Abstract
The ubiquitin-conjugating enzyme Cdc34 (cell division cycle 34) plays an essential role in promoting the G1–S-phase transition of the eukaryotic cell cycle and is phosphorylated in vivo. In the present study, we investigated if phosphorylation regulates Cdc34 function. We mapped the in vivo phosphorylation sites on budding yeast Cdc34 (yCdc34; Ser207 and Ser216) and human Cdc34 (hCdc34 Ser203, Ser222 and Ser231) to serine residues in the acidic tail domain, a region that is critical for Cdc34's cell cycle function. CK2 (protein kinase CK2) phosphorylates both yCdc34 and hCdc34 on these sites in vitro. CK2-mediated phosphorylation increased yCdc34 ubiquitination activity towards the yeast Saccharomyces cerevisiae Sic1 in vitro, when assayed in the presence of its cognate SCFCdc4 E3 ligase [where SCF is Skp1 (S-phase kinase-associated protein 1)/cullin/F-box]. Similarly, mutation of the yCdc34 phosphorylation sites to alanine, aspartate or glutamate residues altered Cdc34–SCFCdc4-mediated Sic1 ubiquitination activity. Similar results were obtained when yCdc34's ubiquitination activity was assayed in the absence of SCFCdc4, indicating that phosphorylation regulates the intrinsic catalytic activity of Cdc34. To evaluate the in vivo consequences of altered Cdc34 activity, wild-type yCdc34 and the phosphosite mutants were introduced into an S. cerevisiae cdc34 deletion strain and, following synchronization in G1-phase, progression through the cell cycle was monitored. Consistent with the increased ubiquitination activity in vitro, cells expressing the phosphosite mutants with higher catalytic activity exhibited accelerated cell cycle progression and Sic1 degradation. These studies demonstrate that CK2-mediated phosphorylation of Cdc34 on the acidic tail domain stimulates Cdc34–SCFCdc4 ubiquitination activity and cell cycle progression.
Keywords: cell cycle, cell division cycle 34 (Cdc34), E2 ubiquitin-conjugating enzyme, phosphorylation, S-phase kinase-associated protein 1 (Skp1)/cullin/F-box (SCF) ubiquitin ligase, yeast
Abbreviations: CDK, cyclin-dependent kinase; CK2, protein kinase CK2; CHX, cycloheximide; DTT, dithiothreitol; 5-FOA, 5-fluoro-orotic acid; GST, glutathione S-transferase; Cdc34, cell division cycle 34; hCdc34, human Cdc34; HECT, homologous to E6-associated protein C-terminus; HEK-293 cells, human embryonic kidney 293 cells; Na PPi, sodium pyrophosphate; ORF, open reading frame; RING, really interesting new gene; Skp1, S-phase kinase-associated protein 1; SCF, Skp1/cullin/F-box; UPS, ubiquitin–proteasome system
INTRODUCTION
The UPS (ubiquitin–proteasome system) is a fundamental regulatory pathway for controlling protein stability that underlies many cellular processes, such as signalling, development and cell cycle progression. Protein ubiquitination, the covalent attachment of the 76-amino-acid polypeptide ubiquitin to substrate proteins, is a cascade of enzymatic steps involving three classes of enzymes. First, the ubiquitin-activating enzyme (E1) forms a high-energy thiolester bond with ubiquitin in an ATP-dependent reaction. Activated ubiquitin is then transferred from the E1 to the active-site cysteine in the ubiquitin-conjugating enzyme (E2) to form a thiolester bond. Finally, the E2, in conjunction with a ubiquitin ligase (E3), transfers ubiquitin to a lysine residue of the target protein to form an isopeptide bond. A lysine residue of the substrate-bound ubiquitin may then function as the site for further ubiquitination generating polyubiquitin chains (reviewed in [1]). Recognition and degradation by the 26S proteasome requires Lys48-linked polyubiquitin chains of at least four ubiquitin moieties [2]. Polyubiquitin chains linked through other lysine residues of ubiquitin have functions independent of proteolysis. For example, Lys63-linked attachment of polyubiquitin activates specific proteins for DNA repair, signal transduction and endocytosis (reviewed in [3,4]).
The class of E3 ubiquitin ligases comprises a diverse group of proteins/multisubunit complexes that contain substrate-specific recognition domains. Control of the E3–substrate protein interaction is a critical mechanism for regulated proteolysis. The two major classes of E3 ligases are defined by the presence of either a HECT (homologous to E6-associated protein C-terminus) domain or a RING (really interesting new gene) domain, which determines the E2's role in ubiquitination. HECT-type E3s accept ubiquitin from the E2 at a catalytic site to form a thiolester intermediate and then catalyse transfer of ubiquitin to the substrate. In contrast, RING-type E3s do not have catalytic activity but recruit the E2 to directly transfer ubiquitin on to the E3-bound substrate protein [1].
The multisubunit SCF [Skp1 (S-phase kinase-associated protein 1)/cullin/F-box] E3 ligase complex is one of the best-characterized RING-type E3s and belongs to the subclass of CRL (Cullin-RING ligases). The invariant core of SCF complexes comprises the scaffold protein Cdc53 (cell division cycle 53)/Cul1, the adaptor protein Skp1 and the RING domain protein Rbx1/Roc1/Hrt1. Rbx1 recruits the E2 ubiquitin-conjugating enzyme Cdc34 (cell division cycle 34), whereas different F-box proteins bind to Skp1 and are important for the recruitment of specific substrates to the complex [5]. F-box proteins typically have a bipartite structure, with an N-terminal F-box motif for Skp1 binding and a C-terminal substrate recognition domain, such as a WD40 domain or a leucine-rich repeat [6–8]. Phosphorylation of the substrate is often required for substrate recognition by the F-box protein and ubiquitin-mediated proteolysis.
The importance of the UPS for cell cycle progression is exemplified by the central role of the ubiquitin-conjugating enzyme Cdc34, which co-operates with the SCF E3 ligase to regulate the levels of key cell cycle regulators. Cdc34's importance for cell cycle progression is evolutionarily conserved from the yeast Saccharomyces cerevisiae to mammalian cells [9–11]. Hence, hCdc34 can complement the temperature-sensitive S. cerevisiae strain cdc34-2, indicating that the human enzyme is functional in yeast [12]. A major substrate of the Cdc34–SCF complex in S. cerevisiae is the CDK (cyclin-dependent kinase) inhibitor Sic1, whose precise ubiquitin-mediated proteolysis is necessary for the G1–S-phase cell cycle transition [13]. G1-phase CDK activity phosphorylates Sic1 on at least six of its nine phosphodegron sites to mediate high-affinity binding to the WD40 domain of the F-box protein Cdc4 [14]. Cdc4 then recruits phosphorylated Sic1 to the SCFCdc4 complex for ubiquitination by Cdc34 [6–8]. This pathway is evolutionarily conserved since hCdc34 targets the CDK inhibitor p27Kip1 for SCFSkp2-mediated ubiquitination to promote G1–S-phase cell cycle progression [11]. Evidence also links Cdc34 to the G2/M-phase transition, where Cdc34 is implicated in the ubiquitin-dependent degradation of the budding yeast inhibitory kinase Swe1 [15], but others have suggested that Cdc34 is not involved in Swe1 ubiquitination and degradation [16]. Cdc34 is also thought to play a role in mitotic spindle function [17]. In addition to roles in the cell cycle, Cdc4 recruits several other critical factors to the SCFCdc4 complex for phosphorylation-dependent, Cdc34-mediated ubiquitination, including the transcription factor GCN4 (general control non-derepressible 4), the Cln-Cdc28 inhibitor/polarization factor Far1 and the cell cycle regulator Cdc6 [18].
Although substrate phosphorylation is a major mechanism for regulating ubiquitination of critical regulatory proteins, post-translational modifications of SCF ligase subunits also regulate the activity of this complex. For example, modification of Cul1 with the ubiquitin-like protein Nedd8 (neddylation) stimulates SCF-dependent ubiquitination in higher eukaryotes, possibly at a step of Cdc34 recruitment [19]. In addition, emerging studies suggest that phosphorylation of Cdc34 and SCF subunits may directly regulate the activity of this complex. For example, CK2 (protein kinase CK2)-mediated phosphorylation of the RING domain protein Rbx2/Roc2/Hrt2 regulates cell cycle progression and the SCF-mediated degradation of IκBα (inhibitory κBα) and p27Kip1 in HeLa cells [20]. In addition, CK2 phosphorylates hCdc34 [21]. Mutation of five potential CK2 phosphorylation sites in the C-terminus of Cdc34 altered its subcellular distribution [21]. CK2 has also been implicated in the binding and phosphorylation of Ubc3B, a gene highly related to Cdc34 (Ubc3) [22]. These studies suggested that phosphorylation of Ubc3B induces its binding to the F-box protein βTrCP and enhances β-catenin degradation. In addition to hCdc34, budding yeast Cdc34 is phosphorylated on serine residues in vivo, although the functional importance of this modification is unknown [10].
Given its pivotal roles in SCF-mediated ubiquitination and control of cell cycle progression, in the present study we investigated the enzymatic and biological functions of Cdc34 phosphorylation. We mapped the in vivo phosphorylation sites on both yCdc34 and hCdc34 and analysed the importance of the yCdc34 phosphorylation sites for SCFCdc4-mediated Sic1 ubiquitination and cell cycle progression. These studies demonstrate that the CK2-mediated phosphorylation of Cdc34 on the acidic C-terminal tail domain regulates the ubiquitination and cell cycle functions of this enzyme.
MATERIALS AND METHODS
Generation of plasmids
The plasmids used in the present study are listed in Table 1. To generate a mammalian expression construct of hCdc34 with an N-terminal FLAG epitope, Cdc34 was amplified by PCR from a human placental cDNA library, digested with BamHI and XhoI and integrated into these sites of pCMV-Tag2 (Stratagene). Point mutations were introduced into hCdc34 of pCMV-hCDC34 to generate the phosphorylation site mutants S231T, S(all4)A (S203A/S222A/S231A/S236A), S203 (S222A/S231A/S236A), S222 (S203A/S231A/S236A), S236 [=3Ala (S203A/S222A/S231A)] and 3Asp by site-directed mutagenesis (QuikChange®; Stratagene) according to the manufacturer's instructions. For bacterial expression, wild-type hCdc34 and the phosphorylation site mutants 3Ala and 3Asp were subcloned from the pCMV constructs into the NdeI and XhoI sites of pET15b (Novagen), encoding an N-terminal His6 sequence. For studies in S. cerevisiae, hCdc34, 3Ala and 3Asp with N-terminal FLAG tags were amplified by PCR, and the NsiI–BglII fragments were cloned into the NsiI and BamHI sites of pMS325 (Table 1). Truncated derivatives of yCdc34 were generated by PCR with pESC1 as a template [23] and cloned into the BglII and PacI sites of pESC1. Point mutations were introduced into yCdc34 of pESC1 to generate the phosphorylation site mutants S(all6)A (S207A/S216A/S263A/S268A/S282A/S292A), S207 (S216A/S263A/S268A/S282A/S292A), S216 (S207A/S263A/S268A/S282A/S292A), S263 (S207A/S216A/S268A/S282A/S292A), S268 (S207A/S216A/S263A/S282A/S292A), S282 (S207A/S216A/S263A/S268A/S292A), S292 (S207A/S216A/S263A/S268A/S282A), 2Ala (S207A/S216A), 2Asp and 2Glu by site-directed mutagenesis (QuikChange®; Stratagene) according to the manufacturer's instructions. To generate the yeast plasmid pMS384-yCdc34(Wt) in which expression of yCdc34 is under the control of its endogenous promoter, the 273 bp CDC34 promoter sequence upstream of the start codon was amplified by genomic PCR and cloned into the XbaI and BglII sites of pRS415 (Stratagene). Wild-type yCdc34 and the phosphorylation site mutants 2Ala, 2Asp and 2Glu were subcloned from the pESC1 constructs into the BglII and XhoI sites of pMS384 (Table 1). The point mutant yCdc34(F72V) [24] was subcloned from pET21(+)yCdc34(F72V) into the BglII and XhoI sites of pMS384. Bacterial expression constructs of His6-tagged yeast Cdc34 and its mutant derivatives were generated by subcloning from the pESC1 constructs into the NdeI and XhoI sites of pET15b. For bacterial expression of His6-tagged Sic1, the ORF (open reading frame) of SIC1 was amplified by genomic PCR and cloned into the BamHI and EcoRI sites of pRSETb (Invitrogen). All of the constructs were verified by DNA sequencing.
Table 1. Plasmids used in the present study.
CMV, cytomegalovirus.
| Plasmid | Description | Source or reference |
|---|---|---|
| pCMV-Tag2b | CMV promoter, N-terminal FLAG tag | Stratagene |
| pCMV-hCdc34(Wt) | Human wild-type Cdc34 | The present study |
| pCMV-hCdc34(S231T) | (S231T) | The present study |
| pCMV-hCdc34[S(all4)A] | (S203A/S222A/S231A/S236A) | The present study |
| pCMV-hCdc34(S203) | (S222A/S231A/S236A) | The present study |
| pCMV-hCdc34(S222) | (S203A/S231A/S236A) | The present study |
| pCMV-hCdc34(S236) | (S203A/S222A/S231A) | The present study |
| pCMV-hCdc34(3Asp) | (S203D/S222D/S231D) | The present study |
| pET15b | N-terminal His6 tag, T7 promoter | Novagen |
| pET15b-hCdc34(Wt) | Human wild-type Cdc34 | The present study |
| pET15b-hCdc34(3Ala) | (S203A/S222A/S231A) | The present study |
| pET15b-hCdc34(3Asp) | (S203D/S222D/S231D) | The present study |
| pMS325 | ARS1/CEN4, LEU2, PNOP1, N-terminal FLAG tag, derivative of pNOPPATA1L | The present study |
| pMS325-hCdc34(Wt) | Human wild-type Cdc34 | The present study |
| pMS325-hCdc34(3Ala) | (S203A/S222A/S231A) | The present study |
| pMS325-hCdc34(3Asp) | (S203D/S222D/S231D) | The present study |
| pESC-TRP | 2μ, TRP1, PGAL1-10, N-terminal FLAG tag | Stratagene |
| pESC1 | Yeast wild-type Cdc34 in pESC-TRP | [23] |
| pESC1(1-252) | Truncation of C-terminal amino acids 253–295 | The present study |
| pESC1(1-200) | Truncation of C-terminal amino acids 201–295 | The present study |
| pESC1(S(all6)A) | (S207A/S216A/S263A/S268A/S282A/S292A) | The present study |
| pESC1(S207) | (S216A/S263A/S268A/S282A/S292A) | The present study |
| pESC1(S216) | (S207A/S263A/S268A/S282A/S292A) | The present study |
| pESC1(S263) | (S207A/S216A/S268A/S282A/S292A) | The present study |
| pESC1(S268) | (S207A/S216A/S263A/S282A/S292A) | The present study |
| pESC1(S282) | (S207A/S216A/S263A/S268A/S292A) | The present study |
| pESC1(S292) | (S207A/S216A/S263A/S268A/S282A) | The present study |
| pESC1(2Ala) | (S207A/S216A) | The present study |
| pESC1(2Asp) | (S207D/S216D) | The present study |
| pESC1(2Glu) | (S207E/S216E) | The present study |
| pET15b-yCdc34(Wt) | Yeast wild-type Cdc34 | The present study |
| pET15b-yCdc34(2Ala) | (S207A/S216A) | The present study |
| pET15b-yCdc34(2Asp) | (S207D/S216D) | The present study |
| pET15b-yCdc34(2Glu) | (S207E/S216E) | The present study |
| pET21(+)-yCdc34(F72V) | N-terminal His6 tag, T7 promoter, (F72V) | [24] |
| pRS415 | CEN6, LEU2 | Stratagene |
| pMS384 | Yeast CDC34 promoter in pRS415 | The present study |
| pMS384-yCdc34(Wt) | Yeast wild-type Cdc34 | The present study |
| pMS384-yCdc34(2Ala) | (S207A/S216A) | The present study |
| pMS384-yCdc34(2Asp) | (S207D/S216D) | The present study |
| pMS384-yCdc34(2Glu) | (S207E/S216E) | The present study |
| pMS384-yCdc34(F72V) | (F72V) | The present study |
| pRSETb | N-terminal His6 tag, T7 promoter | Invitrogen |
| pRSETb-Sic1 | Sic1 | The present study |
Yeast strains and cultures
The S. cerevisiae strains used in the present study are derivatives of YES71 (MATα, ura3-52, trp1Δ63, leu2Δ1, his3Δ, cdc34-2::HIS3 pURA3-yCDC34) [25] and YMS034 (MATa ura3-1 trp1-1 ade2-1 leu2-3, 112 his3-11, 15 can1-100, cdc34::kanMX4 pYX212-CDC34). Strain YMS034 is a derivative of W303-1A and was generated by gene disruption of the CDC34 ORF by a standard PCR-based strategy. Yeast cells were grown and manipulated by established procedures. Derivatives of YES71 and YMS034 cells expressing yeast or human wild-type Cdc34 or the phosphorylation site mutants were generated by counter-selection on medium containing 5-FOA (5-fluoro-orotic acid) [26]. For growth analysis, fresh cultures of cells were grown to an D600 (attenuance) of 0.8 overnight in YPD medium [1% (w/v) yeast extract, 2% (w/v) peptone and 2% (w/v) glucose] at 30 °C. Serial 10-fold dilutions were spotted on to YPD plates in the presence or absence of cycloheximide (0.75–1.25 μg/ml) and incubated at 30 °C for 24–48 h.
Cell cycle analysis by flow cytometry
Fresh cultures of YMS034 expressing wild-type yCdc34 or the phosphorylation site mutants 2Ala, 2Asp and 2Glu were diluted into YPD medium and grown to a D600 of 0.2 at 30 °C before cells were arrested in G1 with 10 μM α-factor for 120 min at 25 °C. To release the cultures from arrest, cells were washed twice with YPD and resuspended in fresh YPD. At the time points indicated, 1 ml aliquots were taken, the cells centrifuged (13000 g for 1 min at room temperature) and fixed in 500 μl of 70% (v/v) ethanol at 4 °C overnight. Fixed cells were centrifuged (13000 g for 1 min at room temperature), washed with 1 ml of FACS buffer (50 mM Tris/HCl, pH 7.5, 150 mM NaCl and 1 mM EDTA), centrifuged, resuspended in 100 μl of PBS containing 0.5 mg/ml RNase A (Sigma) and incubated for 120 min at 30 °C. After addition of 25 μg of proteinase K (Promega) and adjustment to 5 mM CaCl2, the cells were incubated for another 60 min at 42 °C, then washed with 1 ml of PBS, and the DNA stained overnight with 0.1 mg/ml propidium iodide (Molecular Probes) in 100 μl of PBS at 4 °C. Before flow cytometry analysis on a FACSCalibur™ (Becton Dickinson), the cells were diluted with 1 ml of PBS supplemented with 0.01 mg/ml propidium iodide and briefly sonicated. Histograms were generated with CellQuest software (Becton Dickinson).
Expression and purification of recombinant proteins
Escherichia coli BL21(DE3) pLysS cells containing pET15b constructs of human or yeast wild-type Cdc34 and their corresponding phosphosite mutants, pET21(+)-yCdc34(F72V) [24] or pRSETb-SIC1 were grown in LB (Luria–Bertani) medium containing 100 μg/ml ampicillin at 37 °C to a D600 of 0.8 and expression was induced by the addition of IPTG (isopropyl β-D-thiogalactoside) to 0.75 mM. After 3 h at 37 °C cells were harvested and lysed and proteins were purified by affinity chromatography on Ni-NTA (Ni2+-nitrilotriacetate)–agarose (Qiagen) according to the manufacturer's instructions.
To prepare FLAGSCFCdc4 E3 ligase, Sf21 insect cells (2.5×106/ml) were co-infected with recombinant baculoviruses encoding yeast Cdc4p (FLAG), Cdc53p (Myc), Rbx1, Skp1 [FLAG or GST (glutathione S-transferase)] at an MOI (multiplicity of infection) of 5 for 40 h. Immunopurification of FLAGSCFCdc4 complexes from Sf21 insect cell lysates with M2 anti-FLAG beads was performed essentially as described previously [8]. For ubiquitination reactions, FLAGSCFCdc4 complexes were eluted from M2 anti-FLAG beads with FLAG peptide (Sigma) according to the manufacturer's instructions.
In vivo [32P]Pi labelling of Cdc34
For in vivo 32P labelling of hCdc34, HEK-293 cells (human embryonic kidney 293 cells) were seeded at 1.6×106 cells/10 cm dish and transiently transfected with 15 μg of plasmid DNA of either pCMV-Tag2 vector, pCMV-hCDC34 or the following pCMV-hCDC34(S222), pCMV-hCDC34(S236) using FuGENE™ 6 (Boehringer Mannheim, Germany). [32P]Pi labelling, preparation of lysates, immunoprecipitation and phospho amino acid analysis of hCdc34 were performed as described previously [27]. For in vivo 32P labelling of yCdc34, yeast strain W303-1B was transformed with either empty vector (pESC-TRP1, Stratagene), pESC1-Cdc34(Wt) or the mutant derivatives pESC1-Cdc34(1-253), pESC1-Cdc34(1-200), pESC1-Cdc34[S(all6)A], pESC1-Cdc34(S207), pESC1-Cdc34(S216), pESC1-Cdc34(S263), pESC1-Cdc34(S268), pESC1-Cdc34(S282), or pESC1-Cdc34(S292). Then 50 ml cultures (on synthetic complete medium lacking tryptophan) were grown to a D600 of 0.8, the cells were pelleted by centrifugation (10000 g for 1 min at 4 °C) and washed twice in PO4-free YPD medium. Cells were resuspended in 1 ml of PO4-free YPD medium containing 0.75 mCi of [32P]Pi and incubated for 60 min at 30 °C. After washing with cold PBS, cells were lysed in 1 ml of Y-PER lysis buffer (Pierce) supplemented with 1 mM DTT (dithiothreitol), 2 mM PMSF, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 10 μg/ml pepstatin, 30 mM NaF, 20 mM Na PPi (sodium pyrophosphate), 10 mM 2-glycerophosphate and 1 mM vanadate for 10 min. After centrifugation for 20 min at 13000 g, supernatants were subjected to three consecutive rounds of immunoprecipitation with 10 μl of anti-FLAG M2 beads (Sigma) for 90 min at 4 °C each. The beads were combined, washed five times with RIPA buffer [20 mM Tris/HCl, pH 7.6, 300 mM NaCl, 2 mM EDTA, 1% (w/v) Triton X-100, 1% (w/v) sodium deoxycholate and 0.1% (w/v) SDS] supplemented with 1 mM DTT, 2 mM PMSF, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 10 μg/ml pepstatin, 30 mM NaF, 20 mM Na PPi, 10 mM 2-glycerophosphate and 1 mM vanadate and once with 20 mM Hepes (pH 7) and 1 mM DTT. Immunoprecipitates were separated by SDS/PAGE, and products were visualized by autoradiography or by immunoblotting with anti-Cdc34 antibody and ECL® detection (Amersham).
CK2-mediated phosphorylation of Cdc34
For CK2-mediated Cdc34 phosphorylation in vitro, hCdc34 (5 μM) or yCdc34 (10 μM) was mixed with 10, 25 or 100 units of CK2 (New England Biolabs) and incubated in CK2 reaction buffer (20 mM Tris/HCl, pH 7.5, 50 mM KCl and 10 mM MgCl2) supplemented with 100 μM ATP and 10 μCi of [γ-32P]ATP in a total volume of 15 μl for 30 min at 30 °C. Reactions were stopped with SDS-Laemmli buffer and heating at 95 °C. Proteins were separated by SDS/PAGE and visualized by autoradiography and Coomassie Blue staining. To generate phosphorylated Cdc34 for ubiquitination reactions, yCdc34 (15 μM) was incubated in reaction buffer supplemented with 500 units of CK2 and 2 mM ATP in a total volume of 50 μl for 90 min at 30 °C. To prepare mock-treated, unphosphorylated yCdc34, CK2 was omitted from the reaction mixture.
SCF-independent ubiquitin conjugation and SCFCdc4-mediated Sic1 ubiquitination
SCF-independent ubiquitin-conjugating activity of recombinant yCdc34 (1.6 μM) was measured with respect to the formation of ubiquitinated GST–ubiquitin, as described previously [28]. The reaction mixture (18 μl) contained Cdc34, 0.75 μg of GST–ubiquitin and 0.1 μg of purified E1 (Sigma) in ubiquitination buffer (20 mM Tris/HCl, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 2 mM ATP and 0.2 mM DTT). The reaction was performed for 60 min at 30 °C (∼5% of substrate was converted into ubiquitinated GST–ubiquitin) and terminated by adding SDS/PAGE loading buffer and heating for 2 min at 100 °C. After SDS/PAGE, reaction products were visualized by immunoblotting with anti-GST antibodies (Amersham) to detect GST–ubiquitin. Yeast Cdc34 was detected by probing with anti-Cdc34 antibodies.
To prepare phosphorylated Sic1 substrate for Cdc34–SCFCdc4 ubiquitination assays, purified GST–cyclin A–CDK2 complex [29] was immobilized on 10 μl of glutathione–agarose beads (Sigma), and the beads were washed five times with 1 ml of cold PBS and equilibrated in kinase buffer (50 mM Tris/HCl, pH 7.5, 50 mM NaCl, 10 mM MgCl2 and 0.5 mM DTT). To generate phosphorylated 32P-labelled Sic1, 5 μg of recombinant Sic1 and 100 μCi of [γ-32P]ATP were added, and the reaction mixture (40 μl) was incubated for 45 min at 30 °C. To saturate the phosphorylation sites on 32P-labelled Sic1, the kinase reaction was adjusted to 2 mM ATP in a total volume of 50 μl and incubated for another 30 min at 30 °C. After centrifugation (5000 g for 1 min at 4 °C), the supernatant containing phosphorylated 32P-labelled Sic1 was transferred to a fresh test tube.
Ubiquitination reactions (18 μl) contained ∼100 nM SCFCdc4, 80 nM yeast E1 (AG Scientific), 30 μM ubiquitin (Boston Biochem), ∼80 nM phosphorylated [32P]Sic1 and 1.6 μM yeast Cdc34 in UB buffer (50 mM Tris/HCl, pH 7.5, 50 mM potassium acetate, 2.5 mM magnesium acetate, 1 mM DTT and 2 mM ATP). CK2 activity was inhibited by addition of heparin (2 μM). After incubation for 10–60 min at 26 °C, reactions were stopped by boiling with SDS-Laemmli buffer, proteins were separated by SDS/PAGE and visualized by autoradiography.
Protein binding experiments
Binding reactions contained immobilized FLAG–SCFCdc4 on 4 μl anti-FLAG M2 beads, prepared from ∼1×106 Sf21 insect cells as described above. For mock-treated beads, lysates from of uninfected Sf21 insect cells (∼1×106) was employed. After washing five times with 1 ml of Sf21 lysis buffer [30] supplemented with 30 mM NaF, 20 mM Na PPi, 10 mM 2-glycerophosphate and 1 mM vanadate for 10 min, binding reactions were adjusted to 3 μM of recombinant yCdc34 or its derivatives in 75 μl of Sf21 lysis buffer and incubated at 4 °C for 90 min under constant agitation. Beads and associated proteins were then washed five times with 1 ml of Sf21 lysis buffer for 10 min prior to SDS/PAGE and immunoblotting with anti-His5 (His6Cdc34) and anti-Myc antibodies (MycCdc53).
Stability of Sic1 in vivo
Yeast strains (YMS034) were grown to D600 0.35 in YPD medium at 30 °C. Cells were arrested in G1-phase with 10 μM α-factor for 120 min at 25 °C and released by washing with fresh YPD. Samples were taken at the indicated times post release. Cell lysates were analysed by immunoblotting with an anti-Sic1 antibody and an anti-actin antibody to confirm even loading. For quantification, Sic1 levels were normalized according to actin levels and are expressed as remaining Sic1 relative to time point 0 min (100%).
RESULTS
yCdc34 and hCdc34 are phosphorylated on serine residues in the acidic C-terminal tail domain
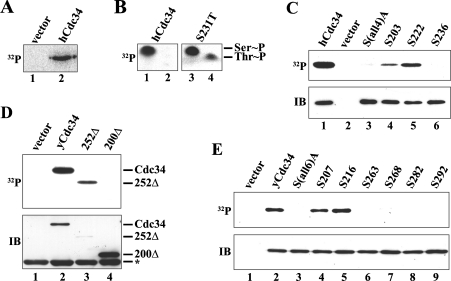
Previous studies in S. cerevisiae have shown that yCdc34 is phosphorylated exclusively on serine residues in vivo [10], whereas studies of hCdc34 suggest candidate serine and threonine phosphorylation sites in the C-terminus of this protein [21]. However, the exact identity of the phosphorylation sites of yCdc34 and hCdc34 as well as their functional importance remains to be defined. Therefore we first sought to identify the in vivo phosphorylation sites on hCdc34 and yCdc34. For hCdc34 phosphorylation analysis, HEK-293 cells were transfected with a FLAG–hCdc34 expression construct, metabolically labelled with [32P]Pi, and hCdc34 was immunoprecipitated with anti-FLAG beads. Autoradiography revealed that hCdc34 was readily phosphorylated in HEK-293 cells in vivo (Figure 1A) and phospho amino acid analysis revealed that phosphorylation occurred exclusively on serine residues (Figure 1B, left panel). Previous deletion studies have suggested that hCdc34 is phosphorylated within the last 36 amino acids of the C-terminus [21]. This region contains four serine residues at positions 203, 222, 231 and 236 (Figure 2), suggesting that they are candidate sites for phosphorylation. To map the phosphorylated serine residue(s) in vivo, we employed two strategies. In the first approach, serine residues 203, 222, 231 and 236 were individually changed to threonine and then analysed by metabolic [32P]Pi labelling and phospho amino acid analysis. The rationale of this approach is that replacement of a serine site that is phosphorylated in vivo with threonine, should yield a phosphothreonine. Replacement of Ser231 with threonine resulted in the detection of both phosphothreonine and phosphoserine (Figure 1B, right panel), indicating that Ser231 is an in vivo phosphorylation site and that hCdc34 was phosphorylated on at least one other serine residue. Replacement of the other serine residues at positions 203, 222 and 236 with threonine did not yield phosphothreonine (results not shown), most probably because only a serine residue at this/these site(s) can act as a phosphoacceptor site. To identify the other phosphorylated serine residue(s), we generated a mutant hCdc34 in which all four serine residues of the C-terminus (203, 222, 231 and 236) were changed to alanine [S(all4)A], to prevent phosphorylation on these sites. This mutant was then used to reintroduce a single serine residue at either site 203, 222 or 236 to generate the following mutants with only one potential phosphoacceptor site: S203 (S222A/S231A/S236A), S222 (S203A/S231A/S236A) and S236 (S203A/S222A/S231A; hereinafter referred to as 3Ala). In vivo 32P labelling of HEK-293 cells transfected with these expression constructs revealed that mutation of all four serine residues to alanine abolished phosphorylation of hCdc34 (Figure 1C, lane 3). Reintroduction of a single serine residue at position 203 or 222 restored phosphorylation of hCdc34 (Figure 1C, lanes 4 and 5), whereas a serine residue at position 236 was not phosphorylated (Figure 1C, lane 6). These results show that hCdc34 is phosphorylated on serine residues at positions 203, 222 and 231 in vivo.
Figure 1. Identification of the in vivo phosphorylation sites on hCdc34 and yCdc34.
(A) HEK-293 cells were transiently transfected with either pCMV-Tag2 (vector) or pCMV-Tag2-hCdc34(Wt) and labelled with [32P]Pi. Human Cdc34 was immunoprecipitated, separated by SDS/PAGE and visualized by autoradiography. (B) Wild-type 32P-labelled hCdc34 (left panel) or its mutant derivative S231T (right panel) were subjected to phospho amino acid analysis. The positions of phosphoserine (Ser∼P) and phosphothreonine (Thr∼P) are indicated. (C) HEK-293 cells were transiently transfected with either pCMV-Tag2 (vector) or pCMV-Tag2 constructs expressing wild-type hCdc34 or its mutant derivatives S(all4)A (S203A/S222A/S231A/S236A), S203 (S222A/S231A/S236A), S222 (S203A/S231A/S236A) and S236 (S203A/S222A/S231A) and subjected to the same treatment as in (A). After SDS/PAGE, proteins were visualized by autoradiography (32P) or by immunoblotting (IB) with an anti-hCdc34 antibody. (D) Yeast cells (W303-1A) were transformed with empty pESC-TRP (vector) or pESC1 constructs expressing either yeast wild-type Cdc34 (yCdc34) or its C-terminal truncation derivatives 252Δ (residues 1–252) and 200Δ (residues 1–200). After labelling with [32P]Pi, cells were lysed, and yCdc34 was immunoprecipitated, separated by SDS/PAGE and visualized by autoradiography (32P) or by immunoblotting (IB) with anti-FLAG antibody. The asterisk indicates the position of the light immunoglobulin chain of the anti-FLAG beads. (E) Yeast cells (W303-1A) were transformed with empty pESC-TRP (vector) or pESC1 constructs expressing either yeast wild-type Cdc34 (yCdc34) or its mutant derivatives S(all6)A (S207A/S216A/S263A/S268A/S282A/S292A), S207 (S216A/S263A/S268A/S282A/S292A), S216 (S207A/S263A/S268A/S282A/S292A), S263 (S207A/S216A/S268A/S282A/S292A), S268 (S207A/S216A/S263A/S282A/S292A), S282 (S207A/S216A/S263A/S268A/S292A) and S292 (S207A/S216A/S263A/S268A/S282A). Cells were subjected to metabolic 32P labelling as in (D), and proteins were visualized by autoradiography (32P) or by immunoblotting (IB) with an anti-Cdc34 antibody.
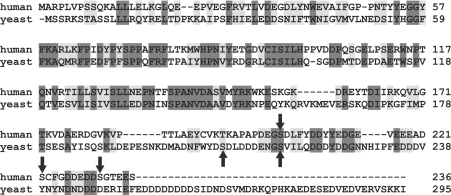
Figure 2. Sequence alignment of yCdc34 and hCdc34.
The amino acid sequences of hCdc34 and yeast Cdc34 were aligned using ClustalW (1.82). Identical residues are shaded in dark grey, conserved residues in light grey, and the in vivo phosphorylation sites are marked by arrows.
We also mapped the phosphorylation sites in yCdc34 to determine if phosphorylation of Cdc34 is evolutionarily conserved from yeast to human. Based on the localization of the phosphorylation sites in hCdc34, we rationalized that the yCdc34 phosphorylation sites, if conserved, may also be localized in the C-terminus. To evaluate this, we first generated two yeast Cdc34 C-terminal deletion constructs, in which either the last 43 (252Δ) or 95 (200Δ) amino acids were deleted. Yeast cells were transformed with plasmids expressing FLAG-tagged wild-type yCdc34, 252Δ or 200Δ, metabolically labelled with [32P]Pi, and yCdc34 was immunoprecipitated with anti-FLAG beads. Autoradiography demonstrated that wild-type yCdc34 and the 252Δ deletion mutant were readily phosphorylated in vivo (Figure 1D, lanes 2 and 3). However, no phosphorylation of the 200Δ deletion mutant was detectable (Figure 1D, lane 4), even though more 200Δ protein was present when compared with wild-type or 252Δ protein. Based on these results we concluded that the yeast Cdc34 phosphorylation sites are located within the C-terminal 95 amino acids and that amino acids 200–252 contain at least one phosphorylation site.
Previous work has shown that yCdc34 is phosphorylated exclusively on serine residues in vivo [10]. Yeast Cdc34 contains six serine residues in the C-terminal 95 amino acids, located at positions 207, 216, 263, 268, 282 and 292. To localize the specific phosphorylation sites, we employed the same strategy as with hCdc34. First site-directed mutagenesis was employed to mutate all six serine residues to alanine [S(all6)A] to prevent phosphorylation on these sites. This mutant was then used to reintroduce a single serine residue at either site 207, 216, 263, 268, 282 or 292 to determine by metabolic 32P labelling if this particular site is phosphorylated in vivo. As expected, wild-type yCdc34 was readily phosphorylated, whereas mutation of all six serine residues to alanine [S(all6)A] abolished phosphorylation (Figure 1E, lanes 2 and 3), confirming that phosphorylation occurs on one or more of these sites. Reintroduction of a single serine residue at position 207 or 216 restored phosphorylation of yCdc34, whereas a serine residue at position 263, 268, 282 or 292 was not phosphorylated (Figure 1E, lanes 6–9). Therefore yeast Cdc34 is phosphorylated on serine residues 207 and 216 in vivo.
Our results demonstrate that both yCdc34 and hCdc34 are phosphorylated within the acidic C-terminal tail domain, a region that is required for the cell cycle functions of both orthologues [31–33], suggesting evolutionary conservation of phosphorylation. Amino acid sequence alignment revealed that phosphorylation site Ser216 of yCdc34 is similar to phosphorylation site Ser203 of hCdc34 (Figure 2).
CK2 phosphorylates hCdc34 and yCdc34 on their in vivo phosphorylation sites
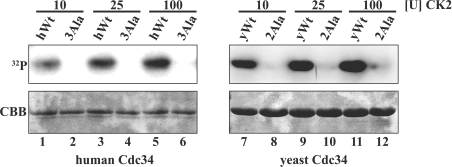
Previous work has implicated CK2 in phosphorylation of hCdc34 [21] and Ubc3B, which is highly homologous with hCdc34 [22]. CK2 is a highly conserved, ubiquitous and pleiotropic serine/threonine protein kinase that prefers aspartate and glutamate residues either N- or C-terminal to the phosphorylated site [34]. Examination of the amino acid sequence surrounding the phosphorylation sites of yCdc34 and hCdc34 indicates the presence of aspartate and glutamate residues (Figure 2). To determine if purified CK2 can phosphorylate hCdc34 and yCdc34, we performed kinase assays in vitro. As shown in Figure 3(A), CK2 efficiently phosphorylated both wild-type hCdc34 (left-hand panel) and yCdc34 (right-hand panel). Moreover, mutation of the phosphorylation sites to alanine residues abrogated the ability of CK2 to phosphorylate either human 3Ala or yeast 2Ala Cdc34s. Therefore CK2 specifically phosphorylates the in vivo phosphorylation sites of both hCdc34 and yCdc34, thus identifying CK2 as a candidate kinase of these enzymes.
Figure 3. CK2 phosphorylates the in vivo phosphorylation sites of hCdc34 and yCdc34.
Purified recombinant human wild-type (hWt, left-hand panel) and yeast wild-type Cdc34 (yWt, right-hand panel) and their respective alanine phosphorylation site mutants [3Ala (S203A/S222A/S231A) and 2Ala (S207A/S216A)] were incubated with increasing amounts of CK2 in the presence of [γ-32P]ATP in vitro. Proteins were separated by SDS/PAGE and visualized by autoradiography (32P) and by Coomassie staining (CBB).
CK2-mediated phosphorylation of yeast Cdc34 increases Cdc34–SCFCdc4-mediated Sic1 ubiquitination in vitro
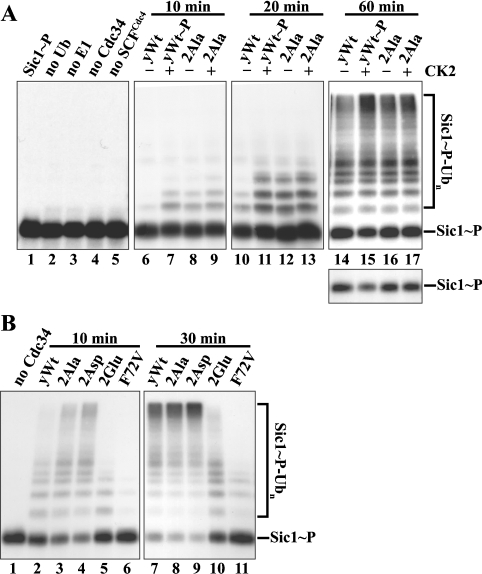
Cdc34 participates with the SCF E3 ligase to mediate substrate ubiquitination in vivo. Hence, CK2-mediated Cdc34 phosphorylation may affect Cdc34–SCF-mediated substrate ubiquitination. To test this possibility, we monitored yCdc34–SCFCdc4-mediated ubiquitination of a major physiological substrate of this complex, the budding yeast CDK inhibitor Sic1, in vitro. We purified yeast SCFCdc4 E3 complexes with anti-FLAG M2 beads from lysate of Sf21 insect cells co-infected with baculoviruses encoding the yeast FLAGCdc4, GSTSkp1, MycCdc53 and Roc1/Rbx1/Hrt1 subunits as described in the Materials and methods section. We compared the SCFCdc4-mediated Sic1 ubiquitination activities of unphosphorylated and CK2-phosphorylated wild-type yCdc34. As a control, and to confirm that any effects are specifically due to CK2-mediated phosphorylation of yCdc34 rather than addition of CK2, we evaluated the effect of CK2 on the ubiquitination activity of the yCdc34 2Ala phosphosite mutant, which is not phosphorylated by CK2 in vitro (Figure 3B). In addition, to eliminate any potential effects of CK2-mediated phosphorylation of other components of the assay such as SCFCdc4 subunits, the assay was performed in two stages. First, yCdc34 was phosphorylated by CK2, and then CK2 was inhibited by adding its known inhibitor, heparin [21]. The phosphorylated yCdc34 was then added to E1, SCFCdc4 and Sic1 to initiate ubiquitination. It was important to inhibit CK2 activity prior to initiation of the ubiquitination assay, since we found that CK2 can phosphorylate the SCFCdc4 subunits, Cdc4 and Skp1 in vitro (results not shown). We performed a time-course experiment in which Sic1 ubiquitination was monitored for 10, 20 and 60 min. As shown in Figure 4(A), phosphorylated, 32P-labelled Sic1 (Sic1 ∼ P, lane 1) was readily ubiquitinated in the presence of E1 ubiquitin-activating enzyme, ATP, ubiquitin, SCFCdc4 and yCdc34, as evidenced by a ladder of Sic1 bands, ranging from mono-ubiquitinated to higher molecular mass polyubiquitinated species (Sic1∼P-Ubn) migrating at the top of the SDS/PAGE gel (Figure 4A, lanes 6–17). Omission of individual components necessary for ubiquitination from the reaction mix prevented Sic1 ubiquitination (lanes 2–5). When compared with unphosphorylated wild-type yCdc34 (lanes 6, 10 and 14), CK2-phosphorylated yCdc34 (lanes 7, 11 and 15) showed enhanced Sic1 ubiquitination at all time points tested, as observed by increased levels of ubiquitinated Sic1 conjugates. After 10 and 20 min of incubation, there was little Sic1 ubiquitination with unphosphorylated yCdc34 (lanes 6 and 10). Conversely, phosphorylated yCdc34 (lanes 7 and 11) showed increased levels of mono- and multi-ubiquitinated Sic1. Following 60 min of incubation, the levels of high-molecular-mass polyubiquitinated Sic1 conjugates migrating at the top of the SDS/PAGE gel were increased with phosphorylated yCdc34 when compared with unphosphorylated yCdc34 (lanes 14 and 15, upper panel). Consistent with an increased Sic1 ubiquitination activity of phosphorylated yCdc34, less unmodified Sic1 substrate remained after 60 min (lanes 14 and 15, lower panel). Conversely, when the phosphosite mutant 2Ala was analysed, there was no difference in Sic1 ubiquitination between untreated (lanes 8, 12 and 16) and CK2-treated (lanes 9, 13 and 17) yCdc34 at any of the time points tested (lanes 8, 9, 12, 13, 16 and 17). These results demonstrate that increased Cdc34–SCFCdc4-mediated ubiquitination of Sic1 is specifically due to CK2-mediated phosphorylation of yCdc34.
Figure 4. The phosphorylation sites of yeast Cdc34 are important for Sic1 ubiquitination in vitro.
(A) Phosphorylation of recombinant yCdc34 by CK2 enhances SCFCdc4-mediated ubiquitination of Sic1. To monitor Sic1 ubiquitination, SCFCdc4 complexes were supplemented with E1, ubiquitin, ATP, phosphorylated, 32P-labelled Sic1 (lane 1), and yeast wild-type Cdc34 (yWt), which was pre-incubated in the absence (–) or presence (+) of CK2. Prior to ubiquitination, CK2 was inhibited by 2 μM heparin. As controls, the phosphosite mutant 2Ala was subjected to the same treatment (lanes 8, 9, 12, 13, 16 and 17) and single components were omitted from the reaction (lanes 2–5). After incubation for 10, 20 or 60 min at 26 °C, proteins were separated by SDS/PAGE and visualized by autoradiography. The lower panel shows a shorter exposure of the unconjugated Sic1-P substrate remaining. (B) Mutation of the phosphorylation sites of recombinant yCdc34 alters SCFCdc4-mediated ubiquitination of Sic1. Yeast wild-type Cdc34 (yWt; lanes 2 and 7), the phosphosite mutants 2Ala (lanes 3 and 8), 2Asp (lanes 4 and 9), 2Glu (lanes 5 and 10) or the point mutant yCdc34(F72V) (lanes 6 and 11) were assayed as in (A), except that incubation times were 10 and 30 min. As a control, yCdc34 was omitted from the reaction (lane 1).
The phosphorylation sites of yeast Cdc34 are important for SCFCdc4-mediated ubiquitination of Sic1
Based on the above finding that phosphorylation of yCdc34 increases SCFCdc4-mediated Sic1 ubiquitination, we sought to further examine the importance of these residues by mutating them to other amino acids, including alanine, aspartate and glutamate. The rationale of this approach is that the negatively charged carboxy side groups of aspartate and glutamate at the phosphorylation sites may mimic the negative charge conferred on phosphorylation. Conversely, alanine contains a neutral side group and thus may mimic unphosphorylated yCdc34. As a negative control, we employed the yCdc34(F72V) mutant, which binds SCFCdc4 with approx. 100-fold higher affinity than wild-type yCdc34 [24] and is extensively impaired in SCFCdc4-mediated Sic1 ubiquitination in vitro (Figure 4B, lanes 6 and 11), as shown previously [24,35]. As evidenced by increased levels of high-molecular-mass polyubiquitinated Sic1 and reduced levels of unmodified Sic1 substrate, the phosphosite mutants 2Ala (lane 3) and in particular 2Asp (lane 4) were more active than wild-type yCdc34 (lane 2). Conversely, the phosphosite mutant 2Glu (lane 5) showed reduced Sic1 ubiquitination. Longer incubation of the reaction (30 min) demonstrated that the 2Glu mutant is able to form high-molecular-mass polyubiquitinated Sic1 conjugates (lane 10), but still not as efficiently as the other yCdc34 variants (lanes 7–9). These results demonstrate that residues Ser207 and Ser216 in the acidic tail domain of yCdc34 are important for Cdc34–SCFCdc4-mediated Sic1 ubiquitination in vitro. An interesting aspect of these results is that ubiquitination activity was not related to the nature of the charge at the phosphorylation site, but rather the nature of the amino acid. Therefore, although the presence of the negatively charged aspartate residue increased activity, the negatively charged glutamate residue decreased activity. Similarly, introduction of an alanine residue with a neutral side group increased activity. This suggests that the charge at the phosphorylation site is not the primary determinant of activity, but rather the conformation induced by phosphorylation or mutation of the phosphorylation site.
The intrinsic SCF-independent activity of yCdc34 is altered by mutation of the phosphorylation sites
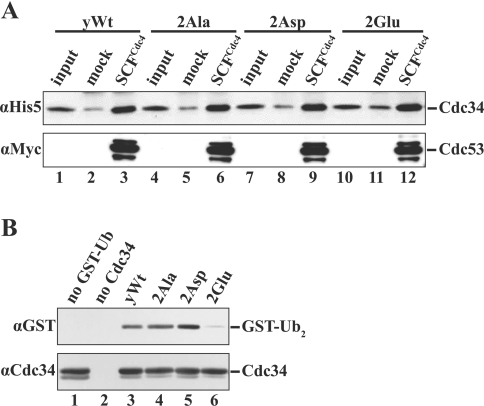
Cdc34 has two major catalytic roles in ubiquitination. First, Cdc34 accepts the activated ubiquitin from the ubiquitin-activating enzyme E1 in a transesterification reaction, and then conjugates ubiquitin to a substrate protein in association with the SCF ubiquitin ligase. Hence, phosphorylation of Cdc34 may regulate substrate ubiquitination by modulating Cdc34's association with the SCF complex. Alternatively, it is possible that phosphorylation may affect the intrinsic catalytic activity of Cdc34 independent of SCF, as has been described previously for the related ubiquitin-conjugating enzyme hHR6A [27]. To examine the first possibility, we investigated if phosphorylation or mutation of the phosphorylation sites affects yCdc34 association with the SCF complex in vitro. These studies revealed that unphosphorylated and CK2-phosphorylated wild-type yCdc34 as well as the phosphosite mutants 2Ala, 2Asp and 2Glu bound SCFCdc4 to a similar level (Figure 5A). These results strongly suggest that phosphorylation of yCdc34 does not alter its SCF-mediated ubiquitination activity through changes in its association with subunits of the SCF complex.
Figure 5. The phosphorylation sites of yeast Cdc34 regulate ubiquitin-conjugating activity independent of SCF E3 ligase in vitro.
(A) Mutation of the phosphorylation sites of recombinant yeast Cdc34 do not affect binding to SCFCdc4 in vitro. Protein-binding studies of yeast wild-type Cdc34 (yWt; lanes 1–3) and the phosphosite mutants 2Ala (lanes 4–6), 2Asp (lanes 7–9) and 2Glu (lanes 10–12) with either SCFCdc4 or mock-treated anti-FLAG beads were performed as described in the Materials and methods section. After extensive washing of the beads, proteins were separated by SDS/PAGE, and His6-tagged yCdc34 was visualized by immunoblotting with anti-His5 antibody (upper panel). As a control for SCFCdc4 loading, blots were reprobed with anti-Myc antibody to detect Myc-tagged Cdc53 (lower panel). (B) The catalytic activity of recombinant yeast wild-type Cdc34 (yWt; lane 3) and the phosphosite mutants 2Ala (lane 4), 2Asp (lane 5) and 2Glu (lane 6) was compared by monitoring the formation of mono-ubiquitinated GST–ubiquitin (GST–Ub2) in the absence of SCF E3 ligase in vitro, as described in the Materials and methods section. As controls, GST–Ub (lane 1) and Cdc34 (lane 2) were omitted from the reaction. Proteins were separated by SDS/PAGE and visualized by immunoblotting with antibodies directed against the GST tag (αGST) present on GST–Ub and yCdc34 (αCdc34) respectively.
To determine if phosphorylation affects yCdc34 activity independent of SCFCdc4, we compared the ubiquitin conjugation activities of the wild-type yCdc34 and the different phosphorylation site mutants in a simplified in vitro ubiquitination reaction, i.e. in the absence of the SCF E3 ligase and a bona fide substrate like the CDK inhibitor Sic1. Cdc34-mediated ubiquitin-conjugation activity was determined based on the formation of mono-ubiquitinated GST–ubiquitin (GST–Ub2), which has previously been used to measure Cdc34 activity in the absence of the SCF E3 ligase [28,35]. Studies of the different yeast Cdc34 phosphosite mutants revealed that mutation of the phosphorylation sites altered Cdc34 activity, as judged by their ability to form mono-ubiquitinated GST–ubiquitin (GST–Ub2). Therefore the 2Asp mutant displayed the highest activity, whereas the 2Glu mutant, displayed, the lowest activity (Figure 5B), correlating to the activity seen when assayed with SCFCdc4 (Figure 4B). The 2Ala Cdc34 mutant however displayed activity similar to wild-type yCdc34 (Figure 5B). These assays could not be performed with CK2-phosphorylated yCdc34, since addition of heparin to inhbit CK2 significantly impacted on ubiquitination activity. These results demonstrate that the ubiquitin-conjugating activity of yCdc34 is affected by mutation of the phosphorylation sites, independently of SCF. These results strongly suggest that alterations in SCF–Cdc34-mediated ubiquitination of Sic1 are not a consequence of changes in yCdc34's association with the E3 ligase but rather a consequence of changes in yCdc34's intrinsic catalytic activity.
Mutation of the yCdc34 phosphorylation sites affects cell cycle progression and Sic1 degradation in S. cerevisiae
The finding that the phosphorylation sites of yCdc34 are important for catalytic activity raised the question of whether phosphorylation is important for the cell cycle functions of this enzyme. To address this issue, we employed the budding yeast S. cerevisiae since Cdc34 is essential for viability and cell cycle functions in this organism and the effect of ectopically expressed yCdc34 can be assessed in the absence of endogenous yCdc34. To evaluate the importance of the yCdc34 phosphorylation sites, we assessed cell cycle progression in cells expressing either wild-type or the mutant yCdc34s where the phosphorylation sites were mutated to either alanine, aspartate or glutamate residues, as with the ubiquitination assays. Centromeric, low copy plasmids expressing either wild-type or mutant yCdc34 under the control of the CDC34 promoter (tester plasmid) were introduced into a yeast strain carrying a chromosomal deletion of the CDC34 gene. Since yCdc34 is essential for cell viability, this strain was kept viable by the presence of a plasmid-encoded wild-type CDC34 gene (maintenance plasmid). After the maintenance plasmid was shuffled out by selection on medium containing 5-FOA [26], the potential effect that mutation of the phosphorylation sites has on essential functions of yCdc34 was then determined in proliferation assays and cell cycle studies.
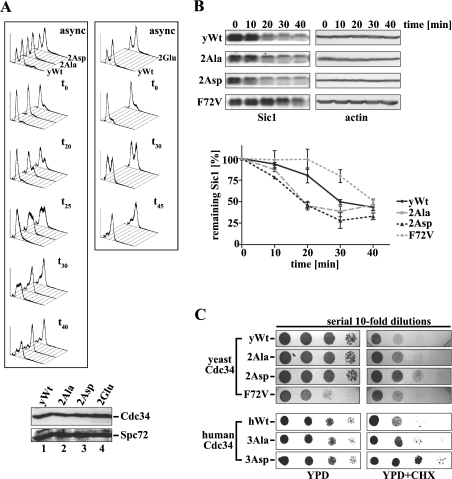
Counter-selection experiments with 5-FOA revealed that cells expressing wild-type or either of the Cdc34 phosphosite mutants were viable (results not shown). Since yCdc34 is critical in promoting G1–S-phase cell cycle progression in yeast, we sought to determine whether phosphorylation of yCdc34 is important for this function. For these studies, we performed a detailed cell cycle analysis of synchronized yeast cells progressing from G1 through S into the G2/M-phase of the cell cycle. Cells expressing either yeast wild-type Cdc34 or the phosphosite mutants were synchronized in G1 by arresting growth with α-factor, and then released into the cell cycle by incubating cells in fresh medium lacking the mating pheromone, a commonly used approach for cell cycle studies in yeast. Cell cycle progression was assessed by analysing DNA content by flow cytometry at different time points following α-factor removal. As shown in Figure 6(A), cells expressing the phosphosite mutants 2Ala and 2Asp progressed faster from G1 through S into the G2/M-phase than cells expressing wild-type yCdc34 (left-hand panel). The accelerated cell cycle progression of 2Ala and 2Asp was evident from 20 to 40 min following re-entry into the cell cycle. Hence, 20 and 25 min after α-factor withdrawal, a significant proportion of cells expressing the phosphosite mutants 2Ala or 2Asp had progressed from G1 through S into the G2/M-phase. Conversely, cells expressing wild-type yCdc34 were still predominantly in the G1- and S-phase. This effect was further evident 30 min after release into the cell cycle when almost all of the cells expressing 2Ala or 2Asp progressed into the G2/M-phase, whereas approx. 50% of cells expressing wild-type yCdc34 were still in the G1/S-phase. In contrast with cells expressing 2Ala and 2Asp, cells expressing the 2Glu phosphosite mutant displayed kinetics indistinguishable from cells expressing wild-type yCdc34 (Figure 6A, right-hand panel). These studies indicate that the phosphorylation sites of yCdc34 play an important role in regulating the kinetics of G1/S-phase cell cycle progression.
Figure 6. Mutation of the yCdc34 phosphorylation sites affects cell cycle progression and Sic1 turnover in vivo.
(A) Exponentially, asynchronously (async) growing yeast cells (YMS034) expressing wild-type yCdc34 (yWt) or the phosphosite mutants 2Ala, 2Asp (left-hand panel) and 2Glu (right-hand panel) were synchronized in G1-phase by α-factor arrest, released into the cell cycle by incubating in fresh medium, and aliquots were withdrawn at the time points indicated (t0=0 min). Cellular DNA was stained with propidium iodide and analysed by flow cytometry. The histograms are representative of at least three independent experiments. Equivalent expression levels of wild-type (lane 1) and mutant yCdc34s (lanes 2–4) were confirmed by immunoblotting of protein lysates of cells with anti-Cdc34 antibody (upper panel) and even loading was confirmed by immunoblotting with anti-Spc72 antibody (lower panel). (B) Yeast strains expressing either wild-type yCdc34 (yWt), 2Ala, 2Asp or F72V were synchronized in G1 of the cell cycle by α-factor arrest. At the indicated times after release, cell extracts were prepared and analysed by immunoblotting with Sic1 antibody (left-hand panel) and actin antibody as a loading control (right-hand panel). Sic1 levels of cells expressing wild-type yCdc34 (solid black line), F72V (dotted grey line) and the phosphosite mutants 2Ala (dashed grey line) and 2Asp (dotted black line) of three independent experiments were quantified (lower panel). The data points represent the means±S.D. (n=3). (C) Mutation of the phosphorylation sites confers increased resistance on cycloheximide (CHX). Serial 10-fold dilutions of yeast cells (YES71) containing either yeast wild-type Cdc34 (yWt) or the phosphosite mutants 2Ala, 2Asp and 2Glu or the point mutant yCdc34(F72V) were spotted on to YPD plates containing no (upper left panel) or 1.25 μg/ml CHX (upper right panel) and then incubated at 30 °C. Yeast cells containing human wild-type Cdc34 (hWt) or the phosphosite mutants 3Ala and 3Asp were subjected to the same analysis (lower panels) except that 0.75 μg/ml CHX was employed.
Since the cells expressing 2Ala and 2Asp yCdc34s displayed accelerated G1–S-phase cell cycle progression (Figure 6A) and increased Cdc34–SCFCdc4 activity towards Sic1 in vitro (Figure 4B), we next sought to determine if this was reflected by increased Sic1 turnover in vivo. Sic1 is stable in early G1 and degraded in a Cdc34-dependent manner as cells progress from G1-phase of the cell cycle into S-phase [13]. Therefore we analysed the rate of Sic1 degradation during the G1–S-phase transition by arresting cells in G1-phase with α-factor and then releasing cells into the cell cycle, as described above. Previous studies have shown that the levels of Sic1 mRNA transcript are constant during transition from G1 to the G2 cell cycle phase [36]. We therefore monitored and quantified the disappearance of Sic1 by immunoblotting (Figure 6B), which is reflective of its rate of ubiquitination and degradation during this cell cycle phase. As a control we assessed the yCdc34(F72V) mutant, which is severely impaired in SCF-mediated Sic1 ubiquitination activity in vitro (Figure 4B) and in vivo [35]. Consistent with previous studies [35], cells expressing the yCdc34(F72V) mutant were delayed in the destruction of Sic1 when compared with cells expressing wild-type yCdc34 (Figure 6B). Conversely, cells expressing the phosphosite mutants 2Ala and 2Asp showed accelerated degradation of Sic1 compared with wild-type cells. These results strongly suggest that the accelerated progression of cells expressing the phosphosite mutants 2Ala and 2Asp from G1 through S into G2/M-phase is due to their increased SCF-mediated ubiquitination and degradation of Sic1.
We also compared overall cell proliferation by drop test analysis. Since the 2Ala and 2Asp mutants displayed differences in cell cycle progression, we compared the growth of these mutants with wild-type cells. This revealed no major differences in the growth rate between cells expressing yeast wild-type Cdc34 and the phosphosite mutants 2Ala and 2Asp under optimal growth conditions (Figure 6C, upper left panel, YPD). Similarly, cells expressing human wild-type Cdc34 or the phosphosite mutants 3Ala and 3Asp displayed the same growth rate (Figure 6C, lower left panel, YPD). Interestingly, the yCdc34(F72V) mutant, which was severely impaired in SCFCdc4-mediated Sic1 ubiquitination in vitro (Figure 4B), displayed a modest slow growth phenotype (Figure 6C), indicating that a large alteration in Cdc34–SCFCdc4 activity is required for observable phenotypic alterations in the growth rate. We therefore sought to determine if the Cdc34 phosphosite mutants affected proliferation under more stringent growth conditions. A previous study has reported that ubiquitin levels are critical for the survival of S. cerevisiae cells in the presence of the translational inhibitor CHX (cycloheximide) and that CHX is a valuable tool to investigate conditional phenotypes in the ubiquitin–proteasome pathway [37]. For example, mutants with low levels of ubiquitin, e.g. caused by a defect in the ubiquitin recycling pathway, display hypersensitivity to CHX, whereas overexpression of ubiquitin or stabilization of ubiquitin in partial-loss-of-function proteasome mutants confers resistance to CHX. We therefore compared the proliferation of yeast cells expressing human or yeast wild-type Cdc34 with that of their corresponding phosphosite mutants in the presence of low concentrations of CHX. As shown in Figure 6(C), the aspartate phosphosite mutants of both yeast Cdc34 (2Asp, upper right panel) and human (3Asp, lower right panel) displayed resistance to CHX (YPD+CHX) when compared with their wild-type counterparts. The alanine phosphosite mutants of hCdc34 and yCdc34 also showed a slightly increased CHX resistance. These results indicate that mutation of the phosphorylation sites of Cdc34 to aspartate or alanine residues confers a growth advantage when ubiquitin levels are reduced [37].
DISCUSSION
In the present study, we identified the in vivo phosphorylation sites of both yCdc34 and hCdc34. Previous work has demonstrated that S. cerevisiae yCdc34 is phosphorylated on serine residue(s) in vivo [10]. We have now mapped the in vivo phosphorylation sites of yCdc34 to Ser207 and Ser216 in the acidic tail domain. Phosphorylation of hCdc34 is dependent on the C-terminal 36 amino acids [21]. These authors suggested that hCdc34 is phosphorylated on five sites (Ser203, Ser222, Ser231, Thr233 and Ser236) in this region, since mutation of these sites to alanine residues abolishes phosphorylation in vivo. Our studies now identify the in vivo phosphorylation sites of hCdc34 on serine residues at positions 203, 222 and 231. Amino acid sequence alignment revealed that phospho-Ser231 in hCdc34 is similar to phospho-Ser216 in yCdc34 (Figure 2), indicating that, at least in part, phosphorylation of Cdc34 is evolutionarily conserved and therefore likely to be functionally important. In support of this idea, a recent study using an evolutionary proteomics approach has demonstrated that the likelihood of phosphorylation of regulatory sites in substrates of the cAMP-dependent protein kinase is strongly correlated with their degree of evolutionary conservation [38]. The idea that Cdc34 phosphorylation is evolutionarily conserved is further supported by our finding that CK2 efficiently and specifically phosphorylates both hCdc34 and yCdc34 on their in vivo phosphorylation sites. CK2 is a highly conserved, ubiquitous and pleiotropic serine/threonine kinase and has been previously implicated in the phosphorylation of hCdc34 and a related human homologue termed Ubc3B [21,22]. The optimal CK2 consensus phosphorylation sequence, S/T-X-X-E/D, consists of either negatively charged side chains of acidic residues (either aspartic or glutamic acid) or phosphoserine or phosphothreonine at position n+3 C-terminal to the phosphoacceptor site (reviewed in [39]). Phosphorylation sites Ser231 of hCdc34 (S-G-T-E) and Ser207 of yCdc34 (S-D-L-D) conform to this consensus. Although sites Ser203 (S-D-L-F) and Ser222 (S-C-F-G) of hCdc34 and site Ser216 of yCdc34 (S-V-I-L) do not perfectly match this consensus, they contain sequence features characteristic of CK2 phosphorylation sites, including acidic residues upstream of the phosphoacceptor site or in position n+1, similar to phosphorylation sites of known CK2 substrates, e.g. EBV (Epstein–Barr virus) ZEBRA, Fragmin, NDPK A, RAD, ER and CUT proteins (reviewed in [40]). CK2 is known to be important for cell cycle progression in both yeast and mammalian cells through phosphorylation of several cell cycle regulators (reviewed in [40]). In S. cerevisiae CK2 is essential for viability [41], while temperature-sensitive alleles of CK2 cause cells to arrest in the G1- and G2/M-phases of the cell cycle at the non-permissive temperature [42]. In mammalian cells, inhibition of CK2 activity by antisense oligonucleotides or microinjection of anti-CK2 antibodies inhibited growth factor-induced proliferation of human fibroblasts, due to arrest in the G0/G1- and G1/Sphases of the cell cycle [39,43,44]. Our results now suggest that a substrate of CK2 in mediating cell cycle progression is Cdc34.
At a mechanistic level, CK2-mediated phosphorylation of Cdc34 may control the function of this enzyme at several levels in vivo, such as regulating its proteolytic turnover, subcellular localization, association with the SCF E3 ligase or its intrinsic catalytic activity. Yeast Cdc34 is a relatively stable protein [10] whose levels remain constant during the cell cycle [45,46], strongly suggesting that phosphorylation does not regulate its proteolytic turnover. A previous study has suggested that CK2-mediated phosphorylation of hCdc34 is important for its nuclear retention [21]. However, our subcellular fractionation studies demonstrate that there is no difference in the subcellular distribution of yeast wild-type Cdc34 and the phosphosite mutants 2Ala, 2Asp and 2Glu (results not shown). The reasons for this variation are unclear, but may be due to different mechanisms regulating yCdc34 and hCdc34. Alternatively, in addition to mutating the in vivo phosphorylation sites Ser203, Ser222 and Ser231, in the studies on hCdc34, Thr233 and Ser236 in the C-terminus were also mutated to alanine [21], which may affect hCdc34 subcellular distribution.
As outlined in the introduction, Cdc34–SCFCdc4-mediated ubiquitination is the rate-limiting step in the proteolysis of Sic1 (reviewed in [47]) and is critical for the G1–S-phase cell cycle transition in S. cerevisiae [7,13]. Yeast Cdc34 binds to the SCF complex through the Cdc53 and Rbx1/Hrt1/Roc1 subunits [8,30,45]. Region 171–209, which includes phosphosite 207 as well as Phe72 in the N-terminus in yCdc34 are important for binding to SCF [45] and Rbx1 [24]. Our in vitro binding studies showed that CK2-mediated phosphorylation or mutation of the phosphorylation sites did not alter Cdc34 binding to SCFCdc4, nor the individual subunits Cdc53 or Rbx1 in vitro (results not shown). This is consistent with studies with hCdc34, which demonstrate that acidic tail domain is dispensable for efficient binding of hCdc34 to the SCF subunits Cul1 and Roc1 [33]. Our in vitro studies indicate that CK2 increases Cdc34–SCFCdc4-mediated Sic1 ubiquitination by phosphorylation of yCdc34. This is further exemplified by studies with the phosphosite mutants. Hence, the yeast phosphosite mutants 2Ala and 2Asp showed increased SCFCdc4-mediated Sic1 ubiquitination activities compared with unphosphorylated wild-type and the phosphosite mutant 2Glu in vitro. When monitored in the absence of SCF ligase, these phosphosite mutants displayed similar changes in their intrinsic catalytic ubiquitin-conjugating activity. An interesting aspect of these results is that their activity did not correlate with the charge conferred by the introduced amino acid. Although the aspartate mutant yielded the highest activity, the glutamate mutant had lowest activity, even though both amino acids have negatively charged carboxy side groups, suggesting that the conformation of the amino acid in addition to the charge is important for regulating Cdc34 activity. Therefore phosphorylation of Cdc34 is likely to lead to conformational changes resulting in increased ubiquitination activity.
Structurally, Cdc34 belongs to the class II ubiquitin-conjugating enzyme family, which consists of a highly conserved N-terminal catalytic domain containing the active-site cysteine and an acidic C-terminal extension (tail domain). Numerous studies have demonstrated that the acidic tail domain of yCdc34 (region 171–295) is essential for viability of S. cerevisiae and for promoting cell cycle progression [31,32,45]. The importance of the acidic tail domain is further exemplified by studies demonstrating that transposition of the yCdc34 tail on to Rad6, an E2 ubiquitin-conjugating enzyme with distinct, non-overlapping functions, confers cell cycle function on Rad6 [31,48]. Similarly, the acidic tail domain of hCdc34 (region 201–236) is critical for complementation of the temperature-sensitive cdc34-2 mutation in S. cerevisiae and in vitro ubiquitination of p27Kip1 [33]. Recent work has shown that Cdc34–SCFCdc4-mediated ubiquitination of Sic1 can be separated into two steps, involving the attachment of the first ubiquitin (mono-ubiquitination), which is rate limiting, and subsequent attachment of further Lys48-linked ubiquitins (polyubiquitination), which is rapid [28]. Interestingly, this work demonstrated that an acidic loop region of yCdc34, comprising residues 103–114 is important for polyubiquitination through Lys48-linked ubiquitins. The authors suggest that the acidic loop plays an important conformational role by positioning Lys48 of Sic1-linked ubiquitin to attack the Cdc34–ubiquitin thiolester during polyubiquitination. Our studies now suggest that phosphorylation of the Cdc34 in the acidic tail domain may also regulate the conformation of ubiquitin-charged Cdc34 to bring it in a more favourable position for ubiquitination.
At a biological level, the importance of Cdc34 phosphorylation is exemplified by the cell cycle studies demonstrating that the yCdc34 2Ala or 2Asp mutants progressed with faster kinetics from G1 through S into the G2/M-phase of the cell cycle than cells expressing wild-type or 2Glu yCdc34. For example, at 25 min following α-factor release, approx. 50% of the cells expressing 2Ala and 2Asp yCdc34s progressed to G2/M-phase, while most of the wild-type and 2Glu-expressing cells were still in G1/S-phase (Figure 6A). The increased kinetics of cell cycle progression of cells expressing the phosphosite mutants 2Ala and 2Asp was associated with an increased rate of Sic1 degradation in vivo (Figure 6B), which correlated with increased SCF-mediated ubiquitination of Sic1 in vitro by these mutants (Figure 4B). Altogether, these results strongly suggest that phosphorylation of Cdc34 is important for regulating SCF-mediated activity in vivo. Previous studies have shown that phosphorylation of hCdc34 increases during the G0/G1–S-phase cell cycle transition [21]. Since the phosphosite mutants 2Ala and 2Asp exhibit increased basal activity compared with wild-type yCdc34, it is likely that cells expressing these mutants progress with faster kinetics since there is no lag in the time required for their stimulation by CK2-mediated phosphorylation following release from α-factor arrest. Interestingly, although the 2Glu mutant was less active than wild-type yCdc34 in SCFCdc4-mediated Sic1 ubiquitination in vitro, cells expressing this mutant displayed the same cell cycle kinetics and growth rate as those expressing wild-type yCdc34. This suggests that the reduction in activity observed for the 2Glu mutant is not rate limiting for cell cycle progression and proliferation and that a larger reduction in yCdc34 activity is required to reduce cell cycle progression. In support of this, the even more severe impairment in SCFCdc4-mediated Sic1 ubiquitination observed with the yCdc34(F72V) mutant in vitro (Figure 4B) [24,35] resulted in only a modest slow growth phenotype (Figure 6B) or was without any effect on proliferation [35], depending on the genetic background of the yeast strain. Previous work in S. cerevisiae has demonstrated that overexpression of different cell-cycle-regulatory genes can induce differential effects on cell cycle phase distribution, ranging from movement of only 3% to >90% of the population to a specific cell cycle stage [49]. In addition, other studies have shown that deletion of cell cycle regulators such as Mdt1 causes a G2/M-phase cell cycle delay without effects on the overall rate of cell proliferation, as evaluated by drop test analysis [1]. Therefore different cell cycle regulators and alterations in their activity can induce variable magnitudes of effects on the cell cycle and proliferation depending on their specific role. Our studies evaluating proliferation by drop test analysis revealed that cells expressing the aspartic acid or alanine phosphosite mutants of both yCdc34 and hCdc34 showed enhanced cell growth under conditions when ubiquitin levels were reduced (presence of cycloheximide) [37]. A correlation between Cdc34 activity and ubiquitin levels has previously been implied by a study demonstrating that overexpression of ubiquitin suppresses the growth defect associated with several different mutant alleles of yeast Cdc34, including cdc34-2 [50]. These results further suggest that mutation of the phosphorylation sites to either alanine or aspartate residues renders Cdc34 more efficient in mediating cell proliferation than wild-type Cdc34. In conclusion, these studies demonstrate that CK2-mediated phosphorylation of Cdc34 is important for regulating the catalytic activity and cell cycle functions of this enzyme.
Acknowledgments
We thank W. Harper (Department of Pathology, Harvard Medical School, Boston, MA, U.S.A.) for the gift of baculoviruses encoding the yeast SCF subunits, M. Goebl (Department of Biochemistry and Molecular Biology, Indiana University School of Medicine, Indianapolis, IN, U.S.A.), D. Skowyra (Edward A. Daisy Department of Biochemistry and Molecular Biology, St Louis University School of Medicine, St Louis, MO, U.S.A.) and M. Ellison (Department of Biochemistry, Institute of Biomolecular Design, University of Alberta, Edmonton, Alberta, Canada) for the gifts of yeast strains, antibodies and plasmids, and A. Hammet and J. Heierhorst (St Vincent Institute of Medical Research, Fitzroy, Melbourne, Victoria, Australia) for critical comments on this paper. M. S. was a recipient of a postdoctoral fellowship (SA1335) provided by the German Research Foundation (Deutsche Forschungsgemeinschaft). This research was supported by grants from the Association for International Cancer Research and in part by the U.S. Department of Defense Breast Cancer Research Program (DAMD17-03-1-0667) and the Multiple Myeloma Research Foundation.
References
- 1.Pickart C., Eddins M. Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 2.Thrower J. S., Hoffman L., Rechsteiner M., Pickart C. M. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun L., Chen Z. J. The novel functions of ubiquitination in signaling. Curr. Opin. Cell Biol. 2004;16:119–126. doi: 10.1016/j.ceb.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Pickart C. M., Fushman D. Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Petroski M., Deshaies R. Function and regulation of cullin–RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2005;6:8–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 6.Bai C., Sen P., Hofmann K., Ma L., Goebl M., Harper J. W., Elledge S. J. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 7.Feldman R., Correll C., Kaplan K., Deshaies R. A complex of Cdc4p, Skp1p, and Cdc53p/Cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 8.Skowyra D., Craig K., Tyers M., Elledge S., Harper J. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 9.Goebl M. G., Yochem J., Jentsch S., McGrath J. P., Varshasky A., Byers B. The yeast cell cycle gene CDC34 encodes a ubiquitin-conjugating enzyme. Science. 1988;241:1331–1335. doi: 10.1126/science.2842867. [DOI] [PubMed] [Google Scholar]
- 10.Goebl M. G., Goetsch L., Byers B. The Ubc3 (Cdc34) ubiquitin-conjugating enzyme is ubiquitinated and phosphorylated in vivo. Mol. Cell. Biol. 1994;14:3022–3029. doi: 10.1128/mcb.14.5.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butz N., Ruetz S., Natt F., Hall J., Weiler J., Mestan J., Ducarre M., Grossenbacher R., Hauser P., Kempf D., Hofmann F. The human ubiquitin-conjugating enzyme Cdc34 controls cellular proliferation through regulation of p27(Kip1) levels. Exp. Cell Res. 2005;303:482–493. doi: 10.1016/j.yexcr.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 12.Plon S. E., Leppig K. A., Do H. N., Groudine M. Cloning of the homolog of the CDC34 cell cycle gene by complementation in yeast. Proc. Natl. Acad. Sci. U.S.A. 1993;90:10484–10488. doi: 10.1073/pnas.90.22.10484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwob E., Bohm T., Mendenhall M., Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell. 1994;79:181–184. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 14.Orlicky S., Tang X., Willems A., Tyers M., Sicheri F. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell. 2003;112:243–256. doi: 10.1016/s0092-8674(03)00034-5. [DOI] [PubMed] [Google Scholar]
- 15.Kaiser P., Flick K., Wittenberg C., Reed S. Regulation of transcription by ubiquitination without proteolysis: Cdc34/SCF(Met30)-mediated inactivation of the transcription factor Met4. Cell. 2000;102:303–314. doi: 10.1016/s0092-8674(00)00036-2. [DOI] [PubMed] [Google Scholar]
- 16.McMillan J. N., Theesfeld C. L., Harrison J. C., Bardes E. S., Lew D. J. Determinants of Swe1p degradation in Saccharomyces cerevisiae. Mol. Biol. Cell. 2002;13:3560–3575. doi: 10.1091/mbc.E02-05-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reymond F., Wirbelauer C., Krek W. Association of human ubiquitin-conjugating enzyme CDC34 with the mitotic spindle in anaphase. J. Cell Sci. 2000;113:1687–1694. doi: 10.1242/jcs.113.10.1687. [DOI] [PubMed] [Google Scholar]
- 18.Patton E. E., Willems A. R., Tyers M. Combinatorial control in ubiquitin-dependent proteolysis: don't Skp the F-box hypothesis. Trends Genet. 1998;14:236–243. doi: 10.1016/s0168-9525(98)01473-5. [DOI] [PubMed] [Google Scholar]
- 19.Kawakami T., Chiba T., Suzuki T., Iwai K., Yamanaka K., Minato N., Suzuki H., Shimbara N., Hidaka Y., Osaka F., et al. NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. 2001;20:4003–4012. doi: 10.1093/emboj/20.15.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim Y.-S., Lee J.-Y., Son M.-Y., Park W., Bae Y.-S. Phosphorylation of threonine 10 on CKBBP1/SAG/ROC2/Rbx2 by protein kinase CKII promotes the degradation of IκBα and p27Kip1. J. Biol. Chem. 2003;278:28462–28469. doi: 10.1074/jbc.M302584200. [DOI] [PubMed] [Google Scholar]
- 21.Block K., Boyer T., Yew P. Phosphorylation of the human ubiquitin-conjugating enzyme, Cdc34, by casein kinase 2. J. Biol. Chem. 2001;276:41049–41058. doi: 10.1074/jbc.M106453200. [DOI] [PubMed] [Google Scholar]
- 22.Semplici F., Meggio F., Pinna L., Oliviero S. CK2-dependent phosphorylation of the E2 ubiquitin conjugating enzyme UBC3B induces its interaction with β-TrCP and enhances β-catenin degradation. Oncogene. 2002;21:3978–3987. doi: 10.1038/sj.onc.1205574. [DOI] [PubMed] [Google Scholar]
- 23.Varelas X., Ptak C., Ellison M. Cdc34 self-association is facilitated by ubiquitin-thiolester formation and is required for its catalytic activity. Mol. Cell. Biol. 2003;23:5388–5400. doi: 10.1128/MCB.23.15.5388-5400.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deffenbaugh A., Scaglione K., Zhang L., Moore J., Buranda T., Sklar L., Skowyra D. Release of ubiquitin-charged Cdc34-S-Ub from the RING domain is essential for ubiquitination of the SCFCdc4-bound substrate Sic1. Cell. 2003;114:611–622. doi: 10.1016/s0092-8674(03)00641-x. [DOI] [PubMed] [Google Scholar]
- 25.Prendergast J. A., Ptak C., Kornitzer D., Steussy C. N., Hodgins R., Goebl M., Ellison M. J. Identification of a positive regulator of the cell cycle ubiquitin-conjugating enzyme Cdc34 (Ubc3) Mol. Cell. Biol. 1996;16:677–684. doi: 10.1128/mcb.16.2.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boeke J. D., Trueheart J., Natsoulis G., Fink G. R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 27.Sarcevic B., Mawson A., Baker R. T., Sutherland R. L. Regulation of the ubiquitin-conjugating enzyme hHR6A by CDK-mediated phosphorylation. EMBO J. 2002;21:2009–2018. doi: 10.1093/emboj/21.8.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petroski M. D., Deshaies R. J. Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin–RING ubiquitin-ligase complex SCF–Cdc34. Cell. 2005;123:1107–1120. doi: 10.1016/j.cell.2005.09.033. [DOI] [PubMed] [Google Scholar]
- 29.Sarcevic B., Lilischkis R., Sutherland R. L. Differential phosphorylation of T-47D human breast cancer cell substrates by D1-, D3-, E-, and A-type cyclin/CDK complexes. J. Biol. Chem. 1997;272:33327–33337. doi: 10.1074/jbc.272.52.33327. [DOI] [PubMed] [Google Scholar]
- 30.Skowyra D., Koepp D., Kamura T., Conrad M., Conaway R., Conaway J., Elledge S., Harper J. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science. 1999;284:662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- 31.Kolman C., Toth J., Gonda D. Identification of a portable determinant of cell cycle function within the carboxyl-terminal domain of the yeast CDC34 (UBC3) ubiquitin conjugating (E2) enzyme. EMBO J. 1992;11:3081–3090. doi: 10.1002/j.1460-2075.1992.tb05380.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ptak C., Prendergast J., Hodgins R., Kay C., Chau V., Ellison M. Functional and physical characterization of the cell cycle ubiquitin-conjugating enzyme CDC34 (UBC3). Identification of a functional determinant within the tail that facilitates CDC34 self-association. EMBO J. 1994;269:26539–26545. [PubMed] [Google Scholar]
- 33.Block K., Appikonda S., Lin H., Bloom J., Pagano M., Yew P. The acidic tail domain of human Cdc34 is required for p27Kip1 ubiquitination and complementation of a cdc34 temperature sensitive yeast. Cell Cycle. 2005;4:1421–1427. doi: 10.4161/cc.4.10.2054. [DOI] [PubMed] [Google Scholar]
- 34.Songyang Z., Lu K., Kwon Y., Tsai L., Filhol O., Cochet C., Brickey D., Soderling T., Bartleson C., Graves D., et al. A structural basis for substrate specificities of protein Ser/Thr kinases: primary sequence preference of casein kinase I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol. Cell. Biol. 1996;16:6486–6493. doi: 10.1128/mcb.16.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petroski M. D., Kleiger G., Deshaies R. J. Evaluation of a diffusion-driven mechanism for substrate ubiquitination by the SCF–Cdc34 ubiquitin ligase complex. Mol. Cell. 2006;24:523–534. doi: 10.1016/j.molcel.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 36.Knapp D., Bhoite L., Stillman D. J., Nasmyth K. The transcription factor Swi5 regulates expression of the cyclin kinase inhibitor p40SIC1. Mol. Cell. Biol. 1996;16:5701–5707. doi: 10.1128/mcb.16.10.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanna J., Leggett D. S., Finley D. Ubiquitin depletion as a key mediator of toxicity by translational inhibitors. Mol. Cell. Biol. 2003;23:9251–9261. doi: 10.1128/MCB.23.24.9251-9261.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Budovskaya Y., Stephan J., Deminoff S., Herman P. An evolutionary proteomics approach identifies substrates of the cAMP-dependent protein kinase. Proc. Natl. Acad. Sci. U.S.A. 2005;102:13933–13938. doi: 10.1073/pnas.0501046102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olsten M. E., Litchfield D. W. Order or chaos?. An evaluation of the regulation of protein kinase CK2. Biochem. Cell. Biol. 2004;82:681–693. doi: 10.1139/o04-116. [DOI] [PubMed] [Google Scholar]
- 40.Meggio F., Pinna L. A. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 2003;17:349–368. doi: 10.1096/fj.02-0473rev. [DOI] [PubMed] [Google Scholar]
- 41.Padmanabha R., Wu J., Hanna D., Glover C. Isolation, sequencing, and disruption of the yeast CKA2 gene: casein kinase II is essential for viability in Saccharomyces cerevisiae. Mol. Cell. Biol. 1990;10:4089–4099. doi: 10.1128/mcb.10.8.4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanna D., Rethinaswamy A., Glover C. Casein kinase II is required for cell cycle progression during G1 and G2/M in Saccharomyces cerevisiae. J. Biol. Chem. 1995;270:25905–25914. doi: 10.1074/jbc.270.43.25905. [DOI] [PubMed] [Google Scholar]
- 43.Pepperkok R., Lorenz P., Jakobi R., Pyerin W. Cell growth stimulation by EGF: inhibition through antisense-oligodeoxynucleotides demonstrates important role of casein kinase II. Exp. Cell Res. 1991;197:245–253. doi: 10.1016/0014-4827(91)90429-x. [DOI] [PubMed] [Google Scholar]
- 44.Lorenz P., Pepperkok R., Ansorge W., Pyerin W. Cell biological studies with monoclonal and polyclonal antibodies against human casein kinase II subunit b demonstrate participation of the kinase in mitogenic signaling. J. Biol. Chem. 1993;268:2733–2739. [PubMed] [Google Scholar]
- 45.Mathias N., Steussy C., Goebl M. An essential domain within Cdc34p is required for binding to a complex containing Cdc4p and Cdc53p in Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:4040–4045. doi: 10.1074/jbc.273.7.4040. [DOI] [PubMed] [Google Scholar]
- 46.Galan J. M., Peter M. Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc. Natl. Acad. Sci. U.S.A. 1999;96:9124–9129. doi: 10.1073/pnas.96.16.9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Glickman M. H., Ciechanover A. The ubiquitin–proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 48.Silver E. T., Gwozd T. J., Ptak C., Goebl M., Ellison M. J. A chimeric ubiquitin conjugating enzyme that combines the cell cycle properties of CDC34 (UBC3) and the DNA repair properties of RAD6 (UBC2): implications for the structure, function and evolution of the E2s. EMBO J. 1992;11:3091–3098. doi: 10.1002/j.1460-2075.1992.tb05381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stevenson L. F., Kennedy B. K., Harlow E. A large-scale overexpression screen in Saccharomyces cerevisiae identifies previously uncharacterized cell cycle genes. Proc. Natl. Acad. Sci. U.S.A. 2001;98:3946–3951. doi: 10.1073/pnas.051013498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prendergast J. A., Ptak C., Arnason T. G., Ellison M. J. Increased ubiquitin expression suppresses the cell cycle defect associated with the yeast ubiquitin conjugating enzyme, CDC34 (UBC3). Evidence for a noncovalent interaction between CDC34 and ubiquitin. J. Biol. Chem. 1995;270:9347–9352. doi: 10.1074/jbc.270.16.9347. [DOI] [PubMed] [Google Scholar]