Abstract
IκB-ζ [inhibitor of NF-κB (nuclear factor κB) ζ] is a nuclear protein that is induced upon stimulation of TLRs (Toll-like receptors) and IL (interleukin)-1 receptor. IκB-ζ harbours C-terminal ankyrin repeats that interact with NF-κB. Our recent studies have shown that, upon stimulation, IκB-ζ is essential for the induction of a subset of inflammatory genes, represented by IL-6, whereas it inhibits the expression of TNF (tumour necrosis factor)-α. In the present study, we investigated mechanisms that determine the different functions of IκB-ζ. We found that co-expression of IκB-ζ and the NF-κB subunits synergistically activates transcription of the hBD-2 (human β-defensin 2) and NGAL (neutrophil gelatinase-associated lipocalin) genes, whereas it inhibits transcription of E-selectin. Reporter analyses indicated that, in addition to an NF-κB-binding site, a flanking C/EBP (CCAAT/enhancer-binding protein)-binding site in the promoters is essential for the IκB-ζ-mediated transcriptional activation. Using an artificial promoter consisting of the NF-κB- and C/EBP-binding sites, transcriptional activation was observed upon co-transfection with IκB-ζ and NF-κB, indicating that these sequences are minimal elements that confer the IκB-ζ-mediated transcriptional activation. Chromatin immunoprecipitation assays and knockdown experiments showed that both IκB-ζ and the NF-κB subunits were recruited to the NGAL promoter and were essential for the transcriptional activation of the hBD-2 and NGAL promoters on stimulation with IL-1β. The activation of the NGAL promoter by transfection of IκB-ζ and NF-κB was suppressed in C/EBPβ-depleted cells. Thus IκB-ζ acts as an essential transcriptional activator by forming a complex with NF-κB on promoters harbouring the NF-κB- and C/EBP-binding sites, upon stimulation of TLRs or IL-1 receptor.
Keywords: CCAAT/enhancer-binding protein (C/EBP), inflammation, inhibitor of nuclear factor κB ζ (IκB-ζ), innate immunity, nuclear factor κB (NF-κB), transcription
Abbreviations: AP-1, activator protein 1; C/EBP, CCAAT/enhancer-binding protein; ChIP, chromatin immunoprecipitation; CSF, colony-stimulating factor; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GST, glutathione S-transferase; hBD-2, human β-defensin 2; HEK-293, human embryonic kidney-293; IκB, inhibitor of nuclear factor κB; IκB-ζ(L), long-form IκB-ζ; IκB-ζ(S), short-form IκB-ζ; IL, interleukin; LPS, lipopolysaccharide; MBP, maltose-binding protein; NF-κB, nuclear factor-κB; NGAL, neutrophil gelatinase-associated lipocalin; RNAi, RNA interference; RT, reverse transcription; siRNA, small interfering RNA; TLR, Toll-like receptor; TNF, tumour necrosis factor
INTRODUCTION
Cells alter gene expression profiles in response to environmental changes. The expression of genes is regulated not only by initiation of transcription, but also by processing of transcripts, transport to the cytoplasm and translation of mRNA. Nevertheless, the initiation of transcription is the major step controlling the expression levels for most genes, and precise regulation of the activities of transcription factors is vital to accomplish specific tasks in cells that are essential to maintain life.
The transcription factor NF-κB (nuclear factor-κB) plays key roles in the innate and adaptive immune systems as well as in apoptosis, cell proliferation and differentiation [1–3]. It consists of homo- and hetero-dimers of five subunits, p65/RelA, RelB, c-Rel, p50/NF-κB1 and p52/NF-κB2. All subunits bind to DNA through the conserved Rel homology domain. The p65, RelB and c-Rel subunits contain a transcriptional activation domain at their C-terminal regions, which is lacking in p50 and p52. In resting cells, NF-κB localizes in the cytoplasm via association with cytosolic IκB (inhibitor of NF-κB) proteins such as IκB-α, -β and -ϵ. Upon stimulation, the cytosolic IκB proteins are phosphorylated and degraded by the ubiquitin–proteasome system. The liberated NF-κB dimer translocates into the nucleus, where it binds to its cognate κB enhancer elements and interacts with the basal transcription machinery or transcriptional coactivators to stimulate gene expression.
The activity of NF-κB is also modulated in the nucleus [1,2,4]. NF-κB activation, in turn, leads to transcription of IκB-α, and re-synthesized IκB-α binds to the nuclear NF-κB to export it to the cytoplasm, thereby attenuating its activity. Reversible acetylation of NF-κB regulates the duration of its action in the nucleus via modulation of its interaction with IκB-α [5]. In addition, phosphorylation of p65 allows its association with coactivators such as CBP [CREB (cAMP-response-element-binding protein)-binding protein]/p300, while unphosphorylated nuclear p65 associates with a histone deacetylase in the nucleus and is unable to bind to DNA [6]. As well as the coactivators or histone deacetylase, other nuclear proteins can interact with NF-κB and modulate its activity positively or negatively. The transcription factor Twist [7] and glucocorticoid receptor [8] interact directly with p65 and repress p65-mediated transcriptional activation. Reportedly, c-myc [9], p202a [10] and prostaglandins [11] also negatively regulate NF-κB in the nucleus. On the other hand, FBI-1 (factor that binds to inducer of short transcripts 1) [12] or TLS (translocated in liposarcoma) [13] have been reported to augment the transcriptional activity of nuclear NF-κB. Bcl-3 also binds to NF-κB in the nucleus, but its effects on the activity of NF-κB remain elusive and appear to differ for different target genes [14–16].
During screening for genes that are induced by pro-inflammatory stimuli, we identified a nuclear protein with ankyrin repeats that is strongly induced in response to bacterial LPS (lipopolysaccharide), which stimulates TLR (Toll-like receptor) 4 [17]. This protein, designated IκB-ζ, is barely detectable in resting cells, but is induced not only by LPS, but also by other microbial components that stimulate TLRs such as peptidoglycan, bacterial lipopeptide and CpG DNA [17–19]. The induction is also observed upon stimulation with IL (interleukin)-1β, but not by another pro-inflammatory cytokine, TNF (tumour necrosis factor)-α [17,20]. The protein IκB-ζ was also identified by other groups as MAIL (molecule possessing ankyrin repeats induced by LPS) [21] or INAP (IL-1-inducible nuclear ankyrin repeat protein) [22]. In a recent study, we showed that the stimulus specificity of IκB-ζ induction is determined at the post-transcriptional level, but not at the transcriptional level, on the basis of the finding that the stability of IκB-ζ mRNA is specifically up-regulated by stimulation with LPS or IL-1β, but not with TNF-α [20].
At least two major IκB-ζ variants are generated by alternative splicing [17,21]. We designated the longer form IκB-ζ(L) and the shorter form IκB-ζ(S), which lacks the N-terminal 99 amino acids of IκB-ζ(L). Both forms of IκB-ζ are induced upon stimulation, and IκB-ζ(L) is the predominant form in macrophages stimulated by LPS [20]. In contrast with the typical cytosolic and constitutively expressed IκB proteins, IκB-ζ is localized in the nucleus, where it preferentially binds, via the C-terminal ankyrin repeats, to the NF-κB p50 subunit rather than to the p65 subunit [17].
Although IκB-ζ was initially characterized as a negative regulator of NF-κB by reporter analyses [17], subsequent studies demonstrated that IκB-ζ also acts as a positive regulator of NF-κB [23]. Analyses using GAL4-fusion proteins of IκB-ζ revealed that its N-terminal region exhibits transcriptional activation activity, after association with the NF-κB p50 subunit. Overexpression of IκB-ζ augmented IL-6 production in response to LPS, but inhibited TNF-α production, indicating specific target gene activity of IκB-ζ. IκB-ζ-deficient mice exhibited chronic inflammation in the skin and on ocular surfaces [19,24], suggesting anti-inflammatory roles for IκB-ζ. However, prominent phenotypes of IκB-ζ-deficient cells were observed when the responses of macrophages following stimulation with LPS were analysed [19]. Peritoneal macrophages from IκB-ζ-deficient mice exhibited severe impairment in production of a subset of inflammatory genes, represented by IL-6, in response to LPS. The impaired induction of the genes was also observed upon stimulation with other microbial ligands for TLRs as well as IL-1β, which elicit the induction of IκB-ζ. This indicated that IκB-ζ is indispensable for the induction of the subset of inflammatory genes activated through TLR/IL-1 receptor signalling pathways. As expected from the preferential binding of IκB-ζ to the p50 subunit [17] and the role of p50 in mediating transcriptional activation via IκB-ζ [23], the genes whose induction was dependent on IκB-ζ were not induced in the p50 subunit (NF-κB1)-deficient cells [19]. Interestingly, IL-6 production in the IκB-ζ-deficient cells was not affected on stimulation with TNF-α, which does not induce IκB-ζ [19].
Thus IκB-ζ exhibits dual roles in NF-κB-mediated transcription, but the mechanism that determines the opposite actions of IκB-ζ on different genes remains to be clarified. In the present study, we analysed the molecular mechanisms of the IκB-ζmediated transcriptional activation. We focused on hBD-2 (human β-defensin 2) and NGAL (neutrophil gelatinase-associated lipocalin), both of which are preferentially induced by stimulators of TLR/IL-1 receptor signalling pathways, rather than by TNF-α [25–30]. We found that the promoters of both genes are synergistically activated by co-transfection of IκB-ζ and NF-κB. Promoter analyses of these genes identified a cis element that is critical for the observed induction. ChIP (chromatin immunoprecipitation) assays and RNAi (RNA interference) experiments indicated that IκB-ζ acts as an essential transcriptional activator by forming a complex with NF-κB on the promoters with an NF-κB-binding site and the cis element, upon stimulation of TLRs or IL-1 receptor.
EXPERIMENTAL
Cell culture, reagents and antibodies
The human embryonic kidney cell line HEK-293 and the human lung adenocarcinoma cell line A549 were cultured in DMEM (Dulbecco's modified Eagle's medium) containing 10% heat-inactivated fetal calf serum, 100 units/ml penicillin and 100 μg/ml streptomycin at 37 °C under 5% CO2. Mouse bone-marrow-derived macrophages were obtained by culturing femoral and tibial bone marrow from sex- and age-matched wild-type (C57BL/6) and IκB-ζ-deficient mice [19] in the same medium supplemented with L929 cell-conditioned medium as a source of macrophage CSF (colony-stimulating factor). LPS from Escherichia coli 0111:B4 was purchased from List Biological Laboratories. IL-1β and PMA were purchased from Genzyme Techne and Sigma–Aldrich respectively. Anti-(human IκB-ζ) polyclonal antibody was raised as follows. A cDNA fragment for amino acids 189–718 of human IκB-ζ was subcloned into pGEX-2T (GE Healthcare) or pMALg [17], and the recombinant protein was expressed in bacteria as a GST (glutathione S-transferase)- or a MBP (maltose-binding protein)-fusion protein. Antiserum was raised following immunization of a rabbit with the MBP-fusion protein and was subjected to affinity purification using Affi-Gel-10 resin (Bio-Rad Laboratories) coupled with the GST-fusion protein. Antibodies against the NF-κB p65 subunit (C-20) and the p50 subunit (NLS) were purchased from Santa Cruz Biotechnology.
Plasmids
Expression plasmids were constructed by subcloning a DNA fragment obtained by PCR into pcDNA3 (Invitrogen) with or without an N-terminal FLAG tag. Reporter plasmids for E-selectin, hBD-2 or NGAL were created by subcloning a human genomic fragment amplified by PCR into pGL3-basic vector (Promega). The mutations introduced into the hBD-2 promoter were as follows: TGGGGTTTCC (NF-κB-binding site 1) to TCGGGTTTCC; GGCATTTTCT (NF-κB-binding site 2) to GCCATTTTCTT; TTTGCATAAG [C/EBP (CCAAT/enhancer-binding protein)-binding site 1] to TAACCATAAG. The mutations in the NGAL promoter were as follows: GGGAATGTCC (NF-κB-binding site) to AATAATGTCC; ATTGCCTCAC (C/EBP-binding site 1) to AAACCCTCAC; GTGCAGAAAT (C/EBP-binding site 2) to GTGCAGGTTT; CTTGCCCAAT (C/EBP-binding site 3) to CAACCCCAAT (mutated nucleotides are underlined in all sequences). Plasmid pNF-κB-Luc harbouring four tandem NF-κB-binding sequences (GGGAATTTCC×4) was purchased from Clontech, and the phRL-TK internal control reporter plasmid expressing Renilla luciferase was from Promega. Plasmid phBD-2-κB-Luc was generated by insertion of a double-stranded oligonucleotide containing two copies of the sequence spanning from −206 to −187 bp of the hBD-2 promoter (5′-AGGGATTTTCTGGGGTTTCC-3′) into the NheI and BglII sites of pNF-κB-Luc, resulting in replacement of the tandem NF-κB-binding sequences. All mutated plasmids were sequenced to confirm correct sequences and orientation.
Quantification of mRNA by real-time RT (reverse transcription)–PCR
HEK-293 cells (5.0×105 cells) were transfected with the indicated expression plasmids by the calcium phosphate method [31]. At 24 h after transfection, total RNA was extracted using TRIzol® (Invitrogen) according to the manufacturer's instructions. Bone-marrow-derived macrophages were stimulated with 20 ng/ml LPS for the indicated periods, and total RNA was extracted. A 5 μg sample of the total RNA was reverse-transcribed using an oligo(dT) primer and the ReverTra AceR reverse transcriptase (Toyobo). Real-time RT–PCR was carried out with the reverse-transcribed cDNAs using SYBR Premix Ex Taq (Takara Bio) by a Light Cycler (Roche Diagnostics). The primers used were 5′-GCCATGAGGGTCTTGT-3′ and 5′-AGCCCTTTCTGAATCCG-3′ for hBD-2, 5′-CAGTTCCGGGAAAGATCA-3′ and 5′-ATGGTGCTAATGTCAGG-3′ for human E-selectin, 5′-AGCGACGAGTACAAGATCCG-3′ and 5′-AGCTGCTCCACCTTCTTCTG-3′ for human C/EBPβ, 5′-GCGAGCGCAACAACATC-3′ and 5′-CGACAGCTCCACCAACTTCT-3′ for human C/EBPδ, 5′-TCGGAGTCAACGGATTT-3′ and 5′-CCACGACGTACTCAGC-3′ for human GAPDH (glyceraldehyde-3-phosphate dehydrogenase), 5′-CGTTTCACCCGCTTTG-3′ and 5′-AATAAGAAGAGGCTCCAG-3′ for mouse NGAL, and 5′-GATGACCCAGATCATGTTTGA-3′ and 5′-GGAGAGCATAGCCCTCGTAG-3′ for mouse β-actin.
Luciferase reporter assay
HEK-293 cells (1.25×105 cells) were transfected with 100 ng of the indicated reporter plasmid together with 5 ng of the expression plasmid for the NF-κB subunit p65 or p50, 100 ng of the IκB-ζ(L) expression plasmid and 2.5 ng of the internal control plasmid phRL-TK using Lipofectamine™ 2000 (Invitrogen) according to the manufacturer's protocol. A549 cells (1.25×105 cells) were transfected with 100 ng of the indicated reporter plasmid together with 10 ng of the expression plasmid for the NF-κB subunit p65 or p50, 10 ng of the IκB-ζ expression plasmid and 2.5 ng of the internal control plasmid phRL-TK using Lipofectamine™ 2000. Cells were lysed 24 h after transfection. To analyse the responses to IL-1β stimulation, A549 cells (1.25×105 cells) were transfected with 200 ng of the hBD-2 reporter plasmids or 50 ng of the NGAL reporter plasmids, together with 2.5 ng of the internal control plasmid phRL-TK. At 24 h after transfection, cells were stimulated with 1 ng/ml IL-1β for 5 h, and the cells were then lysed. The luciferase activity was measured using the dualluciferase reporter system (Promega) according to the manufacturer's instructions. The transfection efficiency was normalized by the Renilla luciferase activity derived from phRL-TK. The data shown are the means±S.E.M. for duplicate samples, representative of at least two independent experiments.
ChIP assay
The ChIP assay was performed according to a described protocol (Upstate Cell Signaling Solutions) with some modifications. A549 cells (5×106 cells) were stimulated with 1 ng/ml IL-1β for 4 h, and fixed with 1% formaldehyde for 10 min at room temperature (25 °C). Cells were washed twice with ice-cold PBS, collected by centrifugation at 500 g for 5 min, and resuspended in the lysis buffer containing 50 mM Tris/HCl (pH 8.1), 1% SDS and 10 mM EDTA supplemented with 1× Complete™ protease inhibitor cocktail (Roche Applied Science) on ice for 20 min. The cell lysate was sonicated 10 times for 30 s with 1 min intervals using a Bioruptor (Cosmo Bio). Debris was removed by centrifugation at 15000 g for 10 min, and the supernatant was diluted 10 times with the buffer containing 16.7 mM Tris/HCl (pH 8.1), 0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 167 mM NaCl, supplemented with 1× Complete™. Extracts were pre-cleared for 1 h with 80 μl of Protein A–Sepharose 4 Fast Flow (GE Healthcare) 50% slurry containing 0.2 mg/ml yeast tRNA (Ambion). Immunoprecipitation was performed at 4 °C overnight with 2 μg each of specific antibodies. Immune complexes were collected with 50 μl of Protein A–Sepharose 4 Fast Flow 50% slurry with yeast tRNA. Sepharose beads were washed sequentially in the following buffers: low-salt wash buffer (20 mM Tris/HCl, pH 8.1, containing 150 mM NaCl, 0.1% SDS, 1% Triton X-100 and 2 mM EDTA), high-salt wash buffer (the same buffer, but with 500 mM NaCl), LiCl wash buffer (10 mM Tris/HCl, pH 8.1, 0.25 M LiCl, 1% sodium deoxycholate, 1% Triton X-100 and 1 mM EDTA), and twice with Tris/EDTA buffer (10 mM Tris/HCl, pH 8.0, and 1 mM EDTA). Immune complexes were extracted from the beads with 1% SDS and 0.1 M NaHCO3, and cross-linking was reversed by heating at 65 °C overnight. Proteins were digested with proteinase K at 50 °C for 1 h, and DNA was purified by phenol/chloroform extraction and precipitated with ethanol. The immunoprecipitated DNA thus obtained was used for quantitative real-time PCR. The primers used were 5′-TTCTTTCCTTTCTGTGGGTGTG-3′ and 5′-CAACTCCTGCGGAAACACTT-3′ for the κB site in the NGAL promoter, 5′-CCATCACCCTCATATCCACC-3′ and 5′- TGTGTCTTCACTCTGCAGCC-3′ for a 3′-gene segment in the NGAL gene and 5′-GTTGTAGTATGCCCCCTAAGAG-3′ and 5′-CTCAGGGCAAACCTGAGTCATC-3′ for the κB site in the IL-8 promoter.
siRNA (small interfering RNA) transfection experiments
Duplexed modified RNA oligonucleotides (Stealth RNAi) were synthesized by Invitrogen. The sequences of the sense strands of the siRNAs were as follows: 5′-UCACACAGUAGGAAGAUCUCAUCCC-3′ for the NF-κB p65 subunit, 5′-GCACGAAUGACAGAGGCGUGUAUAA-3′ for the NF-κB p50 subunit, 5′-UAUGAAGGAACGUGUCACCAUCUGC-3′ for IκB-ζ, 5′-AGCUCCAGGACCUUGUGCUGCGUCU-3′ for C/EBPβ, and 5′-UUUAGUGGUGGUAAGUCCAGGCUGU-3′ for C/EBPδ. Stealth RNAi negative control duplexes (Invitrogen) were used as controls. A549 cells (3.0×105 cells) were transfected with 40 pmol of siRNA using Lipofectamine™ 2000. At 24 h after transfection, the cells were stimulated with 1 ng/ml IL-1β for 4 h. Cell lysate was prepared and subjected to Western blotting analysis with specific antibody. Reacting proteins were visualized by chemiluminescence using Lumi-Lightplus Western blotting substrate (Roche Diagnostics). In reporter analyses, A549 cells (7.5×104 cells) were transfected with 10 pmol of siRNA, together with 200 ng of a reporter plasmid using Lipofectamine™ 2000. At 24–36 h after transfection, the cells were stimulated with 1 ng/ml IL-1β for 5 h or with 10 ng/ml PMA for 18 h. HEK-293 cells (5.0×105 cells) were transfected with 40 pmol of siRNA. At 24 h after transfection, total RNA was extracted, and mRNA expression levels were quantified by real-time RT–PCR. In reporter analyses, HEK-293 cells (1.25×105 cells) were transfected with 10 pmol of siRNA, together with 100 ng of indicated reporter plasmid and 2.5 ng of the internal control plasmid phRL-TK using Lipofectamine™ 2000. At 24 h after transfection, the cells were transfected with 5 ng of the expression plasmid for the NF-κB subunit p65 or p50 and 100 ng of the IκB-ζ(L) expression plasmid. Following further incubation for 24 h, the cells were then lysed, and the luciferase activities were measured. The amount of transfected DNA was kept constant by using an empty vector.
RESULTS
Co-expression of IκB-ζ and the NF-κB subunit increases expression of hBD-2
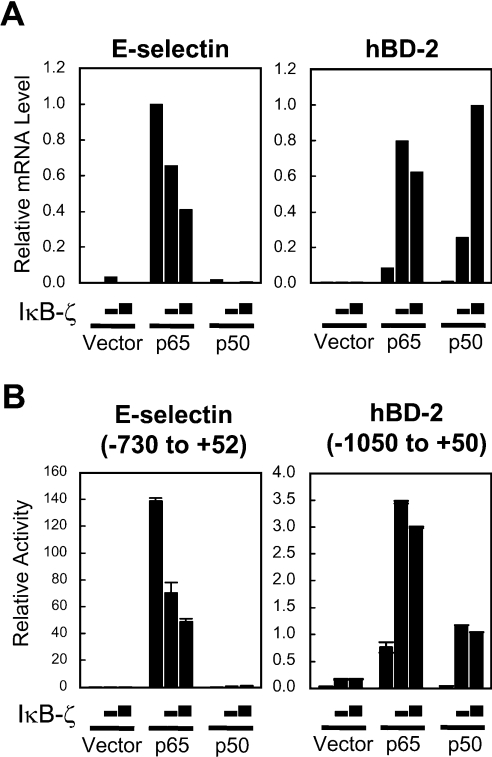
Our previous studies have shown that IκB-ζ has dual opposite functions on expression of different inflammatory genes activated by NF-κB [17,19,23]. To identify genes whose expression is regulated by IκB-ζ, we examined the induction levels of several NF-κB target genes in HEK-293 cells transfected with the NF-κB p65 or p50 subunit and IκB-ζ. Because IκB-ζ(L) is the predominant form in LPS-stimulated cells [20], this form was used in the experiment. Quantitative real-time RT–PCR indicated that expression of E-selectin was strongly induced by transfection of the NF-κB p65 subunit as expected, whereas induction of hBD-2 was modest (Figure 1A). Transfection of the NF-κB p50 subunit or IκB-ζ alone did not elicit significant induction of either genes. Co-transfection of IκB-ζ dose-dependently inhibited the p65 subunit-induced expression of E-selectin, confirming our previous report that IκB-ζ is a negative regulator of the NF-κB-mediated transcription of this gene [17]. In contrast, co-transfection of IκB-ζ augmented expression of hBD-2 induced by the p65 subunit. Interestingly, co-transfection of IκB-ζ and the p50 subunit, either of which alone did not affect the expression, resulted in robust induction of hBD-2. The different actions of IκB-ζ on the two genes are attributable to the different promoter structures, since similar effects of IκB-ζ were observed in the reporter analyses using the E-selectin and hBD-2 promoters (Figure 1B). These results indicate that the actions of IκB-ζ on NF-κB-mediated transcription differ and could exhibit opposite effects against different genes.
Figure 1. Co-transfection of IκB-ζ and the NF-κB subunit up-regulates expression of hBD-2, whereas it inhibits that of E-selectin.
(A) HEK-293 cells were transfected with 60 ng of the expression plasmid for the NF-κB subunit p65 or p50 together with 0.6 or 1.2 μg of the IκB-ζ expression plasmid. The total amount of DNA was kept constant with an empty vector. Total RNA was extracted 24 h after transfection. The expression levels of hBD-2 or E-selectin mRNA were quantified by real-time PCR and were normalized to that of GAPDH. (B) HEK-293 cells were transfected with 100 ng of the E-selectin or hBD-2 promoter reporter plasmid together with 5 ng of the expression plasmid for the NF-κB subunit p65 or p50 and 50 or 100 ng of the IκB-ζ expression plasmid. The total amount of DNA was kept constant with an empty vector. At 24 h after transfection, the cells were lysed and luciferase activities were measured. Results are means±S.D. for duplicate samples, representative of at least two independent experiments.
NF-κB-binding sites in the hBD-2 promoter are essential, but not sufficient, for the IκB-ζ-mediated transcriptional activation
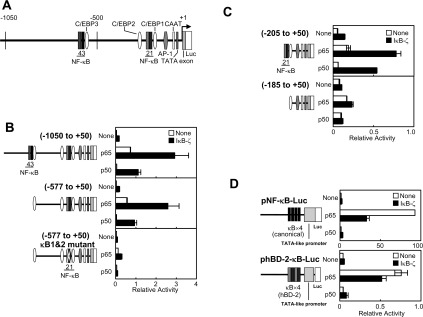
Luciferase reporter analyses indicate that co-transfection of IκB-ζ with the NF-κB subunits activates transcription of the promoter of the hBD-2 gene spanning from −1050 to +50 bp (Figures 1B and 2B; the nucleotide numbering refers to the transcription initiation sites of genes as the base +1). This region of the hBD-2 promoter contains four NF-κB-binding sites, which consists of two tandem consecutive binding sites (Figure 2A).
Figure 2. NF-κB-binding sites in the hBD-2 promoter are essential, but not sufficient, for the IκB-ζ-mediated transcriptional activation.
(A) A schematic illustration of the hBD-2 promoter. (B–D) HEK-293 cells were transfected with indicated reporter plasmids with a 5′-truncation or point mutations of the hBD-2 promoter (B and C), or pNF-κB-Luc harbouring four canonical NF-κB-binding sites or phBD-2-κB-Luc in which the NF-κB-binding sites of the hBD-2 promoter were substituted for those of pNF-κB-Luc (D), together with the expression plasmid for the NF-κB subunit p65 or p50 and the IκB-ζ expression plasmid. The total amount of DNA was kept constant with an empty vector. At 24 h after transfection, the cells were lysed and luciferase activities were measured. Results are means±S.E.M. for duplicate samples, representative of at least two independent experiments.
To identify cis-acting element(s) in the hBD-2 promoter responsible for the IκB-ζ-mediated transcriptional activation, we generated a series of 5′-deletion mutant constructs. The transcriptional activation by IκB-ζ and either the NF-κB p65 or p50 subunit was observed with a promoter fragment beginning at −577 bp, indicating that the distal NF-κB-binding sites (NF-κB-3 and -4 in Figure 2A) are dispensable (Figure 2B). Even a smaller fragment of the promoter, from −205 bp, responded to the co-transfection of IκB-ζ, although the induction levels were lower (Figure 2C). However, further deletion of the proximal NF-κBbinding sites (−185 to +50) severely impaired the IκB-ζ-mediated transcriptional activation, suggesting the importance of the NF-κB-binding sites in this region (NF-κB-1 and -2). To elucidate further the role of the NF-κB-binding sites, we introduced point mutations into the tandem NF-κB-binding sequences. Consistent with the results of the deletion constructs, introduction of mutations at NF-κB-binding sites 1 and 2 in the promoter from −577 to +50 bp significantly damaged not only the induction by p65, but also the augmentation by co-expression of IκB-ζ (Figure 2B, bottom panel). Thus the proximal NF-κB-binding sites 1 and 2 from −205 to −186 bp are essential for the IκB-ζ-mediated transcriptional activation.
To examine whether the sequence of the NF-κB-binding site determines the specific action of IκB-ζ in the observed transcriptional regulation, we constructed a reporter plasmid in which the duplicated sequences of the proximal NF-κB-binding sites 1 and 2 of the hBD-2 promoter were used to replace the tandem canonical NF-κB-binding sequences in the NF-κB reporter plasmid pNF-κB-Luc (phBD-2-κB-Luc). As observed with the E-selectin promoter reporter (Figure 1B), co-transfection of IκB-ζ resulted in the inhibition of p65-mediated activation of the pNF-κB-Luc. Similarly, phBD-2-κB-Luc was activated by p65, but the activity was suppressed by co-transfection of IκB-ζ (Figure 2D). Synergistic transcriptional activation by co-transfection of IκB-ζ, and the p50 subunit was minimal with pNF-κB-Luc or phBD-2-κB-Luc as with the E-selectin promoter reporter. Therefore the NF-κB-binding sites are essential, but the sequences of the sites themselves do not confer the IκB-ζ-mediated transcriptional activation to the promoter.
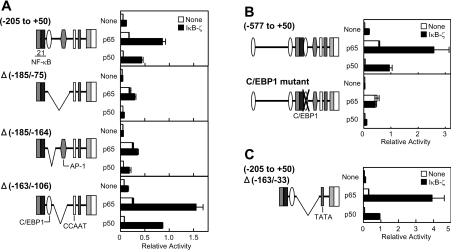
Critical roles for the C/EBP-binding site in the hBD-2 promoter in the IκB-ζ-mediated transcriptional activation
To identify another critical cis element(s) in the hBD-2 promoter for the IκB-ζ-mediated transcriptional activation, we created several internal deletion mutants (Figure 3A). The hBD-2 promoter of −205 to +50 bp responded to the co-expression of IκB-ζ to exhibit the augmented transcription as mentioned above. The response to IκB-ζ was markedly reduced by deleting a region of −185 to −75 bp (Δ−185/−75), which harbours a C/EBP-binding site (C/EBP1 in Figure 2A) and an AP-1 (activator protein 1)-binding site. A mutant with a shorter deletion from −185 to −164 bp (Δ−185/−164), a region containing the C/EBP-binding site, also minimally responded to the co-transfection of IκB-ζ. However, transcription of another deletion mutant lacking −163 to −106 bp (Δ−163/−106), and therefore without the AP-1-binding site, was stimulated upon the co-transfection of IκB-ζ. These results indicate that the region from −185 to −164 bp, which contains a potential C/EBP-binding sequence, GATTTGCATAAGAT, is important for the function of IκB-ζ. To evaluate the role(s) of this sequence, we introduced mutations at conserved nucleotide residues of the C/EBP-binding site. The hBD-2 promoter of −577 to +50 bp harbouring the mutated C/EBP-binding site with a sequence of GATAACCATAAGAT (mutated nucleotides are underlined) did not respond to the co-transfection of IκB-ζ (Figure 3B). Thus the C/EBP-binding site-1 in the hBD-2 promoter is essential for the effects of IκB-ζ.
Figure 3. NF-κB- and C/EBP-binding sites in the hBD-2 promoter are core elements for the IκB-ζ-mediated transcriptional activation.
(A–C) HEK-293 cells were transfected with the hBD-2 promoter reporter plasmids with an internal deletion (A and C) or point mutations in the C/EBP-binding site (B), together with the expression plasmid for the NF-κB subunit p65 or p50 and the IκB-ζ expression plasmid. The total amount of DNA was kept constant with an empty vector. At 24 h after transfection, the cells were lysed and luciferase activities were measured. Results are means±S.E.M. for duplicate samples, representative of at least two independent experiments.
Furthermore, we found that an internal deletion mutant (Δ−163/−33), consisting of only the TATA box and the region of −205 to −163 bp of the hBD-2 promoter, which harbours NF-κB-binding sites 1 and 2 followed by C/EBP-binding site 1, showed the maximum degree of transcriptional activation upon the co-transfection of the NF-κB subunit p65 or p50 and IκB-ζ (Figure 3C). Therefore the NF-κB- and C/EBP-binding sites are core elements for the IκB-ζ-mediated transcriptional activation of the hBD-2 promoter.
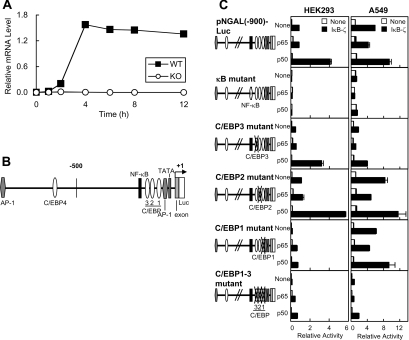
NF-κB- and C/EBP-binding sites are essential for the IκB-ζ-mediated transcriptional activation of the NGAL promoter
Our previous study using microarray analyses showed that a subset of LPS-inducible genes was not induced in IκB-ζ-deficient macrophages [19]. Quantitative RT–PCR showed that NGAL mRNA was induced in LPS-stimulated bone-marrow-derived macrophages of wild-type, but not of IκB-ζ-deficient mice (Figure 4A). Thus NGAL is one of the genes that require IκB-ζ for its induction, and we therefore analysed the promoter of this gene. The NGAL promoter contains an NF-κB-binding site and four C/EBP-binding sites (C/EBP1–4) in addition to AP-1-binding sites (Figure 4B). A reporter plasmid containing the NGAL promoter from −900 to +51 bp [pNGAL(−900)-Luc] was stimulated by co-expression of IκB-ζ and the NF-κB p50 subunit (Figure 4C). We focused on the roles for the NF-κB-binding site and the proximal C/EBP-binding sites (C/EBP1–3), and introduced point mutations into these sites. We found that the IκB-ζ-mediated transcriptional activation was abolished by mutations of the NF-κB-binding site and was substantially decreased by mutations of one of the C/EBP-binding sites (C/EBP1) (Figure 4C, left-hand panels). Introduction of mutations at all the three C/EBP-binding sites (C/EBP1–3) resulted in the minimal induction by IκB-ζ, as the mutation of C/EBP-binding site 1. We carried out similar experiments by using the human lung adenocarcinoma cell line A549, which is very sensitive to IL-1β to elicit induction of NGAL [30] (Figure 4C, right-hand panels). In the cells, transfection of IκB-ζ alone induced the activation of the NGAL promoter, which was abolished by the mutation of the NF-κB-binding site. The induction was severely impaired by the mutation of the C/EBP-binding site 3 and was suppressed further by the mutations of all three C/EBP-binding sites. These results indicate that the NF-κB- and C/EBP-binding sites are also essential for the IκB-ζ-mediated transcriptional activation of the NGAL gene, supporting further the results obtained with the hBD-2 promoter.
Figure 4. NF-κB- and C/EBP-binding sites in the NGAL promoter are essential for the IκB-ζ-mediated transcriptional activation.
(A) Bone-marrow-derived macrophages isolated from wild-type (WT) or IκB-ζ-deficient mice (KO) were stimulated with 20 ng/ml LPS for the periods indicated. Total RNA was extracted, and the expression levels of NGAL mRNA were quantified by real-time RT-PCR and were normalized to that of β-actin. A representative result of two independent experiments is shown. (B) A schematic illustration of the NGAL promoter. (C) HEK-293 cells (left-hand panels) or A549 cells (right-hand panels) were transfected with the NGAL promoter reporter plasmid with or without point mutations at the NF-κB-binding site or the C/EBP-binding site, together with the expression plasmid for the NF-κB subunit p65 or p50 and the IκB-ζ expression plasmid. The total amount of DNA was kept constant with an empty vector. At 24 h after transfection, the cells were lysed, and luciferase activities were measured. Results are means±S.E.M. for duplicate samples, representative of at least two independent experiments.
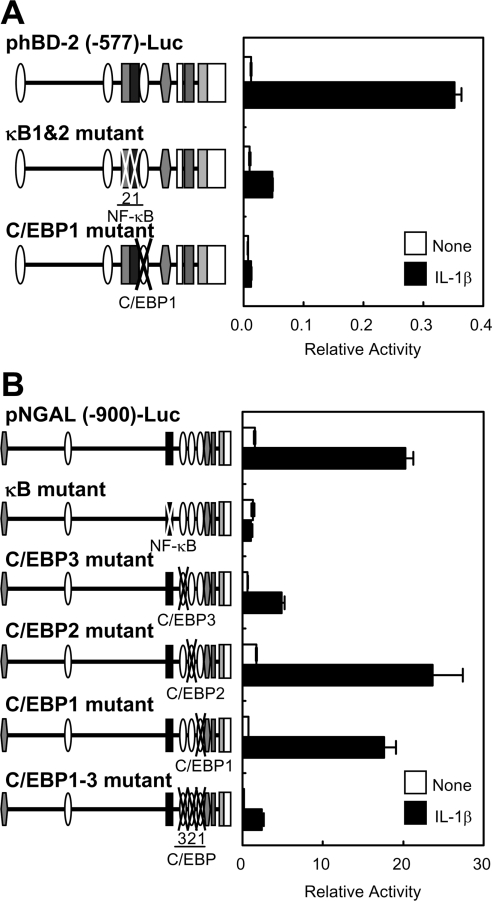
NF-κB- and C/EBP-binding sites are essential for IL-1β-induced transcriptional activation of the hBD-2 and NGAL promoters
Next, we examined whether the NF-κB- and C/EBP-binding sites are also required for induction of the hBD-2 and NGAL genes upon stimulation by IL-1β. Reporter analyses clearly indicated that both NF-κB- and C/EBP-binding sites in the hBD-2 promoter are required for efficient transcriptional activation following stimulation of A549 cells with IL-1β, which elicits induction of IκB-ζ (Figure 5A). As in the hBD-2 promoter, the NF-κB-binding site in the NGAL promoter was also essential for the IL-1β-induced transcription. We found that mutations at C/EBP-binding site 3 substantially reduced the transcriptional induction via IL-1β, and the mutations of all the C/EBP-binding sites suppressed further the induction, as in the IκB-ζ-mediated induction shown in Figure 4C (right-hand panels). Thus the NF-κB and C/EBP-binding sites in the hBD-2 and NGAL promoters are also crucial for activation of these promoters following physiological stimulation (via IL-1β) that leads to induction of IκB-ζ.
Figure 5. NF-κB- and C/EBP-binding sites in the hBD-2 and NGAL promoters are essential for IL-1β-induced transcriptional activation.
A549 cells were transfected with the hBD-2 promoter reporter plasmids (A) or the NGAL promoter reporter plasmids (B) with or without indicated point mutations at the NF-κB-binding site or the C/EBP-binding sites. At 24 h after transfection, the cells were stimulated with 1 ng/ml IL-1β for 5 h. The cells were then lysed, and luciferase activities were measured. Results are means±S.E.M. for duplicate samples, representative of at least two independent experiments.
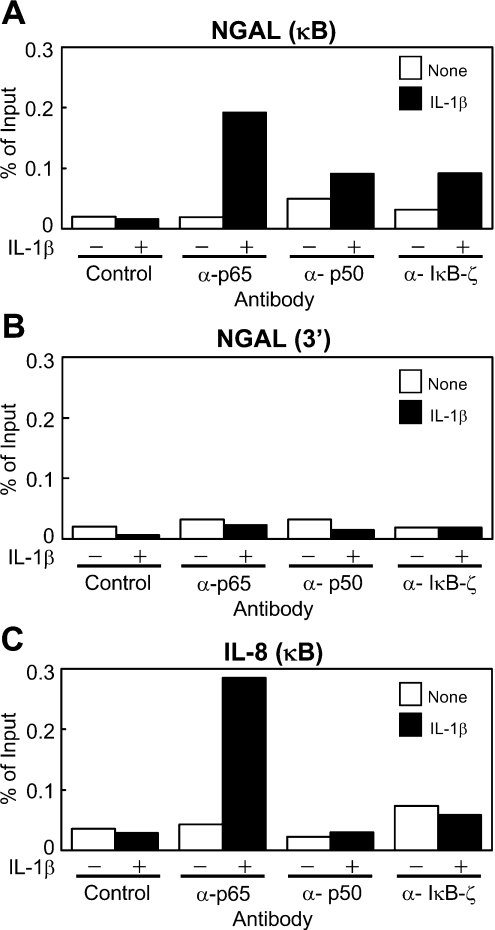
NF-κB and IκB-ζ are recruited to the NGAL promoter upon stimulation with IL-1β
To investigate recruitment of IκB-ζ and/or the NF-κB subunits to the promoters of the genes, we performed ChIP assays using specific antibodies against IκB-ζ or the NF-κB subunits. In unstimulated A549 cells, neither IκB-ζ nor the NF-κB subunits were recruited to the NGAL or IL-8 promoters. However, on stimulation of the cells with IL-1β, the NF-κB p65 subunit was found to be recruited to the NF-κB-binding sites of the NGAL and IL-8 promoters (Figure 6). In contrast, IκB-ζ and the NF-κB p50 subunit were specifically recruited to the NGAL promoter, but not to the IL-8 promoter, in the stimulated cells. None of the three proteins was recruited to the 3′-flanking region of the NGAL gene, thus showing the specificity of the assay. These results demonstrate that upon stimulation with IL-1β, IκB-ζ and both the NF-κB p65 and p50 subunits are recruited to and remain bound at the NF-κB-binding site of the NGAL promoter.
Figure 6. NF-κB subunits p65 and p50, and IκB-ζ are recruited to the NGAL promoter in A549 cells upon stimulation with IL-1β.
(A–C) A549 cells were stimulated with 1 ng/ml IL-1β for 4 h, and ChIP assays were performed with antibodies against p65, p50 or IκB-ζ, or control rabbit IgG. The precipitated DNA containing the κB site of the NGAL promoter (A), the 3′-flanking region of the NGAL gene (B) or the κB site of the IL-8 promoter (C) was assayed by quantitative real-time PCR with specific primers. A representative result of two independent experiments is shown for each.
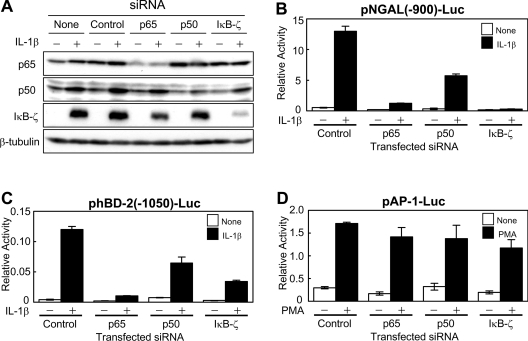
NF-κB and IκB-ζ are essential for transcriptional activation of the NGAL and hBD-2 promoters upon stimulation with IL-1β
We next examined whether NF-κB and/or IκB-ζ are required for the induction of the genes upon stimulation. Transfection of siRNAs for p65, p50 or IκB-ζ specifically knocked down the expression of corresponding endogenous protein, verifying the specificity (Figure 7A). We knocked down the expression levels of the endogenous NF-κB or IκB-ζ in A549 cells by introducing the siRNA, and examined the activities of the NGAL or the hBD-2 promoter reporter on stimulation with IL-1β. The siRNA for the p65 subunit or IκB-ζ strongly repressed induction of both NGAL and hBD-2 promoters in response to IL-1β (Figures 7B and 7C). The siRNA for the p50 subunit of NF-κB also substantially inhibited the activation of both genes. In contrast, none of the siRNAs for the p65 or p50 subunits or IκB-ζ affected PMA-mediated activation of an AP-1 reporter plasmid, pAP-1-Luc, in the same cells (Figure 7D). Thus it was strongly suggested that the NF-κB p65 and p50 subunits and IκB-ζ are essential for the transcriptional activation of the NGAL and the hBD-2 promoters upon stimulation with IL-1β.
Figure 7. NF-κB p65 and p50 subunits and IκB-ζ are essential for the transcriptional activation of the hBD-2 and NGAL promoters upon stimulation with IL-1β.
(A) A549 cells were transfected with siRNA for the p50 subunit, the p65 subunit, IκB-ζ or control. At 24 h after transfection, the cells were stimulated with 1 ng/ml IL-1β for 4 h, and cell extracts were prepared. Immunoblotting analysis was performed using an antibody against the NF-κB p65 subunit or the p50 subunit, IκB-ζ or β-tubulin. (B–D) A549 cells were transfected with siRNA for the p50 subunit, the p65 subunit, IκB-ζ or control, together with indicated reporter plasmids. At 36 h after transfection, the cells were stimulated with 1 ng/ml IL-1β for 5 h (B and C), or, at 24 h after transfection, the cells were stimulated with 10 ng/ml PMA for 18 h (D). The cells were then lysed, and luciferase activities were measured. Results are means±S.D. for duplicate samples, representative of at least three independent experiments.
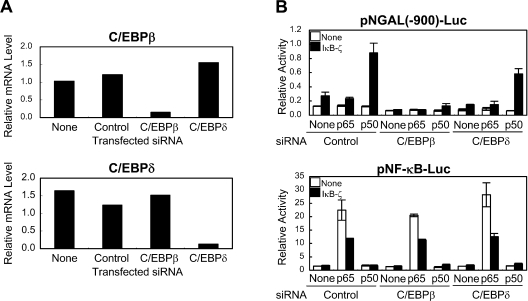
Activation of the NGAL promoter by transfection of the NF-κB p50 subunit and IκB-ζ was suppressed in C/EBPβ-depleted HEK-293 cells
In order to examine whether C/EBPs are required for the IκB-ζ-mediated transcription, we attempted to knock down C/EBPβ and C/EBPδ. Introduction of siRNAs for C/EBPβ and C/EBPδ into HEK-293 cells successfully suppressed their respective mRNAs (Figure 8A). HEK-293 cells treated with siRNA for C/EBPβ or C/EBPδ were transfected with NF-κB and/or IκB-ζ, and we examined their effects on the NGAL promoter. As shown in Figure 8(B), the IκB-ζ-mediated activation of the NGAL promoter was specifically inhibited in the C/EBPβ-depleted cells, whereas the activation was not affected in the C/EBPδ-depleted cells. Another siRNA for C/EBPβ with a different sequence exhibited similar effects (results not shown). Both siRNAs for C/EBPβ and C/EBPδ did not affect the activity on the NF-κB reporter. Thus C/EBPβ is required for the IκB-ζ-mediated transcriptional activation of the NGAL promoter in HEK-293 cells.
Figure 8. Activation of the NGAL promoter by transfection of the NF-κB p50 subunit and IκB-ζ was suppressed in C/EBPβ-depleted HEK-293 cells.
(A) HEK-293 cells were transfected with siRNA for C/EBPβ, C/EBPδ or control. Total RNA was extracted 24 h after transfection. The expression levels of C/EBPβ or C/EBPδ mRNA were quantified by real-time RT–PCR and were normalized to that of GAPDH. A representative result of two independent experiments is shown. (B) HEK-293 cells were transfected with siRNA for C/EBPβ, C/EBPδ or control, together with the indicated reporter plasmids. After 24 h, cells were transfected again with the indicated expression plasmids. Following further incubation for 24 h, the cells were lysed, and luciferase activities were measured. Results are means±S.D. for duplicate samples, representative of at least two independent experiments.
DISCUSSION
Our previous studies have shown that IκB-ζ exhibits dual opposite activities on the expression of different subsets of inflammatory genes [17,19,23]. Consistent with these results, the present study also showed that exogenous expression of IκB-ζ augmented the NF-κB-mediated expression of hBD-2, whereas it inhibited that of E-selectin (Figure 1). In addition to IκB-ζ(L) used in this study, another isoform, IκB-ζ(S), exhibited qualitatively similar effects (results not shown). IκB-ζ alone promoted marginal levels of the transcription of the hBD-2 and NGAL genes, and it required NF-κB for robust transcriptional activation in HEK-293 cells. As expected from this requirement for NF-κB, reporter analyses indicated that NF-κB-binding sites in both promoters are essential for the IκB-ζ-mediated transcriptional activation (Figures 2 and 4C, left-hand panels). The sequence of the NF-κB-binding sites, however, was not sufficient for the activation, and a C/EBP-binding site was also essential (Figures 2, 3 and 4C, left-hand panels). Although NGAL transcription was induced by transfection of IκB-ζ alone in A549 cells, NF-κB is probably involved in the induction since the NF-κB-binding site, as well as a C/EBP-binding site, is also essential for the induction as in the hBD-2 promoter (Figure 4C, right-hand panels). Furthermore, in addition to IκB-ζ, NF-κB is essential for the induction of NGAL transcription in A549 cells (Figure 7). Endogenous nuclear NF-κB (possibly the p50 subunit) levels might be higher in this adenocarcinoma cell line, which is highly sensitive to IL-1β.
Since robust transcriptional activation by IκB-ζ was observed on an artificial small promoter consisting of the NF-κB- and C/EBP-binding sites and TATA box (Figure 3C), these two sites appear to constitute the minimal element that allows the IκB-ζ-mediated transcriptional activation. Since IκB-ζ acted as a negative regulator of NF-κB-mediated activation on the promoters constituted only with the NF-κB-binding sites of the hBD-2 promoter (Figure 2D), the presence of the C/EBP-binding site could convert the action of IκB-ζ. It should be noted that promoters of genes that exhibit IκB-ζ-dependent induction harbour NF-κB- and C/EBP-binding sites, including IL-6, the IL-12 p40 subunit, granulocyte/macrophage CSF and granulocyte CSF.
Among the three proximal C/EBP-binding sites in the NGAL promoter, different sites seemed to be used in HEK-293 cells and A549 cells for the IκB-ζ-mediated inductions (Figure 4). This may reflect that, in the two cell lines, IκB-ζ and NF-κB could form stoichiometrically different transcription complexes with different sizes, which would utilize C/EBP-binding sites located at different distances from the NF-κB-binding site. Multiple C/EBP-binding sites in the NGAL promoter might guarantee the maximum induction in both cell types.
Crucial roles for both NF-κB- and C/EBP-binding sites were also demonstrated in the induction of the hBD-2 and NGAL genes in response to IL-1β (Figure 5). IκB-ζ expression, which is elicited by IL-1β, was essential for the IL-1β-dependent induction of the two genes (Figure 7). In addition to the NF-κB p50 subunit, the p65 subunit was also recruited to the NGAL promoter upon IL-1β stimulation, and was required for the expression of the NGAL gene (Figures 6 and 7). Although preferential binding of IκB-ζ to the p50 subunit has been shown previously [17], the active transcription complex on the genes appeared to contain IκB-ζ and both p50 and p65 subunits of NF-κB. It is noteworthy that the NF-κB-binding sites in the hBD-2 and NGAL promoters are not canonical, as revealed by the fact that their respective p65 subunit-mediated inductions are much weaker than that of the E-selectin promoter or the promoter in pNF-κB-Luc (Figures 1, 2D and 4C, see induction in the absence of IκB-ζ). IκB-ζ binding to NF-κB might alter the preferred DNA-binding sequences of the p65/p50 heterodimer from the canonical NF-κB-binding sequences to non-canonical sequences flanked by a C/EBP-binding site.
Although the recruitment of IκB-ζ and the NF-κB subunits was shown by the ChIP analyses, we could not detect the binding of the IκB-ζ-containing complex to the DNA fragment of the hBD-2 promoter by EMSA (electrophoretic mobility-shift assay) using lysate from cells transfected with IκB-ζ and NF-κB (results not shown). This suggests that the complex is very fragile or that it requires another protein(s) for DNA binding. The C/EBP family of proteins could be such candidates, since NF-κB and the C/EBP family of proteins associate directly via their Rel homology domain and the basic leucine-zipper domain [32–34]. In fact, the IκB-ζ-mediated activation of the NGAL promoter was severely impaired in C/EBPβ-depleted HEK-293 cells (Figure 8). However, in vivo studies using C/EBP-deficient mice have provided results contrary to this hypothesis. The expression of IL-6, an IκB-ζ target gene, following LPS stimulation is unchanged in macrophages from wild-type and C/EBPβ- [35] or C/EBPϵ- [36] knockout mice. In addition, no defect in cytokine production has been detected in macrophages from C/EBPδ-knockout mice [37]. Our preliminary studies indicated that the IκB-ζ-mediated activation of the hBD-2 and NGAL promoter in A549 cells was not significantly affected by depletion of either C/EBPβ or C/EBPδ. Thus other proteins could be involved, in other cell types, in the transcriptionally active complex consisting of IκB-ζ and NF-κB that binds to the promoters harbouring the NF-κB- and C/EBP-binding sites.
The NGAL promoter with the multiple C/EBP-binding sites was induced by transfection of C/EBPβ and C/EBPδ alone (results not shown). As C/EBPδ is one of the IκB-ζ target genes [19], the induction of NGAL could be sustained by C/EBPδ in a late phase. Although the C/EBPδ-mediated induction was completely abolished by the triple mutations of C/EBP-binding sites 1, 2 and 3 in the NGAL promoter (results not shown), marginal levels of IκB-ζ-mediated induction was observed with the mutant (Figures 4 and 5), suggesting that the recognition sites of the IκB-ζcontaining complex and C/EBPs overlap, but are not identical.
Despite intensive studies on the molecular mechanisms of IκB-ζ-mediated transcription, its physiological roles in vivo remain to be analysed. In addition to several cytokines, the present studies indicated that the hBD-2 and NGAL genes are targets for IκB-ζ-mediated transcription. Consistent with the induction spectrum of IκB-ζ, both the hBD-2 and NGAL genes are induced preferentially by IL-1β, but not by TNF-α in A549 human lung adenocarcinoma cells, keratinocytes and other cells [25–30]. A recent report has also shown critical roles for IκB-ζ in IL-1β-dependent induction of the genes [38]. Since both genes encode inducible antimicrobial proteins expressed in various epithelial tissues [39,40], one of the functions of IκB-ζ is likely to be to activate genes that play roles in elimination of infected bacteria. This concept is consistent with the observation that IκB-ζ is induced by microbial ligands of various TLRs, but not by double-stranded RNA, a ligand for TLR3, which is an intracellular receptor for RNA viruses [41].
There is accumulating evidence that both NF-κB- and C/EBP-binding sites are important for regulation of many immune responses and acute-phase response genes such as IL-1β [42], IL-6 [32] and granulocyte CSF [43], in addition to hBD-2 and NGAL. Recent studies also showed that NF-κB- and C/EBP-binding sites are required for induction of IL-6 and 24p3, a mouse homologue of NGAL, on co-stimulation with IL-17 and TNF-α [44,45]. The induction probably involves IκB-ζ, because co-stimulation with IL-17 and TNF-α induces expression of IκB-ζ [20,46]. Nonetheless, not all promoters harbouring both NF-κB- and C/EBP-binding sites are targets for IκB-ζ. For example, the IL-8 promoter contains both sites and is activated synergistically by overexpression of NF-κB and C/EBPβ [32]. IκB-ζ, however, is not recruited to the IL-8 promoter and acts negatively on expression, as revealed by the ChIP assay (Figure 6) and mRNA quantification (results not shown). Probably, the order, distance and the intervening sequence between the NF-κB- and C/EBP-binding sites, as well as the sequence of each binding site and/or chromatin structures, would affect transcriptional activation by IκB-ζ. Comprehensive analyses of genes whose transcription is regulated by IκB-ζ should provide more information on the common promoter structures of these genes. Furthermore, identification of the transcription complex containing IκB-ζ and biochemical characterization of the complex would be invaluable towards understanding the sophisticated mechanisms for regulation of a series of inflammatory reactions.
Acknowledgments
This work was supported in part by grants-in-aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan (to T.M., K.T. and S.Y.), and grants from the Naito Foundation (to T.M.), the Mochida Memorial Foundation for Medical and Pharmaceutical Research (to T.M.), the Kaibara Foundation (to T.M.) and Japan Foundation of Applied Enzymology (to T.M.).
References
- 1.Li Q., Verma I. M. NF-κB regulation in the immune system. Nat. Rev. Immunol. 2002;2:725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 2.Hayden M. S., Ghosh S. Signaling to NF-κB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 3.Bonizzi G., Karin M. The two NF-κB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 2004;25:280–288. doi: 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 4.Chen L. F., Greene W. C. Shaping the nuclear action of NF-κB. Nat. Rev. Mol. Cell Biol. 2004;5:392–401. doi: 10.1038/nrm1368. [DOI] [PubMed] [Google Scholar]
- 5.Chen L., Fischle W., Verdin E., Greene W. C. Duration of nuclear NF-κB action regulated by reversible acetylation. Science. 2001;293:1653–1657. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- 6.Zhong H., May M. J., Jimi E., Ghosh S. The phosphorylation status of nuclear NF-κB determines its association with CBP/p300 or HDAC-1. Mol. Cell. 2002;9:625–636. doi: 10.1016/s1097-2765(02)00477-x. [DOI] [PubMed] [Google Scholar]
- 7.Sosic D., Richardson J. A., Yu K., Ornitz D. M., Olson E. N. Twist regulates cytokine gene expression through a negative feedback loop that represses NF-κB activity. Cell. 2003;112:169–180. doi: 10.1016/s0092-8674(03)00002-3. [DOI] [PubMed] [Google Scholar]
- 8.Almawi W. Y., Melemedjian O. K. Negative regulation of nuclear factor-κB activation and function by glucocorticoids. J. Mol. Endocrinol. 2002;28:69–78. doi: 10.1677/jme.0.0280069. [DOI] [PubMed] [Google Scholar]
- 9.You Z., Madrid L. V., Saims D., Sedivy J., Wang C. Y. c-Myc sensitizes cells to tumor necrosis factor-mediated apoptosis by inhibiting nuclear factor κB transactivation. J. Biol. Chem. 2002;277:36671–36677. doi: 10.1074/jbc.M203213200. [DOI] [PubMed] [Google Scholar]
- 10.Ma X. Y., Wang H., Ding B., Zhong H., Ghosh S., Lengyel P. The interferon-inducible p202a protein modulates NF-κB activity by inhibiting the binding to DNA of p50/p65 heterodimers and p65 homodimers while enhancing the binding of p50 homodimers. J. Biol. Chem. 2003;278:23008–23019. doi: 10.1074/jbc.M302105200. [DOI] [PubMed] [Google Scholar]
- 11.Poligone B., Baldwin A. S. Positive and negative regulation of NF-κB by COX-2: roles of different prostaglandins. J. Biol. Chem. 2001;276:38658–38664. doi: 10.1074/jbc.M106599200. [DOI] [PubMed] [Google Scholar]
- 12.Lee D. K., Kang J. E., Park H. J., Kim M. H., Yim T. H., Kim J. M., Heo M. K., Kim K. Y., Kwon H. J., Hur M. W. FBI-1 enhances transcription of the nuclear factor-κB (NF-κB)-responsive E-selectin gene by nuclear localization of the p65 subunit of NF-κB. J. Biol. Chem. 2005;280:27783–27791. doi: 10.1074/jbc.M504909200. [DOI] [PubMed] [Google Scholar]
- 13.Uranishi H., Tetsuka T., Yamashita M., Asamitsu K., Shimizu M., Itoh M., Okamoto T. Involvement of the pro-oncoprotein TLS (translocated in liposarcoma) in nuclear factor-κB p65-mediated transcription as a coactivator. J. Biol. Chem. 2001;276:13395–13401. doi: 10.1074/jbc.M011176200. [DOI] [PubMed] [Google Scholar]
- 14.Franzoso G., Bours V., Park S., Tomita-Yamaguchi M., Kelly K., Siebenlist U. The candidate oncoprotein Bcl-3 is an antagonist of p50/NF-κB-mediated inhibition. Nature. 1992;359:339–342. doi: 10.1038/359339a0. [DOI] [PubMed] [Google Scholar]
- 15.Bours V., Franzoso G., Azarenko V., Park S., Kanno T., Brown K., Siebenlist U. The oncoprotein Bcl-3 directly transactivates through κB motifs via association with DNA-binding p50B homodimers. Cell. 1993;72:729–739. doi: 10.1016/0092-8674(93)90401-b. [DOI] [PubMed] [Google Scholar]
- 16.Nolan G. P., Fujita T., Bhatia K., Huppi C., Liou H. C., Scott M. L., Baltimore D. The Bcl-3 proto-oncogene encodes a nuclear IκB-like molecule that preferentially interacts with NF-κB p50 and p52 in a phosphorylation-dependent manner. Mol. Cell. Biol. 1993;13:3557–3566. doi: 10.1128/mcb.13.6.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamazaki S., Muta T., Takeshige K. A novel IκB protein, IκB-ζ, induced by proinflammatory stimuli, negatively regulates nuclear factor-κB in the nuclei. J. Biol. Chem. 2001;276:27657–27662. doi: 10.1074/jbc.M103426200. [DOI] [PubMed] [Google Scholar]
- 18.Eto A., Muta T., Yamazaki S., Takeshige K. Essential roles for NF-κB and a Toll/IL-1 receptor domain-specific signal(s) in the induction of IκB-ζ. Biochem. Biophys. Res. Commun. 2003;301:495–501. doi: 10.1016/s0006-291x(02)03082-6. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto M., Yamazaki S., Uematsu S., Sato S., Hemmi H., Hoshino K., Kaisho T., Kuwata H., Takeuchi O., Takeshige K., et al. Regulation of Toll/IL-1-receptormediated gene expression by the inducible nuclear protein IκBζ. Nature. 2004;430:218–222. doi: 10.1038/nature02738. [DOI] [PubMed] [Google Scholar]
- 20.Yamazaki S., Muta T., Matsuo S., Takeshige K. Stimulus-specific induction of a novel nuclear factor-κB regulator, IκB-ζ, via Toll/Interleukin-1 receptor is mediated by mRNA stabilization. J. Biol. Chem. 2005;280:1678–1687. doi: 10.1074/jbc.M409983200. [DOI] [PubMed] [Google Scholar]
- 21.Kitamura H., Kanehira K., Okita K., Morimatsu M., Saito M. MAIL, a novel nuclear IκB protein that potentiates LPS-induced IL-6 production. FEBS Lett. 2000;485:53–56. doi: 10.1016/s0014-5793(00)02185-2. [DOI] [PubMed] [Google Scholar]
- 22.Haruta H., Kato A., Todokoro K. Isolation of a novel interleukin-1-inducible nuclear protein bearing ankyrin-repeat motifs. J. Biol. Chem. 2001;276:12485–12488. doi: 10.1074/jbc.C100075200. [DOI] [PubMed] [Google Scholar]
- 23.Motoyama M., Yamazaki S., Eto-Kimura A., Takeshige K., Muta T. Positive and negative regulation of nuclear factor-κB-mediated transcription by IκB-ζ, an inducible nuclear protein. J. Biol. Chem. 2005;280:7444–7451. doi: 10.1074/jbc.M412738200. [DOI] [PubMed] [Google Scholar]
- 24.Shiina T., Konno A., Oonuma T., Kitamura H., Imaoka K., Takeda N., Todokoro K., Morimatsu M. Targeted disruption of MAIL, a nuclear IκB protein, leads to severe atopic dermatitis-like disease. J. Biol. Chem. 2004;279:55493–55498. doi: 10.1074/jbc.M409770200. [DOI] [PubMed] [Google Scholar]
- 25.Liu A. Y., Destoumieux D., Wong A. V., Park C. H., Valore E. V., Liu L., Ganz T. Human β-defensin-2 production in keratinocytes is regulated by interleukin-1, bacteria, and the state of differentiation. J. Invest. Dermatol. 2002;118:275–281. doi: 10.1046/j.0022-202x.2001.01651.x. [DOI] [PubMed] [Google Scholar]
- 26.Tsutsumi-Ishii Y., Nagaoka I. Modulation of human β-defensin-2 transcription in pulmonary epithelial cells by lipopolysaccharide-stimulated mononuclear phagocytes via proinflammatory cytokine production. J. Immunol. 2003;170:4226–4236. doi: 10.4049/jimmunol.170.8.4226. [DOI] [PubMed] [Google Scholar]
- 27.Liu L., Roberts A. A., Ganz T. By IL-1 signaling, monocyte-derived cells dramatically enhance the epidermal antimicrobial response to lipopolysaccharide. J. Immunol. 2003;170:575–580. doi: 10.4049/jimmunol.170.1.575. [DOI] [PubMed] [Google Scholar]
- 28.Sorensen O. E., Thapa D. R., Rosenthal A., Liu L., Roberts A. A., Ganz T. Differential regulation of β-defensin expression in human skin by microbial stimuli. J. Immunol. 2005;174:4870–4879. doi: 10.4049/jimmunol.174.8.4870. [DOI] [PubMed] [Google Scholar]
- 29.Pioli P. A., Weaver L. K., Schaefer T. M., Wright J. A., Wira C. R., Guyre P. M. Lipopolysaccharide-induced IL-1β production by human uterine macrophages up-regulates uterine epithelial cell expression of human β-defensin 2. J. Immunol. 2006;176:6647–6655. doi: 10.4049/jimmunol.176.11.6647. [DOI] [PubMed] [Google Scholar]
- 30.Cowland J. B., Sorensen O. E., Sehested M., Borregaard N. Neutrophil gelatinase-associated lipocalin is up-regulated in human epithelial cells by IL-1β, but not by TNF-α. J. Immunol. 2003;171:6630–6639. doi: 10.4049/jimmunol.171.12.6630. [DOI] [PubMed] [Google Scholar]
- 31.Chen C., Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsusaka T., Fujikawa K., Nishio Y., Mukaida N., Matsushima K., Kishimoto T., Akira S. Transcription factors NF-IL6 and NF-κB synergistically activate transcription of the inflammatory cytokines, interleukin 6 and interleukin 8. Proc. Natl. Acad. Sci. U.S.A. 1993;90:10193–10197. doi: 10.1073/pnas.90.21.10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stein B., Cogswell P. C., Baldwin A. S., Jr Functional and physical associations between NF-κB and C/EBP family members: a Rel domain-bZIP interaction. Mol. Cell. Biol. 1993;13:3964–3974. doi: 10.1128/mcb.13.7.3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LeClair K. P., Blanar M. A., Sharp P. A. The p50 subunit of NF-κB associates with the NF-IL6 transcription factor. Proc. Natl. Acad. Sci. U.S.A. 1992;89:8145–8149. doi: 10.1073/pnas.89.17.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanaka T., Akira S., Yoshida K., Umemoto M., Yoneda Y., Shirafuji N., Fujiwara H., Suematsu S., Yoshida N., Kishimoto T. Targeted disruption of the NF-IL6 gene discloses its essential role in bacteria killing and tumor cytotoxicity by macrophages. Cell. 1995;80:353–361. doi: 10.1016/0092-8674(95)90418-2. [DOI] [PubMed] [Google Scholar]
- 36.Tavor S., Vuong P. T., Park D. J., Gombart A. F., Cohen A. H., Koeffler H. P. Macrophage functional maturation and cytokine production are impaired in C/EBPϵ-deficient mice. Blood. 2002;99:1794–1801. doi: 10.1182/blood.v99.5.1794. [DOI] [PubMed] [Google Scholar]
- 37.Poli V. The role of C/EBP isoforms in the control of inflammatory and native immunity functions. J. Biol. Chem. 1998;273:29279–29282. doi: 10.1074/jbc.273.45.29279. [DOI] [PubMed] [Google Scholar]
- 38.Cowland J. B., Muta T., Borregaard N. IL-1β-specific up-regulation of neutrophil gelatinase-associated lipocalin is controlled by IκB-ζ. J. Immunol. 2006;176:5559–5566. doi: 10.4049/jimmunol.176.9.5559. [DOI] [PubMed] [Google Scholar]
- 39.Raj P. A., Dentino A. R. Current status of defensins and their role in innate and adaptive immunity. FEMS Microbiol. Lett. 2002;206:9–18. doi: 10.1111/j.1574-6968.2002.tb10979.x. [DOI] [PubMed] [Google Scholar]
- 40.Flo T. H., Smith K. D., Sato S., Rodriguez D. J., Holmes M. A., Strong R. K., Akira S., Aderem A. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 2004;432:917–921. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- 41.Alexopoulou L., Holt A. C., Medzhitov R., Flavell R. A. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y., Rom W. N. Regulation of the interleukin-1β (IL-1β) gene by mycobacterial components and lipopolysaccharide is mediated by two nuclear factor-IL6 motifs. Mol. Cell. Biol. 1993;13:3831–3837. doi: 10.1128/mcb.13.6.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Natsuka S., Akira S., Nishio Y., Hashimoto S., Sugita T., Isshiki H., Kishimoto T. Macrophage differentiation-specific expression of NF-IL6, a transcription factor for interleukin-6. Blood. 1992;79:460–466. [PubMed] [Google Scholar]
- 44.Ruddy M. J., Wong G. C., Liu X. K., Yamamoto H., Kasayama S., Kirkwood K. L., Gaffen S. L. Functional cooperation between interleukin-17 and tumor necrosis factor-α is mediated by CCAAT/enhancer-binding protein family members. J. Biol. Chem. 2004;279:2559–2567. doi: 10.1074/jbc.M308809200. [DOI] [PubMed] [Google Scholar]
- 45.Shen F., Hu Z., Goswami J., Gaffen S. L. Identification of common transcriptional regulatory elements in interleukin-17 target genes. J. Biol. Chem. 2006;281:24138–24148. doi: 10.1074/jbc.M604597200. [DOI] [PubMed] [Google Scholar]
- 46.Shen F., Ruddy M. J., Plamondon P., Gaffen S. L. Cytokines link osteoblasts and inflammation: microarray analysis of interleukin-17- and TNF-α-induced genes in bone cells. J. Leukocyte Biol. 2005;77:388–399. doi: 10.1189/jlb.0904490. [DOI] [PubMed] [Google Scholar]