2 PH reveals additional α-heterochromatin. Su(UR)ES does not affect the viability, fertility, or morphological characters of the imago, and has semidominant expression in the heterozygote and distinct maternal effect. The results obtained provide evidence that the processes leading to DNA underreplication in IH and PH are affected by the same genetic mechanism.
In 1932 E. Heitz came up with the term “heterochromatin” for the chromosomal regions that remain condensed after mitosis and are revealed as grains of densely stained material in the interphase nucleus (1). Notwithstanding years of intensive investigations, the mysteries of the organization and functions of heterochromatin have not yet been unveiled. According to the hypotheses of the day, heterochromatin in somatic cells is formed in early development as a result of packaging chromosomal material by specific protein complexes, which result in epigenetically inherited “silence” of heterochromatin domains (see reviews in refs. 2–7).
In Drosophila melanogaster, the pericentric regions of mitotic autosomes, X chromosome and the whole Y are heterochromatic. In the cells with polytene chromosomes, these regions are underrepresented and look like diffuse (β) and dense (α) material without banding pattern and fuse to a common chromocenter. DNA underrepresentation could be the consequence of underreplication of densely packaged chromosome material during polytenization or the consequence of elimination of DNA sequences (ref. 8; see references in ref. 6). Direct evidence for any of these possibilities is not available. We use here the term “underreplication” as more widely distributed in the literature. These features of heterochromatin organization in the polytene tissues might have been associated with a feature of its primary structure, namely being poor in coding material and rich in repeats. However, there are many regions in the euchromatin arms of D. melanogaster chromosomes that do not contain highly repetitive DNA, but are underreplicated in the course of polytenization. Incomplete unusual polytenization leads to the formation of breaks or, as many authors call them (9–11), “weak points” in such regions. Not being homologous, weak point regions often contact one another (“ectopic conjugation”). Ectopic contacts occasionally may be so frequent that the chromosome is constantly twisted or forms loops. Bridges in 1935 indicated numerous loops, helices, ectopic nodes, and contacts in the proximal half of chromosome 2L of Drosophila, which sometimes makes it difficult to have well-spread chromosomes and to map bands. For the similarity that they bear with pericentric heterochromatin (PH), weak point regions are called intercalary heterochromatin (IH) (12) or “fragile” sites (ref. 13, for more details see ref. 6).
The genetic content of some IH regions is known: there is a histone gene cluster in 39E, the gene complex BX-C in 89E. Interestingly, the frequency of breaks in IH and, therefore, the degree of underreplication lowers under conditions when position effect variegation is suppressed—at higher temperature and with additional Y chromosome (14). As has been shown recently, such conditions are favorable for the “improvement” of the banding pattern in the eu-heterochromatin junction regions in the basements of chromosome arms so that the regions reveal a banding arrangement usually looking like β-heterochromatin (15). These facts are indicative of the overlapping control of the processes of polytenization in IH and PH and allows the genes affecting the processes of replication of silent heterochromatic domains to be predicted. Herein we present a detailed description of the effects of the genetic factor that suppresses underreplication in PH and IH. We had found it earlier in the strain In(1)scV2, mapped to chromosome 3 and called Su(UR)ES (a suppressor of underreplication) (16).
MATERIALS AND METHODS
Drosophila Stocks.
In(1)scV2 contains an inversion in the X chromosome with breakpoints in 1B2–3 and 20F (in the middle of the nucleolar organizer). Two strains from different stocks were studied (courtesy of A. Garcia-Bellido, Madrid, Spain, and the Bowling Green Stock Center, Bowling Green, OH).
w Su(UR)ES is a strain derived from In(1)scV2 by chromosomal substitution, with only chromosome 3 retained from the initial stock.
In(1)sc4 and In(1)scL8 with breaks in 1B3–4 and distal PH, and 1B2–3 and proximal PH, respectively. Df(3L)vin and Df(3L)clu are deletions in the 68A–68C region (courtesy of S. Nokkala, Turku, Finland and the Bloomington Stock Center, Bloomington, IN).
Oregon-R is a wild-type stock.
w Cy/L D/Sb is a marker stock; chromosomes with D and Cy carry inversions that suppress cross.
Recombination mapping was carried out by using the ru h th st cu sr e ca strain. The strains and markers are described in the reference book (17).
Larvae were grown on a standard medium at 25°C.
Cytological Methods.
Polytene chromosome slides were prepared from third instar larval salivary glands stained with acetic orsein and squashed in 55% lactic acid according to a routine technique. To reveal densely stained α-heterochromatin in pericentric regions, the preparations were stored at room temperature for several days; after that only the densest items remained stained.
To determine the frequency of breakage at weak points, 100–120 nuclei were analyzed in each region under study (10–12 nuclei for each of 8–10 individuals). The most polytenized (thickest) chromosomes, in which breaks occurred with the highest frequency, were selected for analysis. The following types of weak point morphology were taken into account: full or partial breaks splitting >30% of chromosome diameter, ectopic fibers, and band shift (11).
The frequencies of IH and telomeres participating in ectopic conjugation were counted by analysis of weakly or moderately squashed salivary gland cell nuclei (15–30 nuclei in each of 6–8 larvae). The ectopic contact was assumed if there was a continuous chromatin thread joining any two nonhomologous bands of the polytene chromosome (for more details, see ref. 11).
Southern Blot Hybridization.
Genomic DNA was isolated from 100 pairs of salivary glands of third instar larvae or 150 fly heads as proposed by (18) with some modifications and was hydrolized by EcoRI, HindIII. Clones (Table 1) were labeled with α32-P and hybridized after Sambrook et al. (24). Three (3,108, 3,144, and rosy) or two (a histone repeat fragment and rosy) clones were hybridized simultaneously. For the hybridization signals to be of the same intensity, the specific activity of each probe was selected on an individual basis depending on which size and copy number in the genome. The filters with genomic DNA hydrolyzed by HindIII were reused for hybridization with 28S rDNA after removal of hybridized probes. This procedure was rerun, and probe aDm23–24 was hybridized. Hybridization intensity was measured on a spectrophotometer Hitachi 557. DNA representation of the clone under study was calculated as a ratio of hybridization intensity in salivary glands to that in adult heads after normalization on rosy.
Table 1.
DNA clones used
| Clone | Details | Courtesy of |
|---|---|---|
| Fragment of histone repeat | 1.35-kb EcoRI/HindIII fragment including part of the H1 gene (290 bp) and histone spacer (1,060 bp) cloned into pBR322 (19) | S. S. Bogachev |
| rosy | 4.2-kb EcoRI/HindIII fragment of rosy, cloned into pUC19 from Carnegie 20 (20) | |
| 28S rDNA | 0.9-kb-HindIII fragment of 28S rRNA gene re-cloned into pUC19 from λ-clone (in phage λ L47.1) the map perfectly matches that in (21) | N. A. Tchurikov |
| 3108 | 3.3-kb EcoRI fragment of Ubx 5’ exon cloned into pφX (22) | T. Kajimura |
| 3144 | 7.0-kb EcoRI fragment of Ubx 3’ exon cloned into pφX (22) | S. Munroe |
| aDm23-24 | DNA of 359-bp satellite cloned into pBR322 (23). | A. Lohe |
RESULTS
The Cytological Phenotype of Su(UR)ES.
We have observed the following cytological phenotype in the salivary gland polytene chromosomes of larvae homozygous for Su(UR)ES:
(i) The weak points known for normal strains are missing in all IH regions. The frequencies of weak points in some IH regions, that normally are the most liable to break, are presented in Table 2 (1–8). In all strains studied, except those homozygous for Su(UR)ES, weak points occur with a frequency of 80–100% in these regions. In Su(UR)ES, all IH regions are represented by one or a few solid bands without a sign of break. For example, breaks occur with an extremely high frequency in regions 42B and 39DE of the polytene chromosomes of normal strains. However, in the Su(UR)ES strain these bands are very big and condensed (Fig. 1).
Table 2.
Frequency of weak points in salivary gland polytene chromosomes of larvae with different genotypes
| N | Genotype | Frequency
of breaks in region
(%)
|
|||||
|---|---|---|---|---|---|---|---|
| 11A | 19E | 39E | 42B | 75C | 89E | ||
| 1 | In(1)scV2, permanent preparations, 1972 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | In(1)scV2, from Spain, 1997 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 | In(1)scV2, from USA, 1997 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | w; Su(UR)ES, derived from scV2 | 0 | 2 | 0 | 0 | 0 | 0 |
| 5 | w/+; Su(UR)ES/Df(3L)vin5 by crossing w; Su(UR)ES to Df(3L)vin5/TM6 | 0 | 0 | 0 | 0 | 0 | 0 |
| 6 | Oregon-R | 92 ± 2.6 | 84 ± 3.8 | 99 ± 1.0 | 81 ± 3.7 | 92 ± 2.6 | 78 ± 4.0 |
| 7 | In(1)sc4 | 95 ± 2.0 | 95 ± 2.0 | 100 | 82 ± 3.7 | 93 ± 3.6 | 83 ± 3.6 |
| 8 | In(1)scL8 | 97 ± 1.7 | 89 ± 3.1 | 97 ± 1.7 | 85 ± 3.6 | 96 ± 1.9 | 89 ± 3.1 |
| 9 | w/+; Su(UR)ES/+ by crossing w; Su(UR)ES to Oregon-R | 37 ± 5.6 | 49 ± 5.7 | 100 | 25 ± 4.5 | 38 ± 5.1 | 13 ± 3.6 |
| 10 | w/+; +/Su(UR)ES by crossing Oregon-R to w; Su(UR)ES | 66 ± 4.7 | 73 ± 4.8 | 100 | 32 ± 4.7 | 72 ± 4.4 | 27 ± 4.5 |
| 11 | w/+; +/D, + by crossing Oregon-R to w; Su(UR)ES/D, + | 91 ± 2.8 | 78 ± 4.4 | 100 | 76 ± 4.4 | 87 ± 3.6 | 80 ± 4.6 |
| 12 | w/+; D, +/+ by crossing w; Su(UR)ES/D, + to Oregon-R | 77 ± 4.2 | 59 ± 4.5 | 100 | 54 ± 2.1 | 73 ± 4.4 | 51 ± 3.7 |
| 13 | w/+; Su(UR)ES/+ by crossing w; Su(UR)ES/D, + to Oregon-R | 63 ± 4.1 | 56 ± 4.5 | 97 ± 2.0 | 29 ± 4.2 | 51 ± 4.5 | 15 ± 3.3 |
| 14 | w/+; +/Su(UR)ES by crossing Oregon-R to w; Su(UR)ES/D, + | 62 ± 4.3 | 50 ± 4.6 | 98 ± 2.1 | 31 ± 3.8 | 65 ± 3.9 | 25 ± 3.7 |
| 15 | Df(3L)clu/+ by crossing Df(3L)clu/TM6 to Oregon-R | 80 ± 3.8 | 79 ± 4.0 | 100 | 49 ± 5.1 | 81 ± 3.5 | 44 ± 5.2 |
| 16 | +/Df(3L)clu by crossing Oregon-R to Df(3L)clu/TM6 | 86 ± 3.5 | 76 ± 4.5 | 100 | 54 ± 4.9 | 80 ± 4.1 | 48 ± 4.7 |
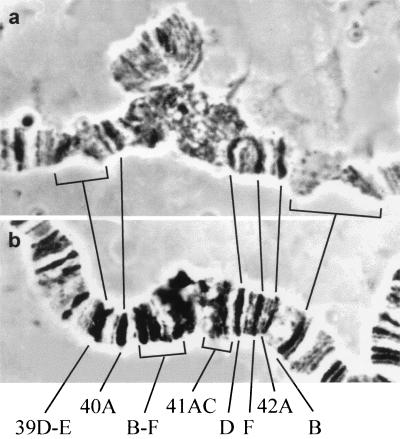
Figure 1.
Morphology of pericentric regions of chromosome 2 in Oregon-R (a) and Su(UR)ES(b). Brackets in a indicate weak points in regions 39DE and 42B of normal chromosomes. In mutants these regions are seen as large dense bands. The segment between 40A and 41D is diffuse β-heterochromatin in Oregon-R chromosomes (a), while the mutants display distinct bands in 40B-F and 41A-C (b).
(ii) The frequency of ectopic pairing between IH themselves, and between telomeres or PH is decreased abruptly. For comparison, these frequencies are presented on the histogram (Fig. 2) as for various regions of IH in In(1)scV2, Su(UR)ES, and some other strains with inversions in which break points are located similarly, but there is no Su(UR)ES.
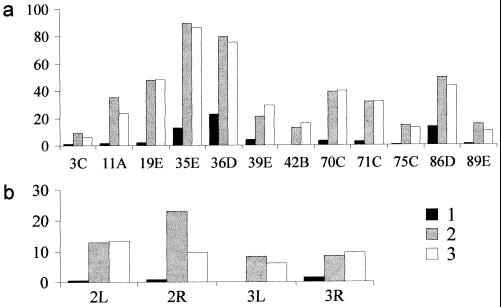
Figure 2.
Frequencies of ectopic conjugation of IH regions (a) and telomere associations (b) in scV2 (1, ▪), sc4 (2, ░⃞) and scL8 (3, □) strains. Abscissa: IH regions (a) telomere regions (b). Ordinates: frequency of participation of the region in ectopic pairing (%).
In the absence of ectopic pairing, Su(UR)ES polytene chromosomes can be perfectly spread on the preparations, they display a very distinct banding pattern all along; the regions in which ectopic contacts and loops are a common occurrence become accessible by fine analysis. A correlation between the respective frequencies of ectopic pairing and breaks had been noted earlier. The causes of this association are not clear and are discussed in detail in refs. 6 and 11.
(iii) Clear bands are seen in the regions that in normal strains form net-like β-heterochromatin in the basements of chromosome arms adjoining the chromocenters in the Su(UR)ES homozygotes. These bands are depicted on Bridges’ maps, but cannot be seen on Lefevre’s map or on standard squashes. So, in all the stocks studied to date the span between bands 40A and 41D is usually filled with diffuse β-heterochromatin, whereas in Su(UR)ES there are blocks of dense bands instead (Fig. 1).
(iv) The most surprising thing about the Su(UR)ES phenotype is additional material in PH. Between arms 3L and 3R appears a region as long as whole chromosome 4 and showing a typical banding pattern (Fig. 3); it is as thick as the euchromatic part of chromosome, which suggests complete polytenization of a normally underreplicated region of PH. We call this region, as if “surfacing” in the Su(UR)ES strain, “Plato Atlantis.” This additional chromosomal fragment shows alternating bands and interbands, which is typical of euchromatin. The thick bands consist of a very dense material, looking like α-heterochromatin, which remains stained after storage in 55% lactic acid for a long time (Fig. 4d). α-heterochromatin can be seen as a tiny grain in the chromocenters of normal strains (Fig. 4a), whereas the amount of α-heterochromatin is sharply increased in the Su(UR)ES strain, especially so between chromosome 2 arms (Fig. 4c).
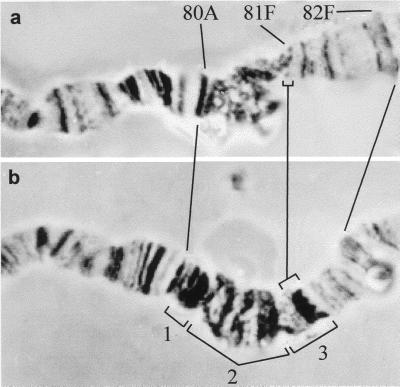
Figure 3.
Morphology of pericentric heterochromatin of chromosome 3 in Oregon-R (a) and Su(UR)ES strains (b). Su(UR)ES chromosome carries three subblock banded fragments between regions 80C and 81F that replace diffuse β-heterochromatin of the normal chromosome. The subblocks are denoted by brackets 1, 2, and 3.
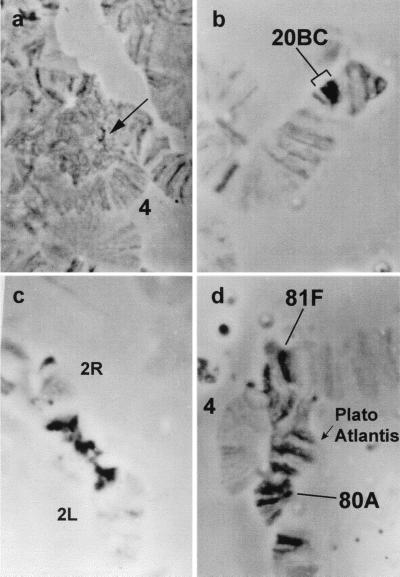
Figure 4.
Additional dense material in centromeric regions of polytene chromosomes. Salivary glands were stained with acetic orsein and squashed in 55% lactic acid, then kept at room temperature. Stain wears off after treatment, only the densest structures remain stained, which are in a very small sector of the chromocenter in Oregon-R (arrow in a). This heterochromatic material is much more abundant in Su(UR)ES strain: region 20BC of the X chromosome (b); large blocks of the pericentric region of chromosome 2 (c); the dense bands of “Plato Atlantis” (d); 4, chromosome 4.
Suppression of DNA Underreplication in Polytene Chromosomes of Su(UR)ES Homozygotes.
The cytological phenotype in the Su(UR)ES strain includes several characteristic changes in polytene chromosome morphology, but all of them are caused by to the same phenomenon, namely suppression of DNA underreplication in the heterochromatic regions, both PH and IH.
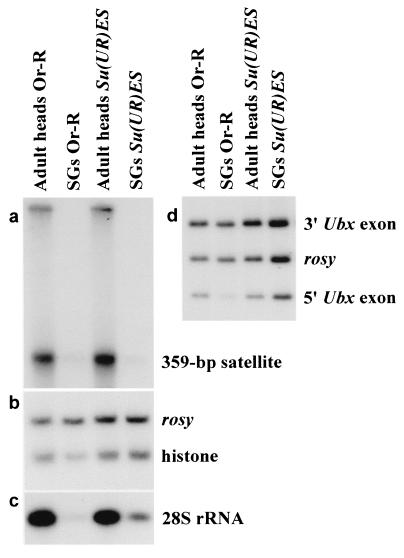
To verify this conclusion, we compared representation of some DNA fragments in polytene chromosomes of Su(UR)ES and the normal Oregon-R strains by Southern blot hybridization. As had been anticipated, the copy number of 359-bp satellite from deep heterochromatin of the X chromosome, histone genes cluster, Ubx and 28S rDNA, was low in the polytene chromosomes of Oregon-R larval salivary glands (Fig. 5), whereas stronger hybridization signals were for Su(UR)ES in all cases, except for 359-bp satellite. Quantitative assessment of underreplication suppression was carried out by densitometry of the bands: peak areas were calculated in each lane and then divided by that of “rosy” (Table 3). The comparison of the figures for Oregon-R and Su(UR)ES showed that the latter carries DNA in 2.4–5 times as much copies as the former does (Table 3). 3′Ubx exon DNA has the same copy number in the chromosomes of both strains, because it keeps away from the “weak point;” we omitted quantitative analysis of 359-bp satellite DNA from consideration because the hybridization signal in both strains was very weak (Fig. 5a).
Figure 5.
Southern blot analysis of DNA in wild-type (Oregon-R) and Su(UR)ES strains. DNA from adult heads and larval salivary glands (SG) was extracted as described in Materials and Methods and hydrolized by HindIII. The same blot was hybridized with probes containing 359-bp satellite (a), histone gene (b), and DNA of 28S rRNA gene (c). Two probes from the 3′Ubx and 5′Ubx exons were hybridized to other blot. DNA of the rosy gene was used as a standard (d).
Table 3.
Representation of DNA sequences in larval salivary gland cells compared to that in adult head cells
| Strain | DNA source | Ratio of DNA qualities
(%)
|
|||
|---|---|---|---|---|---|
| Histone/rosy | 28S rDNA/ rosy | 3′Ubx/ rosy | 5′Ubx/rosy | ||
| Oregon-R | Adult heads | 0.77 (100) | 4.10 (100) | 1.43 (100) | 0.57 (100) |
| Salivary glands | 0.30 (39) | 0.18 (4) | 1.33 (93) | 0.14 (24) | |
| w; Su(UR)ES | Adult heads | 0.76 (100) | 2.70 (100) | 1.53 (100) | 0.51 (100) |
| Salivary glands | 0.71 (93) | 0.57 (21) | 1.52 (99) | 0.56 (110) | |
The results obtained provide further support to the idea that weak points disappear from Su(UR)ES because of restoration of DNA polytenization in these regions.
Genetic and Cytological Localization of Su(UR)ES.
A distinct cytological phenotype allowed the Su(UR)ES locus to be mapped on the cytological and genetic maps.
The locus was first located in chromosome 3 between the ru and th genes; then recombinants were obtained for the ru–h and h–th regions; crossover chromosomes were rendered homozygous and a cytological analysis was performed on the salivary gland chromosomes of the homozygotes. Crossovers between ru and h did not reveal weak points, i.e., phenotypically they were Su(UR)ES. Among 59 crossovers with chromosomes recombinant in the h and th interval, 29 had normal phenotype, and 30 were Su(UR)ES. Thus, the locus in question should be located in the midway between h and th, which is 34.8 cM on the genetic map. As was determined in a special experiment, Su(UR)ES does not affect crossing over frequency in chromosome 3 (data not shown).
Two deficiencies, Df(3L)vin5 and Df(3L)vin4, were mapped by us removing 68A3–68F and 68B5–68F, respectively. Because phenotypically Su(UR)ES/Df(3L)vin5 heterozygotes are Su(UR)ES, and Su(UR)ES/Df(3L)vin4 heterozygotes look like typical Su(UR)ES/+ heterozygotes, the Su(UR)ES gene was located in the region 68A3–68B4.
Heredity of the Trait “Suppression of Underreplication.”
It was because of a cytological phenotype that we found the Su(UR)ES mutation in the In(1)scV2 strain. The phenotype can be observed in two strains with this inversion, supplied from different sources. The mutation has been in this strain for at least 25 years, because the phenotype “suppression of underreplication” was observed on the permanent preparations prepared as early as 1972 and since have been stored in our laboratory (Table 2, lines 1–3). As we became aware, the phenotype is not associated with the X chromosome of scV2, but with chromosome 3, which has no visible cytological disturbances. The mutants are normal with respect to morphology, viability, and fertility, C-heterochromatin staining does not reveal abnormalities in the karyotype (16). To have this phenotype displayed, one would only have to achieve homo- or hemizygosity for the factor located in chromosome 3 at 34.8 cM (Table 2, lines 1–5). Chromosomes of Su(UR)ES/+ heterozygotes have an intermediate phenotype. No such thing as “Plato Atlantis” surfacing in PH of chromosome 3 or a banding pattern in the basements of the chromosome 2 appear; weak points occur in IH. However, IH regions behave differently (Table 2, lines 9 and 10). In region 39E, breaks occur with a frequency of ≈100%, as in normal strains. In 42B and 89E, they are rare, although still more frequent than in Su(UR)ES homozygotes. In 11A, 19E, and 75C, the break frequency in heterozygotes takes on an intermediate value between those for the normal strains and the homozygotes for Su(UR)ES.
Break frequencies are different in reciprocal crosses and, on the whole, are lower in the individuals whose mothers had Su(UR)ES (Table 2, lines 9 and 10). These observations were confirmed in reciprocal crosses between Su(UR)ES and other normal strains, thus favoring maternal effect.
To check this suggestion, we calculated the frequencies of breaks in the progeny of mating of Su(UR)ES/D, Su(UR)ES+ females with Oregon-R males (the chromosome D is linked with inversion In(3LR) allowing genotype identification on polytene chromosome squashes). Among the progeny, Su(UR)ES+/D, Su(UR)ES+ larvae do not carry the mutant Su(UR)ES gene in any dose (Table 2, line 12). Nevertheless, lower weak point frequencies are in their polytene chromosomes than in those of the wild type, which confirms the conclusion about maternal effect of the mutation Su(UR)ES. In the larvae of the same genotype obtained from Oregon-R mothers, the weak point frequencies do not differ from those observed on the wild type (compare lines 6 and 11 in Table 2). The Su(UR)ES/+ larvae obtained from this mating show reduced frequencies of weak points (Table 2, lines 13 and 14).
An interesting peculiarity of the Su(UR)ES phenotype was found in individuals heterozygous for normal chromosome 3L and Df(3L)clu without a small fragment 68A1–2-68B4 where Su(UR)ES is located. Small but significant differences in weak point frequencies in two chromosome regions, 42B and 89E, were found between Oregon-R wild-type and the Su(UR)ES+/Df progeny from direct and reciprocal matings (Table 2, lines 15 and 16). The data show that one dose of the product of the Su(UR)ES+ gene is not sufficient for the phenotype to be normal. However, the effect of the deletion is weaker than the effect of one mutant dose of Su(UR)ES (compare lines 13 and 14 with 15 and 16 in Table 2).
DISCUSSION
Because the factor suppressing heterochromatin underreplication in polytene chromosomes is subject to recombination mapping and the interval of its chromosomal location is narrow from the cytological point of view, the Su(UR)ES phenotype must therefore be caused by a mutation of the gene, whose normal product is involved in the processes leading to underreplication of both PH and IH. Because Su(UR)ES was first found in the In(1)scV2 stock developed by radiation mutagenesis, the mutation may also have been induced by radiation damaging. The mutation shows semidominant expression in the heterozygote and displays maternal effect. These facts suggest that the Su(UR)ES+ product should be active in early embryogenesis, and the dependence of the phenotype on zygotic gene dosage provides evidence that it remains as active later on.
One dose of Su(UR)ES+ in the heterozygote Su(UR)ES+/Df is enough for its phenotype to be nearly normal, whereas the Su(UR)ES/Df and Su(UR)ES/Su(UR)ES+ heterozygotes are mutants. It means that the damage caused by the mutation to the locus has a stronger effect than removal of the locus by deletion. Summing up the data, Su(UR)ES is a mutation that is expressed semidominantly in the heterozygote, has maternal effect, does not affect viability. As far as we know, Su(UR)ES remains to be an only example of mutation affecting polytenization of heterochromatic regions.
There are lethal mutations in the region 68A3-B4, where Su(UR)ES is located, therefore, there could be lethal Su(UR)ES alleles, too.
The supertight packaging of chromosome material can inhibit or delay completion of replication, which could account for imperfect polytenization of chromosomes in heterochromatin regions. The product of the mutant Su(UR)ES allele might reduce the “packaging ratio” and so facilitate DNA replication in heterochromatin. As is known, heterochromatin domains are formed in early embryogenesis, hence a reason for the maternal effect of the Su(UR)ES mutation. However, other explanations are also possible: Su(UR)ES affects the cell cycle, in particular, S phase.
The results obtained provide evidence that the heterochromatin regions are organized very dynamic ally, with perfectly banded euchromatic regions turning into β-heterochromatin. Such phenomena had earlier been observed after exposure to different temperatures during development and variation of heterochromatin in nucleus, as well as in ovarian pseudonurse cells containing polytene chromosomes (refs. 25 and 26 and D.E.K., A.A.A., E.S.B., and I.F.Z., unpublished data). A mutation-induced transformation of β-heterochromatin into euchromatin is being described.
As far as a matter of consistency of heterochromatin region organization is concerned, there is an appealing fact: heterochromatin regions respond differently to the mutant product of Su(UR)ES. Complete polytenization only covers a small PH region of the mutant chromosome 3 to let “Plato Atlantis” emerge; most PH is missing from salivary gland cells. Additional, but still incomplete polytenization takes place in the nucleolar organizer in PH of the X chromosome. All the regions of the weak-points are polytenized in the Su(UR)ES homozygotes, judging by their being no breaks. As regards regions 39E and 89E, it is a fact confirmed by Southern blot hybridization. However, in the heterozygotes for this mutation, some regions retain the “mutant phenotype,” i.e., a low break frequency, whereas others recover and break with normal frequency. These results may suggest that normally such regions are packaged differently, although some other feature of heterochromatin region organization might account for the differences.
At the moment, the most important conclusion that can be drawn from this work is that the same mechanism controls DNA replication of heterochromatic regions, whether pericentric or intercalary. This conclusion additionally favors the progressively developed concept of recent years that the mechanisms of epigenetically inherited silencing of coding genes and highly repetitive sequences of pericentric regions, are similar.
The concept has two groups of facts to rely on:
(i) Cytogenetic observations of coordinated changes in IH and PH at a varying temperature and amount of heterochromatin in the nucleus (14) and, additionally, data on a parallel reduction in DNA underreplication in IH and PH of polytene chromosomes of pseudonurse cells of the otu mutants and decreasing of position effect variegation (25, 26).
(ii) Molecular genetic data provide evidence for the existence of overlapping mechanisms of developmental repression of homeotic genes in Drosophila, silencing in yeast and spreading of repression from pericentric heterochromatin into euchromatin under position effect variegation. It is the formation of different multimeric protein complexes repressing gene activity and maintaining silencing during development that is believed to underlie all these systems of silencing (see references in refs. 2–7).
Noteworthy, homeotic gene BX-C is located in region 89E of typical IH, in which breaks occur as a result of underreplication of a particular sequence of this complex gene (18). Thus, other IH regions might contain repressed genes regulated in a similar fashion, in which case new model systems could be added to the existing ones for studying epigenetically inherited gene repression.
Acknowledgments
This paper is dedicated to the memory of Nikolaj Petrovich Dubinin, who died March 26, 1998. We are very grateful to Professors S. Nokkala and A. Garcia-Bellido, and the Bloomington and Bowling Green Stock Centers for supplying us with Drosophila stocks, and Drs. S. S. Bogachev, N. A. Tchurikov, T. Kajimura, S. Munroe, and A. Lohe for granting us with the clones. We are grateful to L. G. Dubinia for help in treatment of this paper. This work was supported by grants from the Russian Foundation for Basic Research (Grants 96–04-50142, 96–15-97749) and Frontier Genetic Program of Russian Federation.
ABBREVIATIONS
- IH
intercalary heterochromatin
- PH
pericentric heterochromatin
References
- 1.Heitz E. Planta. 1932;28:571–630. [Google Scholar]
- 2.Lohe A R, Hilliker A J. Curr Opin Genet Dev. 1995;5:746–755. doi: 10.1016/0959-437x(95)80007-r. [DOI] [PubMed] [Google Scholar]
- 3.Eissenberg J C, Elgin S C R, Paro R. In: Chromatine Structure and Gene Expression. Elgin S C R, editor. Oxford: IRL; 1995. pp. 147–171. [Google Scholar]
- 4.Henikoff S. In: Gene Silencing in Higher Plants and Related Phenomena in Other Eukaryotes. Meyer P, editor. Berlin: Springer; 1995. pp. 193–208. [Google Scholar]
- 5.Elgin S C R. Curr Opin Genet Dev. 1996;6:193–202. doi: 10.1016/s0959-437x(96)80050-5. [DOI] [PubMed] [Google Scholar]
- 6.Zhimulev I F. Adv Genet. 1997;37:1–566. doi: 10.1016/s0065-2660(08)60341-7. [DOI] [PubMed] [Google Scholar]
- 7.Karpen G H. Curr Opin Genet Dev. 1994;4:281–291. doi: 10.1016/s0959-437x(05)80055-3. [DOI] [PubMed] [Google Scholar]
- 8.Karpen G H, Spradling A C. Cell. 1990;63:97–107. doi: 10.1016/0092-8674(90)90291-l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bridges C B. J Hered. 1935;26:60–64. [Google Scholar]
- 10.Lefevre G., Jr . In: The Genetic and Biology of Drosophila. Ashburner M, Novitsky E, editors. 1a. London: Academic; 1976. pp. 31–66. [Google Scholar]
- 11.Zhimulev I F, Semeshin V F, Kulichkov V A, Belyaeva E S. Chromosoma. 1982;87:197–228. [Google Scholar]
- 12.Kaufmann B P. Proc Natl Acad Sci USA. 1939;25:571–577. doi: 10.1073/pnas.25.11.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laird C D. Am Zool. 1989;29:569–591. [Google Scholar]
- 14.Zhimulev I F, Belyaeva E S, Bolshakov V N, Mal’ceva N I. Chromosoma. 1989;98:378–387. doi: 10.1007/BF00292391. [DOI] [PubMed] [Google Scholar]
- 15.Koryakov D E, Belyaeva E S, Alekseyenko A A, Zhimulev I F. Chromosoma. 1996;105:310–319. doi: 10.1007/BF02524649. [DOI] [PubMed] [Google Scholar]
- 16.Belyaeva E S, Alekseyenko A A, Moshkin Yu M, Koryakov D E, Zhimulev I F. Genetika. 1998;34:1–9. doi: 10.1073/pnas.95.13.7532. (in Russian). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindsley D L, Zimm G G. The Genome of Drosophila melanogaster. New York: Academic; 1992. [Google Scholar]
- 18.Lamb M M, Laird C D. Chromosoma. 1987;95:227–235. doi: 10.1007/BF00294779. [DOI] [PubMed] [Google Scholar]
- 19.Mirkovitch J, Mirault M-E, Laemmli U K. Cell. 1984;39:223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- 20.Spradling A C, Rubin G M. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- 21.Tautz D, Hancock J M, Webb D A, Tautz C, Dover G A. Mol Biol Evol. 1988;5:336–376. [Google Scholar]
- 22.Bender W, Akam M, Karch F, Beachy P A, Peifer M, Spierer P, Lewis E B, Hogness D S. Science. 1983;221:23–29. doi: 10.1126/science.221.4605.23. [DOI] [PubMed] [Google Scholar]
- 23.Lohe A R, Brutlag D L. Proc Natl Acad Sci USA. 1986;83:696–700. doi: 10.1073/pnas.83.3.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 25.Mal’ceva N I, Gyurkovics H, Zhimulev I F. Chromosome Res. 1995;3:191–200. doi: 10.1007/BF00710713. [DOI] [PubMed] [Google Scholar]
- 26.Mal’ceva N I, Belyaeva E S, King R C, Zhimulev I F. Dev Genet. 1997;20:163–174. doi: 10.1002/(SICI)1520-6408(1997)20:2<163::AID-DVG9>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]