Abstract
Scythe was originally identified as a novel Reaper-binding anti-apoptotic protein, although the mechanisms of its functions remain largely obscure. Our previous analysis revealed that Scythe can bind to a proteasomal subunit via N-terminal domains and that the domains are required for appropriate development of Xenopus embryos. In the present study, we show evidence that the N-terminus of Scythe interacts with XEF1AO, a maternal form of Xenopus laevis EF1A that was suggested to be a potential inducer of apoptosis in vertebrates, and that the binding enhances the poly-ubiquitin modification and subsequent degradation of XEF1AO. Scythe is required for degradation of XEF1AO, since immunodepletion of Scythe from embryonic extracts stabilized XEF1AO significantly. Furthermore, we show that apoptosis induced by accumulation of XEF1AO can be suppressed by co-expression of the full-length form of Scythe. These observations indicate that the proteolytic regulation of XEF1AO, mediated through Scythe, is essential to prevent inappropriate accumulation of XEF1AO and resulting apoptotic events during the course of Xenopus development.
Keywords: 26S proteasome, apoptosis, Bcl-2-associated athanogene (BAG), elongation factor 1α (EF1A), ubiquitin
Abbreviations: BAG, Bcl-2-associated athanogene; DEVD, Asp-Glu-Val-Asp; DTT, dithiothreitol; EF1A, elongation factor 1α; GST, glutathione S-transferase; HA, haemaggulutinin; Hsp, heat shock protein; IP, immunoprecipitation; MALDI–TOF-MS, matrix-assisted laser-desorption ionization–time-of-flight MS; MG132, benzoyl-Leu-Leu-Leu-aldehyde; PMF, peptide mass fingerprinting; UBL, ubiquitin-like; UBA, ubiquitin-associated; XEF1AO, Xenopus laevis elongation factor 1α oocyte form
INTRODUCTION
Ubiquitin is a covalent modifier that produces a polyubiquitin chain functioning as a degradation signal [1,2]. In this pathway, substrate recognition, ubiquitination and subsequent proteolysis are key steps in the selective degradation of various cellular proteins [2–6]. Previously, functional co-operation between ubiquitination and ubiquitin-domain proteins has been suggested [7–9]. Indeed, it has been reported that several proteins with a UBL (ubiquitin-like domain), a UBA (ubiquitin-associated) domain and a BAG (Bcl-2-associated athanogene) domain, such as HR23 (homologue of yeast Rad 23), PLIC (protein linking integrin-associated protein to the cytoskeleton) and BAG-1, act as adaptor molecules that mediate substrate ubiquitination and degradation. However, their specific target proteins and functional diversity in higher eukaryotes remain unknown.
Scythe was originally identified as a novel chaperone- and Reaper-binding anti-apoptotic protein from Xenopus oocyte extracts [10–12]. Scythe contains a BAG domain (as an heat shock protein 70-binding region) in its C-terminus and a UBL domain in its N-terminal region, but the function of the latter domain has remained largely elusive. It has been reported that complete immunodepletion of Scythe from extracts prevented Reaper-induced apoptosis [10]. Furthermore, a truncated variant of Scythe lacking more than half of the N-terminal portion (known as the C312 fragment) induced apoptosis even in the absence of Reaper [10,13]. In our previous studies [13,14], we found that the N-terminus of Scythe contains at least two ubiquitin-homology sequences, named Domain I and Domain II, that are redundantly involved in the interaction with proteasomal ubiquitin receptor subunit Xrpn10c. Furthermore, we also found that forced expression of a Scythe mutant lacking the ubiquitin homology regions in Domain I and Domain II caused severe defects in normal Xenopus development [13]. Our results indicate that the N-terminal domain of Scythe acts as an essential segment linking the ubiquitin/proteasome machinery to proper embryonic development, although the exact mechanism by which the mutated form of Scythe induced developmental abnormality has remained completely elusive to date.
In the present study, we show that XEF1AO (Xenopus laevis elongation factor 1α oocyte form), a maternal form of EF1A that was suggested to be a potential inducer of apoptosis in vertebrates [15,16], interacts with Scythe via its N-terminal domain. The binding enhanced polyubiquitin modification of XEF1AO, which can be targeted by the proteasome-mediated proteolytic pathway. Furthermore, we show that the accumulation of XEF1AO induced apoptosis in Xenopus embryos, which can be suppressed by co-expression of the full-length form of Scythe. These observations clearly indicate that the proteolytic regulation of XEF1AO mediated by Scythe and the proteasome pathway is crucial for regulation of apoptosis during the course of Xenopus embryonic development.
EXPERIMENTAL
Plasmid construction
The full-length cDNAs of Scythe and XEF1AO were amplified by PCR from Xenopus cDNA libraries prepared from stage-25 embryos. The PCR products of Scythe and XEF1AO were digested with SalI/NotI and EcoRI/SalI respectively, and inserted into pCI-neo-3T7 and pCI-neo-2S vectors for expression in cultured cells [17]. Note that pCI-neo-3T7 and pCI-neo-2S expression vectors contain three repeats of a T7 tag sequence or two repeats of an S peptide sequence respectively at the N-terminal regions of their products. The truncated and mutated versions of Scythe were constructed by PCR and cloned into appropriate vectors. Vectors were used for experiments after verification of the sequence of the inserted DNA.
Expression of proteins in Xenopus embryos
Full-length and their mutant derivatives of Scythe and XEF1AO cDNA were subcloned into the RN3 vector [18], and the mRNAs were synthesized in vitro using mMESSAGEmMACHINE (Ambion, Austin, TX, U.S.A.). The synthesized mRNAs were dissolved in RNase-free water, and the indicated amounts of the mRNAs in a volume of 9.2 nl were injected into the blastomeres of two-cell-stage Xenopus embryos. Total amounts of injected mRNAs were adjusted by control mRNA. Embryos were cultured in a 0.2×MMR (Marc's modified Ringer's) solution at 20°C. At the indicated stages, each embryo was individually harvested, crushed in PBS, and centrifuged (20000 g, 20min, 4°C) to collect the cytoplasm fraction. An aliquot of the fraction (100 μl) was used for immunoprecipitation with an anti-FLAG antibody and subsequently subjected to Western blot analysis.
Caspase assays
Embryos were harvested at the indicated times and stored at −35°C until use. Each single embryo was crushed in 50 μl of PBS and then centrifuged at 10000 g for 5 min, and the supernatants were collected. To measure caspase activity, 20 μl of each supernatant was incubated with 180 μl of assay buffer [50 mM Hepes, pH 7.5, 100 mM NaCl, 0.1% CHAPS, 10 mM DTT (dithiothreitol), 1 mM EDTA and 10% (v/v) glycerol] containing 200 μM fluorescent peptide substrate, Ac-DEVD-MCA (N-acetyl-Asp-Glu-Val-Asp-4-methylcoumaryl-7-amide), at 25°C for 1 h. The reaction was stopped by the addition of 20 μl of 10% (w/v) SDS. Fluorescence due to MCA (7-amido-4-methylcoumarin) was measured in a fluorspectrophotometer (excitation at 380 nm, emission at 460 nm).
Immunological analysis
For IP (immunoprecipitation) analysis, cultured cells were washed with ice-cold PBS and lysed with IP buffer containing 20 mM Tris/HCl (pH 7.5), 150 mM NaCl, 5 mM EDTA and 1% (v/v) Nonidet P-40. The lysate was sonicated for 1 s and centrifuged at 20000 g for 20 min at 4°C, and the resulting supernatant was incubated with 3 μl of anti-FLAG M2–agarose beads (Sigma Chemical Co., St Louis, MO, U.S.A.) or 10 μl of S-protein agarose beads (Novagen, Madison, WI, U.S.A.) for 2 h at 4°C. After the beads had been washed four times with IP buffer, the precipitated immunocomplexes were subjected to SDS/10%-(w/v)-PAGE.
For Western blotting, the whole cell lysate and immunoprecipitates were separated by SDS/PAGE and transferred on to nitrocellulose membranes (Bio-Rad Laboratories, Richmond, CA, U.S.A.). The membranes were immunoblotted with anti-T7 (Invitrogen, San Diego, CA, U.S.A.), anti-S peptide (Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A.), anti-ubiquitin (FK2, Medical and Biological Laboratories, Nagoya, Japan) and anti-Flag M2 (Sigma) antibodies and then incubated with horseradish peroxidase-conjugated antibody against mouse or rabbit immunoglobulin (Amersham Life Science, Little Chalfont, Bucks, U.K.), followed by detection with enhanced chemiluminescence Western blotting detection reagents (Amersham Biosciences).
PMF (peptide mass fingerprinting)
Proteins on the SDS/PAGE gel were excised, destained with 100 mM ammonium bicarbonate containing 50% (v/v) acetonitrile, and were dehydrated completely by evaporation using a SpeedVac vacuum centrifuge, before 100 μl of 100 mM DTT solution in 100 mM ammonium bicarbonate was added, and the proteins were reduced for 1 h at 56°C. The DTT solution was replaced with 100 μl of 100 mM iodoacetamide in 100 mM ammonium bicarbonate. After 45 min incubation at 37°C in the dark with occasional vortex-mixing, the gel pieces were washed with 100 μl of 100 mM ammonium bicarbonate for 10 min, and dehydrated by addition of acetonitrile following vacuum centrifugation. Proteins were digested in gels with modified trypsin (Sigma) in 100 mM ammonium bicarbonate. The resulting peptides were desalted using ZipTip-C18 (Millipore, Bedford, MA, U.S.A.) and mixed with 10 μg/ml α-cyano-4-hydroxycinnamic acid (Nacalai Tesque, Kyoto, Japan) in 0.3% trifluoroacetic acid and 50% (v/v) acetonitrile. The PMFs of proteins were obtained by MALDI–TOF-MS (matrix-assisted laser-desorption ionization–time-of-flight MS) on Voyager™ Biospectrometry Workstation DE-PRO (PerSeptive Biosystems, Framingham, MA, U.S.A.). A database search was performed using Mascot (http://www.matrixscience.com/search_form_select.html).
Preparation of recombinant proteins
To express recombinant HA (haemaggulutinin)-tagged XEF1AO, the XEF1AO gene was subcloned into the pGEX6P1-5HA vector and transformed into Escherichia coli BL21 (DE3). The GST (glutathione S-transferase) fusion protein was expressed in E. coli, and the extracts were applied to glutathione-immobilized agarose beads (Amersham Biosciences). After extensive washing with buffer A (40 mM Tris/HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.01% Nonidet P-40 and 0.5% DTT), the beads were suspended in an appropriate volume of buffer A containing PreScission protease (Amersham Biosciences), and the mixture was incubated for 16 h at 4°C to allow the protease to cleave the GST tag. The protein thus obtained was then used as purified 5HA–XEF1AO. For preparation of His6-tagged recombinant Scythe full-length protein, the gene was subcloned into the pGEX-His vector and transformed into E. coli BL21 (DE3). The proteins were purified as described above and further applied to Ni2+-nitrilotriacetate– agarose beads (Qiagen, Germany) and eluted by 300 mM imidazole in 20 mM Tris/HCl (pH 7.5). The eluted proteins were then dialysed against buffer B (10 mM Tris/HCl, pH 7.5, and 150 mM NaCl). The proteins thus prepared were then used as purified His6–Scythe.
Cell-free degradation assay of XEF1AO
Immunodepletion of Scythe protein from the extract was performed as follows. Protein A–Sepharose™ beads (Amersham Biosciences) was equilibrated with 26 S buffer [10 mM Tris/HCl, pH 7.5, 150 mM NaCl, 10% (v/v) glycerol, 0.5% Tween-20, 10 mM MgCl2 and 5 mM ATP], and a 10 μl volume of beads was incubated with 10 μg of anti-Scythe IgG or control IgG at 25°C for 5 min. After the beads had been extensively washed with 26 S buffer, they were incubated with 200 μl of Xenopus embryo extracts harvested at stage 18. After 30 min of incubation at 4°C, the beads were removed by brief centrifugation (1000 g, 50s, 4°C). The immunodepletion procedures were repeated at least twice to ensure efficient removal of endogenous Scythe protein. The depleted extracts were diluted to 10 μg of protein/μl with 26 S buffer and then used for the stabilization assay for 5HA–XEF1AO. The stabilization assay for 5HA–XEF1AO in the extract was performed as follows; 500 ng of purified 5HA–XEF1AO protein and 20 μl of diluted extracts were mixed and then incubated for 1 h at 37°C or 24 h at 25°C in the presence or absence of a proteasome inhibitor MG132. Reactions were stopped by adding 1% (w/v) SDS, and the amount of 5HA–XEF1AO was measured by Western blotting.
Mammalian cell culture and transfection
HeLa cells were cultured in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% heat-inactivated calf serum at 37°C. HeLa cells were transiently transfected with the indicated plasmids using Metafectene (Biontex, Munich, Germany) according to the protocol supplied by the manufacturer. The total amount of plasmid DNA was adjusted to 2 μg with an empty vector. The cells were incubated for 24 h after transfection, and then harvested and subjected to Western blot analysis.
RESULTS AND DISCUSSION
Scythe interacts with XEF1AO
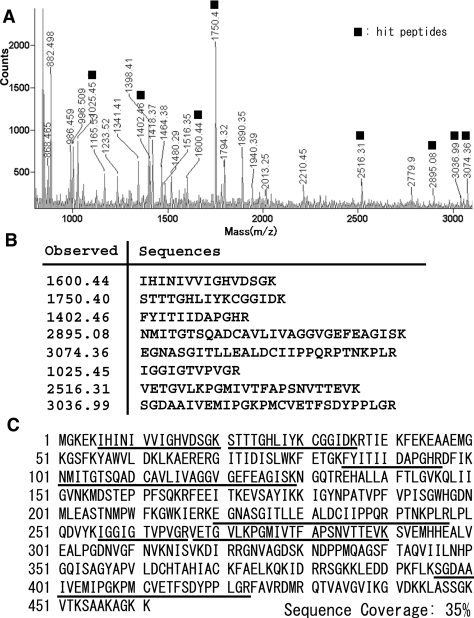
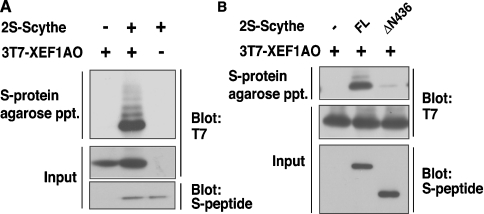
Although Scythe was identified as a Reaper-binding anti-apoptotic protein in Xenopus egg extracts [10], the genuine orthologue of Reaper in vertebrates has not been identified [19], and little is known about other interacting targets that are able to regulate apoptosis. To identify the potential apoptosis-inducing target protein(s) of Scythe in Xenopus embryos, we performed affinity purification of Scythe-binding protein with an S-tagged precipitation system. By using PMF analysis, we identified eight peptides that correspond exactly to the sequence of XEF1AO, a maternal form of Xenopus EF1A (Figure 1). To verify that Scythe and XEF1AO actually interact in vivo, we generated a 2S-tagged Scythe protein and co-expressed it with 3T7-tagged XEF1AO in HeLa cells. We confirmed that Scythe interacted with XEF1AO in vivo (Figure 2A). Furthermore, it was found that the N-terminal 436 amino acids of Scythe (the region covering Domain I and Domain II) are essential for its interaction with XEF1AO, as the deletion of this region abolished the association (Figure 2B). Thus we concluded that XEF1AO is a novel Scythe-binding protein. Although the involvement of XEF1AO in apoptotic regulation had not been clarified in developing Xenopus embryos, mammalian homologues of XEF1AO have been identified as apoptotic modulators. Indeed, it has been reported that the accumulation of eEF1α (eukaryotic EF1α) protein, one of the mammalian homologues of XEF1AO, is linked to induction of apoptosis in various tissues and cultured cells [15,16,20,21]. Thus we hypothesized that XEF1AO plays a crucial role in Scythe-mediated regulation of apoptosis.
Figure 1. Identification of Scythe-associated proteins by PMF.
(A) A spectrum obtained by MALDI–TOF-MS. Signals hit with XEF1AO peptides are indicated by closed boxes. (B) Peptides that exactly correspond to the sequence of XEF1AO are presented. (C) Amino acid sequence of XEF1AO. The regions identified by PMF [shown in (B)] are underlined. These peptides cover 35% of the XEF1AO sequence.
Figure 2. Scythe interacts with XEF1AO in vivo.
(A) Full-length 2S-tagged Scythe and 3T7-tagged XEF1AO were co-expressed in HeLa cells. Tagged proteins were affinity-purified with S-protein–agarose beads, and the co-precipitates were immunoblotted with antibodies against the S-peptide and T7-tag. (B) Either the full-length form (FL) or an N-terminal 436-amino-acid-truncated mutant (ΔN436) of 2S-tagged Scythe was co-expressed with 3T7-tagged XEF1AO in HeLa cells, and co-immunoprecipitates with S-protein–agarose were immunoblotted as described in (A).
Excess accumulation of XEF1AO induces caspase activation in Xenopus embryos
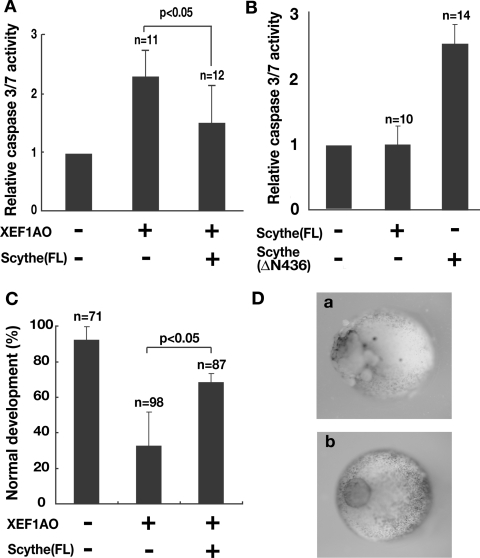
To determine whether the accumulation of XEF1AO causes apoptosis induction in developing embryos, we microinjected mRNA encoding FLAG-tagged XEF1AO into fertilized eggs of X. laevis. Apoptotic cell death generally involves activation of a family of proteases known as caspases [22]. As shown in Figure 3(A), the accumulation of XEF1AO protein indeed induced a significant increase in the cleavage activity of Ac-DEVD-MCA, a well-known substrate of execution caspases, caspase 3, caspase 6 and caspase 7, whereas the expressions of exogenous proteins, such as β-galactosidase and green fluorescent protein, under the same conditions did not stimulate the activity and developmental abnormalities. Interestingly, activation of caspase 3/7 by XEF1AO was observed only after developmental stage 12. Embryos at stages earlier than this did not show caspase activation or any sign of cell death, despite significant accumulation of exogenous XEF1AO protein. This finding implies that cells, including oocytes, fertilized eggs and embryos up to stage 12, are insensitive to the accumulation of XEF1AO. Taking into account the fact that XEF1AO is a maternally expressed protein that is replaced by a somatic form of EF1A, XEF1AS, around developmental stage 12 [23–25], these results suggest that XEF1AO must be appropriately removed from the embryo at a specific stage to prevent induction of apoptotic events after stage 12. Our results show that excess accumulation of XEF1AO results in inappropriate caspase activation in developing embryos after gastrulation to neurulation.
Figure 3. Apoptosis induced by expression of XEF1AO was suppressed by co-expression of Scythe in Xenopus embryos.
(A) mRNA (1 ng) for FLAG-tagged XEF1AO and 4 ng of full-length (FL) Scythe or control mRNAs were microinjected into Xenopus embryos. Embryos were harvested at stage 12 and used for DEVDase assays. The results are shown as means±S.D for the indicated number of embryos. (B) A deletion mutant of Scythe that lacks the N-terminal 436 amino acids caused increased activation of caspase 3/7 in Xenopus embryos. Scythe mRNA (5 ng) encoding T7-tagged Scythe (FL) and Scythe (ΔN436) were microinjected into 2-cell-stage Xenopus embryos. After injection the embryos were incubated for 18 h and then harvested for DEVDase assays. The data represent relative DEVDase activity compared with that of non-injected 2-cell stage embryos and are shown as means±S.D. (C) Ectopic expression of FLAG-tagged XEF1AO by injection of 1 ng of mRNA caused detectable developmental abnormality. In contrast, co-expression of T7–Scythe rescued the abnormality. The results are shown as means±S.D. for the indicated numbers of embryos and are indicative of three independent experiments. Note that some embryos expressing XEF1AO developed to normal viable tailbud embryos. (D) (a) An example of an early stage 12 embryo expressing XEF1AO that began to indicate a first sign of apoptotic cell death. (b) Normal embryos that were injected with control or full-length Scythe mRNA.
Scythe lacking an XEF1AO-binding site induces caspase activation
Since we found that the N-terminal 436 amino acids of Scythe are essential for interaction with XEF1AO (Figure 2B), we next tried to determine whether Scythe lacking N-terminal 436 amino acids (ΔN436) influences apoptosis induction in the developing Xenopus embryo. We injected mRNA encoding the full-length and ΔN436 forms of Scythe proteins into a blastomere of two-cell-stage embryos and harvested the embryos at 18 h after injection for DEVDase assay. We found that the expression of Scythe ΔN436 significantly activated caspase 3/7 (Figure 3B), whereas that of full-length Scythe scarcely activated caspase 3/7. Activation of caspase 3/7 by the expression of Scythe ΔN436 became obvious at stage 12, which corresponds to the time point of down-regulation of XEF1AO in normal embryos. The activity levels of caspase 3/7 became highest at stage 24 in Scythe ΔN436-expressing embryos. The morphology of the embryos expressing Scythe ΔN436 at this stage is identical with that of embryos expressing the apoptosis-inducing fragment of Scythe C312. In contrast, embryos at stages earlier than stage 9 showed little increase in caspase activity or morphological disorder even though they expressed comparable levels of Scythe ΔN436 mutant protein.
Apoptosis induced by XEF1AO can be suppressed by Scythe
Since the N-terminus of Scythe is required for its association with XEF1AO and is also essential for apoptosis regulation, we examined whether full-length Scythe can suppress apoptotic induction via modulating XEF1AO function. To this end, we investigated the effect of co-expression of Scythe on XEF1AO-induced apoptosis in Xenopus embryos. Although caspase activation (Figure 3A) and signs of cell death (Figures 3C and 3D, panel a) were evident after stage 12 in embryos expressing XEF1AO, simultaneous expression of Scythe with XEF1AO suppressed caspase activation and reduced the number of apoptotic embryos (Figures 3A and 3C). These results show that inappropriate apoptosis induction caused by accumulation of XEF1AO can be suppressed by the co-expression of Scythe. Thus, we concluded that XEF1AO is directly regulated by a Scythe-mediated pathway to modulate apoptotic events during the course of Xenopus embryogenesis.
XEF1AO associated with Scythe is polyubiquitinated in vivo
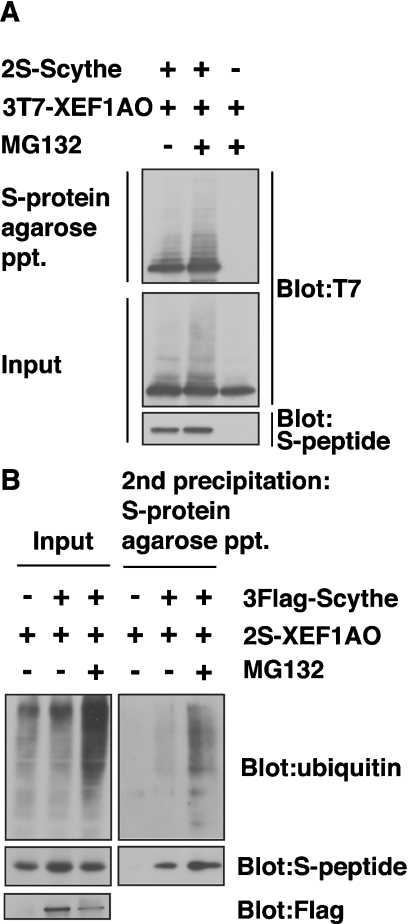
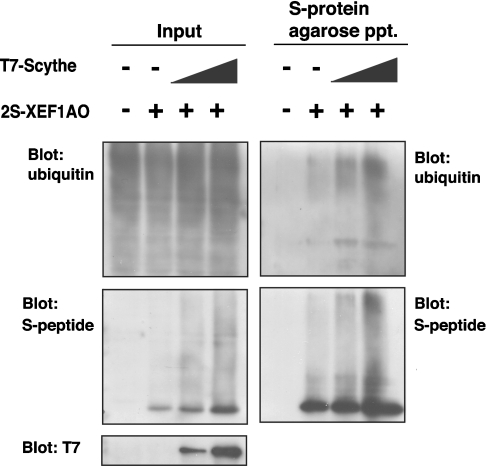
We subsequently noticed that the S-tagged XEF1AO protein co-immunoprecipitated with FLAG-tagged Scythe was detected as multiple ladders (Figures 2A and 4A). The modified forms of XEF1AO were significantly up-regulated when cells were treated with MG132, a potent proteasome inhibitor (Figure 4A). These observations suggest that the multiple modification signals are originated from polyubiquitinated XEF1AO and that the polyubiquitinated form of this protein is susceptible to proteasomemediated degradation. To examine the direct ubiquitin modification of XEF1AO, we performed two-step pull-down experiments (Figure 4B). For the first pull-down, we immunoprecipitated FLAG–Scythe using an anti-FLAG antibody, and the co-precipitated materials including 2S-tagged XEF1AO were treated with a buffer containing 1% (w/v) SDS. Denaturated precipitates were subsequently diluted and precipitated with S-protein agarose (second precipitation). The results indicated clearly that ubiquitin molecules were co-precipitated with 2S–XEF1AO from the MG132-treated cell extracts even after the complete denaturation of 2S–XEF1AO (Figure 4B, second precipitation), suggesting that polyubiquitin moieties are covalently linked to 2S–XEF1AO, which was co-precipitated with FLAG–Scythe. Thus, it can be concluded that the polyubiquitin modification of XEF1AO is closely linked with Scythe- and 26S proteasome-mediated degradation events.
Figure 4. XEF1AO is a Scythe-associated protein that is polyubiquitinated in a Scythe-dependent manner in cultured cells.
(A) 2S-tagged full length Scythe and 3T7-tagged XEF1AO were co-expressed in HeLa cells. Cells were incubated with or without 20 μM MG132 for 6 h before being harvested as indicated. Tagged proteins were affinity-purified with S-protein–agarose beads, and bound proteins were immunoblotted with either anti-S-peptide or anti-T7-tag antibodies. (B) XEF1AO, which was associated with Scythe, is covalently linked with ubiquitin. FLAG-tagged Scythe and 2S-tagged XEF1AO were expressed as in (A). Tagged proteins were immunoprecipitated with anti-FLAG M2– agarose beads, and the precipitates were eluted by SDS denaturation buffer. The eluted samples were diluted with RIPA buffer that did not contain SDS, and then the samples were affinity-purified with S-protein–agarose beads (second precipitation). Bound proteins were subjected to Western blot analysis with anti-ubiquitin, anti-S-peptide and anti-FLAG antibodies. ppt, precipitation.
Ubiquitin modification of XEF1AO is enhanced by Scythe
We found that the polyubiquitin modification of XEF1AO is largely dependent on its association with Scythe. As shown in Figure 5, when cells were treated with MG132, a significant increase in ubiquitinated XEF1AO, concomitant with an increase in the amount of co-existing Scythe protein, was observed. These results suggest that Scythe enhanced the polyubiquitin modification of XEF1AO and that the ubiquitination is linked closely with proteasome-mediated protein degradation events.
Figure 5. Scythe enhances polyubiquitination of XEF1AO.
T7-tagged full-length Scythe and 2S-tagged XEF1AO were co-expressed in HeLa cells. Cells were treated with 20 μM MG132 for 6 h before harvesting. Cells were lysed with RIPA buffer containing 1% (w/v) SDS, and the extracts were diluted with RIPA buffer that did not contain SDS. Then the samples were affinity-purified with S-protein–agarose beads, and bound proteins were immunoblotted with anti-ubiquitin, anti-S-peptide and anti-T7 antibodies. ppt, precipitation.
XEF1AO is degraded through a Scythe-mediated pathway in Xenopus egg extracts
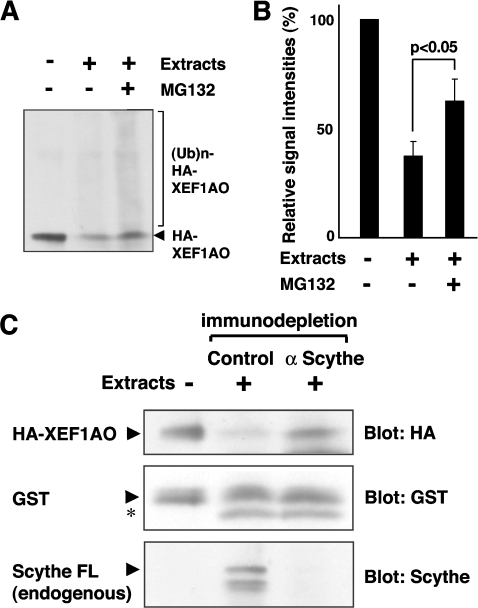
To obtain definite evidence supporting our finding that XEF1AO is degraded by a proteasome- and Scythe-mediated pathway, we established a cell-free assay system using extracts prepared from stage 18 Xenopus embryos. In this system, bacterially expressed 5HA-tagged XEF1AO is highly unstable (Figure 6A), indicating that the degradation machinery for XEF1AO is fully active. It should be noted that the protein biosynthesis occurred at negligible levels in the extracts. We found that HA–XEF1AO is stabilized by the addition of MG132, in part as a high-molecular-mass polyubiquitinated form (Figures 6A and 6B), suggesting that XEF1AO is indeed metabolized via a 26S proteasome- and ubiquitin-dependent degradation pathway.
Figure 6. Scythe is essential for the proteasome-mediated degradation of XEF1AO in Xenopus egg extract.
(A) Bacterially expressed HA–XEF1AO was mixed with Xenopus egg extracts prepared from stage 18 embryos in the presence or absence of 100 μM MG132 as indicated. The mixtures were incubated at 37°C for 1 h and the reaction was stopped by the addition of SDS. The samples were immunoblotted using an anti-HA antibody to measure the amount of XEF1AO. (B) The intensity of the signals derived from XEF1AO and its polyubiquitinated forms in (A) was measured by Scion image and quantitatively represented. The results are shown as the means of duplicates for one out of three independent experiments. (C) Purified HA–XEF1AO was mixed with Xenopus egg extracts at stage 18 that had been immunodepleted either by anti-Scythe IgG or by control IgG. The mixtures were incubated at 25°C for 24 h, and the reaction was stopped by the addition of SDS, followed by immunoblotting with anti-HA antibody to measure the stability of HA–XEF1AO. As a control, bacterially produced GST was added in an equal amount to each sample and blotted with anti-GST antibody, and its stability was compared with that of HA–XEF1AO. The results of anti-Scythe immunoblotting confirmed appropriate immunodepletion of endogenous Scythe protein from the extracts. Arrows indicate the full-length form of corresponding proteins, and the asterisk (*) indicates a non-specific band. FL, full length.
To determine whether Scythe contributes to the degradation of XEF1AO in the extracts, we established a specific antibody against Scythe protein. This antibody was able to immunodeplete endogenous Scythe protein efficiently from the extracts (Figure 6C). We found that HA–XEF1AO was significantly stabilized in the extracts when Scythe was removed (Figure 6C). In contrast, HA–XEF1AO remained highly unstable in the control immunodepletion extracts (Figure 6C). These results indicate clearly that the stability of XEF1AO is specifically modulated by the existence of Scythe, and that Scythe is essential for the down-regulation of XEF1AO protein in extracts derived from stage-18 embryos. To determine whether Scythe itself is sufficient to restore degradation of XEF1AO in the depleted extracts, we added bacterially expressed full-length Scythe protein back into the Scythe-depleted extracts. In this case, we found that there was no restoration in XEF1AO-degrading activity by the addition of recombinant Scythe, implying the possibility that the factor(s) that is essential for the degradation of XEF1AO is co-depleted by anti-Scythe immunodepletion (results not shown). In conclusion, the stability of XEF1AO is specifically modulated by Scythe and its associated protein(s). Defects in this degradation pathway result in stabilization of XEF1AO, and excess accumulation of this protein in embryos at later developmental stages induces inappropriate caspase activation, as shown in Figure 3(A).
Possible role of Scythe in apoptotic regulation
Apoptosis is a physiological form of cell death that is essential for the correct development of multicellular organisms and is precisely controlled by multiple mechanisms [26–28]. It has been reported that components of the protein synthesis pathway play essential roles in regulation of apoptosis [19]. For example, several lines of evidence suggest non-canonical apoptotic functions of EF1A [15,20,21,29]. Indeed, it has been reported that increased EF1A transcripts correlated with p53-induced apoptosis in erythroleukemic cells, and the EF1A gene seems to be directly activated by p53 upon the activation of programmed cell death [15]. In a cardiomyocyte cell line, it has been shown that eEF1A levels undergo a rapid increase upon treatment with apoptosis inducers [16]. Furthermore, antisense treatment of the EF1A gene protects cells against apoptotic stimuli, suggesting that the up-regulation of EF1A plays an important role in execution of the apoptotic programme [20,21]. In vertebrates, several related proteins of the EF1A family have been identified [23–25]. XEF1AS is a somatic form of EF1A expressed in adult cells and later stages of Xenopus embryos, whereas XEF1AO is a germline-specific species that is expressed maternally [24]. Replacement of the maternal form of EF1A by its somatic form at the mid-blastula to neurulation stage is thought to be caused by a switch in gene activity that occurs in early embryonic development [23]. However, the exact mechanism of EF1A down-regulation is currently obscure, and the developmental consequence with failure to degrade XEF1AO at a specific stage has not been adequately examined.
In the present study, we show that XEF1AO binds to Scythe via the N-terminal domain of Scythe and that the binding enhanced the polyubiquitin modification and subsequent degradation of XEF1AO. We have also presented evidence that the accumulation of XEF1AO induces apoptosis in Xenopus embryos, which can be suppressed by co-expression of the full-length form of Scythe. Furthermore, we have shown that an N-terminal deletion mutant of Scythe that fails to bind to XEF1AO induces apoptosis, with concomitant activation of execution caspases. These observations indicate that the proteolytic regulation of XEF1AO mediated by Scythe and the ubiquitin pathway is crucial for regulation of apoptosis during the course of Xenopus embryonic development. In agreement with our findings, Desmots et al. [30] reported recently that knock out of the mouse Scythe gene results in developmental abnormalities and extensive cell death.
Scythe belongs to a family of BAG proteins. It has been reported that the BAG domain of Scythe regulates Hsp70 (heat-shock protein 70)-mediated protein folding and that Scythe-mediated inhibition of Hsp70 is reversed by Reaper, an apoptosis-inducing protein [12]. BAG-1, another UBL-BAG domain protein, has been reported to function as a physical link between the Hsc70 (heat-shock cognate 70)/Hsp70 chaperone system and ubiquitination machineries [8,31]. In a similar way, it is possible that Scythe links the ubiquitination machinery to its apoptotic substrates. In accordance with this idea, we found that polyubiquitin modification of XEF1AO is actually enhanced by Scythe association (Figure 5). The results of the present study suggest that XEF1AO ubiquitination and subsequent degradation occur, at least in part, through a specific interaction with Scythe proteins and thus regulate the embryonic apoptotic pathway. Accordingly, determination of the mechanisms by which Scythe recognizes and ubiquitinates the XEF1AO protein at specific developmental stages is an important challenge for future studies.
Acknowledgments
We thank Dr K. Tanaka, Dr K. Iwai and Dr F. Inagaki for their helpful support and discussions. This work was supported, in part, by grants from the Ministry of Education, Culture, Science and Technology of Japan.
References
- 1.Hershko A., Ciechanover A., Varshavsky A. The ubiquitin system. Nat. Med. 2001;6:1073–1081. doi: 10.1038/80384. [DOI] [PubMed] [Google Scholar]
- 2.Finley D., Ciechanover A., Varshavsky A. Ubiquitin as a central cellular regulator. Cell. 2004;116:S29–S32. doi: 10.1016/s0092-8674(03)00971-1. [DOI] [PubMed] [Google Scholar]
- 3.Voges D., Zwickl P., Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 4.DeMartino G. N., Slaughter C. A. The proteasome, a novel protease regulated by multiple mechanisms. J. Biol. Chem. 1999;274:22123–22126. doi: 10.1074/jbc.274.32.22123. [DOI] [PubMed] [Google Scholar]
- 5.Wilkinson C. R., Seeger M., Hartmann-Petersen R., Stone M., Wallace M., Semple C., Gordon C. Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat. Cell Biol. 2001;3:939–943. doi: 10.1038/ncb1001-939. [DOI] [PubMed] [Google Scholar]
- 6.Shimada M., Kanematsu K., Tanaka K., Yokosawa H., Kawahara H. Proteasomal ubiquitin receptor RPN-10 controls sex determination in Caenorhabditis elegans. Mol. Biol. Cell. 2006;17:5356–5371. doi: 10.1091/mbc.E06-05-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kleijnen M. F., Shih A. H., Zhou P., Kumar S., Soccio R. E., Kedersha N. L., Gill G., Howley P. M. The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol. Cell. 2000;6:409–419. doi: 10.1016/s1097-2765(00)00040-x. [DOI] [PubMed] [Google Scholar]
- 8.Demand J., Alberti S., Patterson C., Hohfeld J. Cooperation of a ubiquitin domain protein and E3 ubiquitin ligase during chaperone/proteasome coupling. Curr. Biol. 2001;11:1569–1577. doi: 10.1016/s0960-9822(01)00487-0. [DOI] [PubMed] [Google Scholar]
- 9.Hara T., Kamura T., Kotoshiba S., Takahashi H., Fujiwara K., Onoyama I., Shirakawa M., Mizushima N., Nakayama K. I. Role of the UBL–UBA protein KPC2 in degradation of p27 at G1 phase of the cell cycle. Mol. Cell. Biol. 2005;25:9292–9303. doi: 10.1128/MCB.25.21.9292-9303.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thress K., Henzel W., Shillinglaw W., Kornbluth S. Scythe: a novel reaper-binding apoptotic regulator. EMBO J. 1998;17:6135–6143. doi: 10.1093/emboj/17.21.6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thress T., Evans E. K., Kornbluth S. Reaper-induced dissociation of a Scythe-sequestered cytochrome c-releasing activity. EMBO J. 1999;18:5486–5493. doi: 10.1093/emboj/18.20.5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thress K., Song J., Morimoto R. I., Kornbluth S. Reversible inhibition of Hsp70 chaperone function by Scythe and Reaper. EMBO J. 2001;20:1033–1041. doi: 10.1093/emboj/20.5.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kikukawa Y., Minami R., Shimada M., Kobayashi M., Tanaka K., Yokosawa H., Kawahara H. Unique proteasome subunit Xrpn10c is a specific receptor for the antiapoptotic ubiquitin-like protein Scythe. FEBS J. 2005;272:6373–6386. doi: 10.1111/j.1742-4658.2005.05032.x. [DOI] [PubMed] [Google Scholar]
- 14.Kawahara H., Kasahara M., Nishiyama A., Ohsumi K., Goto T., Kishimoto T., Saeki Y., Yokosawa H., Shimbara N., Murata S., et al. Developmentally regulated, alternative splicing of the Rpn10 gene generates multiple forms of 26S proteasomes. EMBO J. 2000;19:4144–4153. doi: 10.1093/emboj/19.15.4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kato M. V., Sato H., Nagayoshi M., Ikawa Y. Up-regulation of the elongation factor-1α gene by p53 in association with death of an erythroleukemic cell line. Blood. 1997;90:1373–1378. [PubMed] [Google Scholar]
- 16.Ruest L.-B., Marcotte R., Wang E. Peptide elongation factor eEF1A-2/S1 expression in cultured differentiated myotubes and its protective effect against caspase-3-mediated apoptosis. J. Biol. Chem. 2002;277:5418–5425. doi: 10.1074/jbc.M110685200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takeuchi T., Yokosawa H. ISG15 modification of Ubc13 suppresses its ubiquitin-conjugating activity. Biochem. Biophys. Res. Commun. 2005;336:9–13. doi: 10.1016/j.bbrc.2005.08.034. [DOI] [PubMed] [Google Scholar]
- 18.Lemaire P., Garrett N., Gurdon J. B. Expression cloning of Siamois, Xenopus homeobox gene expressed in dorsal-vegetal cells of blastulae and able to induce a complete secondary axis. Cell. 1995;81:85–94. doi: 10.1016/0092-8674(95)90373-9. [DOI] [PubMed] [Google Scholar]
- 19.Colon-Ramos D. A., Irusta P. M., Gan E. C., Olson M. R., Song J., Morimoto R. I., Elliott R. M., Lombard M., Hollingsworth R., Hardwick J. M., et al. Inhibition of translation and inhibition of apoptosis by Bunyaviral nonstructural proteins bearing sequence similarity to reaper. Mol. Biol. Cell. 2003;14:4162–4172. doi: 10.1091/mbc.E03-03-0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen E., Proestou G., Bourbeau D., Wang E. Rapid up-regulation of peptide elongation factor EF-1α protein levels is an immediate early event during oxidative stress-induced apoptosis. Exp. Cell Res. 2000;259:140–148. doi: 10.1006/excr.2000.4952. [DOI] [PubMed] [Google Scholar]
- 21.Duttaroy A., Bourbeau D., Wang X.-L., Wang E. Apoptosis rate can be accelerated or decelerated by overexpression or reduction of the level of elongation factor 1α. Exp. Cell Res. 1998;238:168–176. doi: 10.1006/excr.1997.3819. [DOI] [PubMed] [Google Scholar]
- 22.Chinnaiyan A., Dixit V. M. The cell-death machine. Curr. Biol. 1996;6:555–562. doi: 10.1016/s0960-9822(02)00541-9. [DOI] [PubMed] [Google Scholar]
- 23.Dje M. K., Mazabraud A., Viel A., Maire M., Denis H., Crawford E., Brown D. D. Three genes under different developmental control encode elongation factor 1-α in Xenopus laevis. Nucleic Acids Res. 1990;18:3489–3493. doi: 10.1093/nar/18.12.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abdallah B., Hourdry J., Krieg P. A., Denis H., Mazabraud A. Germ cell-specific expression of a gene encoding eukaryotic translation elongation factor 1α (eEF-1α) and generation of eEF-1α retropseudogenes in Xenopus laevis. Proc. Natl. Acad. Sci. U.S.A. 1991;88:9277–9281. doi: 10.1073/pnas.88.20.9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deschamps S., Morales J., Mazabraud A., le Maire M., Denis H., Brown D. D. Two forms of elongation factor 1α (EF-1αO and 42Sp50), present in oocytes, but absent in somatic cells of Xenopus laevis. J. Cell Biol. 1991;114:1109–1111. doi: 10.1083/jcb.114.6.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Green D. R., Reed J. C. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 27.Hensey C., Gautier J. Programmed cell death during Xenopus development: a spatio-temporal analysis. Dev. Biol. 1998;203:36–48. doi: 10.1006/dbio.1998.9028. [DOI] [PubMed] [Google Scholar]
- 28.White K., Grether M. E., Abrams J. M., Young L., Farrell K., Steller H. Genetic control of programmed cell death in Drosophila. Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- 29.Lamberti A., Caraglia M., Longo O., Marra M., Abbruzzese A., Arcari P. The translation elongation factor 1A in tumorigenesis, signal transduction and apoptosis. Amino Acids. 2004;26:443–448. doi: 10.1007/s00726-004-0088-2. [DOI] [PubMed] [Google Scholar]
- 30.Desmots F., Russell H. R., Lee Y., Boyd K., McKinnon P. J. The reaper-binding protein Scythe modulates apoptosis and proliferation during mammalian development. Mol. Cell. Biol. 2005;25:10329–10337. doi: 10.1128/MCB.25.23.10329-10337.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luders J., Demand J., Hohfeld J. The ubiquitin-related BAG-1 provides a link between the molecular chaperones Hsc70/Hsp70 and the proteasome. J. Biol. Chem. 2000;275:4613–4617. doi: 10.1074/jbc.275.7.4613. [DOI] [PubMed] [Google Scholar]