Abstract
Centerin [SERPINA9/GCET1 (germinal centre B-cell-expressed transcript 1)] is a serpin (serine protease inhibitor) whose expression is restricted to germinal centre B-cells and lymphoid malignancies with germinal centre B-cell maturation. Expression of centerin, together with bcl-6 and GCET2, constitutes a germinal centre B-cell signature, which is associated with a good prognosis in diffuse large B-cell lymphomas, but the molecular basis for this remains to be elucidated. We report here the cloning, expression and molecular characterization of bacterial recombinant centerin. Biophysical studies demonstrated that centerin was able to undergo the ‘stressed to relaxed’ conformational change which is an absolute requirement for protease inhibitory activity. Kinetic analysis showed that centerin rapidly inhibited the serine protease trypsin (ka=1.9×105 M−1·s−1) and also demonstrated measurable inhibition of thrombin (ka=1.17×103 M−1·s−1) and plasmin (ka=1.92×103 M−1·s−1). Centerin also bound DNA and unfractionated heparin, although there was no functionally significant impact on the rate of inhibition. These results suggest that centerin is likely to function in vivo in the germinal centre as an efficient inhibitor of a trypsin-like protease.
Keywords: centerin, germinal centre B-cell-expressed transcript 1 (GCET1), lymphoma, protease, serine protease inhibitor (serpin), trypsin
Abbreviations: DLBCL, diffuse large B-cell lymphoma; GAG, glycosaminoglycan; GCET1, germinal centre B-cell-expressed transcript 1; HRP, horseradish peroxidase; MENT, myeloid and erythroid nuclear termination stage-specific protein; PEG, poly(ethylene glycol); RCL, reactive centre loop; serpin, serine protease inhibitor; SI, stoichiometry of inhibition; Z, benzyloxycarbonyl
INTRODUCTION
Centerin, also known as GCET1 (germinal centre B-cellexpressed transcript 1) or SERPINA9, is a serpin (serine protease inhibitor) expressed by germinal centre B-cells and lymphoid malignancies with germinal centre B-cell maturation [1,2].
Centerin, together with bcl-6 and GCET2, constitutes a germinal centre B-cell signature identified in molecular profiling studies of DLBCL (diffuse large B-cell lymphoma) [3]. This gene expression pattern, compared with other signatures (activated B-cell and Type III), was associated with a greater rate of overall survival after chemotherapy [3].
Centerin's expression is specific to germinal centre B-cells and germinal centre B-cell-derived lymphomas [1,2]. Northern-blot analysis has shown no expression in heart, brain, placenta, lung, liver, skeletal muscle, kidney and pancreas [1]. Expression analysis of a range of human cancer cell lines only identified centerin in Raji (Burkitt's lymphoma), HL-60 (promyelocytic) [1], DHL16 (activated DLBCL cell) [2] and Nalm-6 (pre-B-cell) [2] cells.
The centerin gene maps to the A clade serpin cluster on chromosome 14q32.1, which also contains α1-antitrypsin and α1-antichymotrypsin together with seven other serpins [4]. The centerin amino acid sequence contains motifs typical of an active serpin, including a serpin signature sequence and RCL (reactive centre loop) hinge region suggestive of functional inhibitory activity. The predicted active-site scissile bond (Arg–Ser) indicates that centerin should preferentially inhibit trypsin-like serine proteases, but currently there is no published data on a physiological target or on the serpin's role in germinal centre biology.
The studies using recombinant centerin presented here show that it has the biophysical characteristics of an inhibitory serpin. Furthermore, we have shown that centerin inhibits the trypsin-like serine proteases trypsin, thrombin and plasmin and is able to bind heparin and DNA.
MATERIALS AND METHODS
Reagents
Restriction enzymes and T4 DNA ligase were purchased from New England Biolabs (Genesearch, Arundel, QLD, Australia). Bovine trypsin, bovine chymotrypsin, human thrombin and bovine Factor Xa were purchased from Sigma–Aldrich (Castle Hill, NSW, Australia). Human leucocyte elastase and human leucocyte cathepsin G were obtained from Athens Research Technology (Athens, GA, U.S.A.) and human plasmin was purchased from Haematologic Technologies (Dural, NSW, Australia). Recombinant human cathepsins L and V and recombinant MENT (myeloid and erythroid nuclear termination stage-specific protein) were obtained from Poh Chee Ong and Wei Wan Dai (Department of Biochemistry and Molecular Biology, Monash University). Unless otherwise stated, all general chemicals were purchased from Sigma–Aldrich. Colorimetric and fluorogenic assays were performed using a Victor 2 1420 multilabel counter (PerkinElmer, Rowville, VIC, Australia).
Cloning, protein expression and purification of recombinant centerin
RNA was extracted from Burkitt's Lymphoma Raji cells using the Qiagen RNeasy kit (Doncaster, VIC, Australia). Total RNA (2 μg) was reverse transcribed using Superscript III (Invitrogen, Mt Waverley, VIC, Australia) with 5 pmol/l oligo(dT) to obtain Raji cDNA. Primers were designed for centerin according to its published sequence [1]. The sense primer was designed to anneal 5′ of the first α-helix, excluding the secretion signal and a GC-rich region of sequence. A BamHI restriction tail was added to facilitate cloning (GGATCCTCCACAAAGAGCACCCCT). The antisense primer was designed to anneal 3′ to the stop codon (GAAATGGCCTGTTAACTGATGGGATCC). Centerin cDNA was amplified from the Raji cDNA using the above primers with Platinum-pfx polymerase (Invitrogen). The centerin cDNA was then subcloned into the BamHI restriction site of the pET-His(3a) bacterial expression vector (Novagen, San Diego, CA, U.S.A.) to produce an N-terminally hexahistidine-tagged protein. Correct orientation and sequence were confirmed by nucleotide sequencing. pET-His(3a)-centerin was transformed into Escherichia coli host BL21(DE3) pLysS cells and recombinant protein production was induced with 0.2 mM IPTG (isopropyl β-D-thiogalactoside) during the late-exponential growth phase for 4 h at 37 °C. Cells were harvested by centrifugation (1500 g) and resuspended in lysis buffer (50 mM sodium phosphate, pH 7.2, 100 mM NaCl and 10 mM imidazole) to which 1 mg/ml lysozyme, 20 μg/ml PMSF, protease inhibitor cocktail (Sigma, Castle Hill, NSW, Australia) (1:1000), 25 μg/ml DNase and 5 mM 2-mercaptoethanol were added. The lysate was incubated on ice for 30 min. The cells were freeze/thawed three times in liquid nitrogen and a 37 °C water bath. Soluble protein was then obtained after centrifugation (35000 g). Soluble protein was bound to a nickel-HisTrap column (GE Healthcare, Castle Hill, NSW, Australia) in 50 mM sodium phosphate (pH 7.2), 100 mM NaCl, 10 mM imidazole and 5 mM 2-mercaptoethanol and His–centerin was eluted using a 0.2 M imidazole step gradient. The protein was further purified using Mono S ion-exchange chromatography (GE Healthcare) with a 0.125-1 M NaCl gradient in 20 mM Hepes (pH 7.0) and 5 mM 2-mercaptoethanol.
CD analysis
CD analysis was performed on a Jasco 820s spectropolarimeter (Jasco, Easton, MD, U.S.A.). Changes in protein secondary structure were monitored by measuring the change in ellipticity at 222 nm using a 0.05 cm-path-length cuvette and a protein concentration of 20 μg/ml in 20 mM Hepes (pH 7.0) and 0.2 M NaCl. Thermal unfolding experiments were performed by heating at a rate of 1 °C/min from 25 to 90 °C. Native His–centerin and trypsin RCL-cleaved His–centerin were analysed. RCL-cleaved His–centerin was obtained by incubating His–centerin and trypsin at a molar ratio of 2.5:1 at 37 °C for 30 min prior to addition of PMSF (1:1000) and separation of His–centerin from trypsin on mono-S ion-exchange chromatography. RCL cleavage was confirmed by SDS/PAGE.
MS
Purified protein (50 μg) was sent to the Department of Biochemistry and Molecular Biology, Monash University to be analysed by MALDI (matrix-assisted laser-desorption ionization) MS by Dr Shane Reeve.
Centerin binding to DNA
Linear pHilD2 DNA (1.5 μg) was incubated with increasing concentrations of recombinant His–centerin (0–20 μg) at room temperature (22 °C) for 20 min in 20 mM Hepes (pH 7.0) and 0.2 M NaCl. DNA (1.5 μg) was also incubated with 5 μg of His–centerin in the presence of increasing concentrations of NaCl (0–0.3 M) at room temperature for 20 min. DNA loading dye (5×) was then added to the reaction and this was separated on a 1% agarose gel by electrophoresis. The DNA was visualized with ethidium bromide and UV light.
GAG (glycosaminoglycan) binding to His–centerin
His–centerin (5 μg) was incubated with unfractionated heparin (0–50 units/ml) (Pfizer, Perth, WA, Australia) or chondroitin sulfate (0–10 μg/ml) (Sigma–Aldrich) at room temperature for 20 min in 20 mM Hepes (pH 7.0) and 0.2 M NaCl. Then 5× acid-native PAGE loading dye was added to the samples prior to running them on a 10% acid-native PAGE gel at 80 V [5]. Gels were then stained with Coomassie Blue.
Heparin–agarose column
His–centerin (200 μg) was bound to a heparin–agarose column (GE Healthcare) in 10 mM sodium phosphate (pH 7.4). Centerin was eluted with a 0.125–1 M NaCl gradient.
Complex formation assays
The ability of His–centerin to inhibit the serine proteases bovine chymotrypsin and trypsin, human cathepsin G, human leucocyte elastase, human plasma thrombin, bovine Factor Xa and human plasmin was investigated by SDS/PAGE. Each serine protease (1 μM) was incubated with either 1 or 5 μM His–centerin in 25 mM Tris (pH 8.0), 100 mM NaCl and 10 mM CaCl2. Reactions were incubated at 37 °C for 30 min and then placed on ice before SDS/PAGE reduced loading buffer was added. The ability of His–centerin to inhibit the cysteine proteases cathepsins L and V was investigated by SDS/PAGE as described previously [6]. Briefly, 1 μM of each cysteine protease was incubated with either 1 or 5 μM His–centerin in 0.1 M acetate (pH 5.5), 1 mM EDTA, 0.1% (w/v) Brij-35 and 10 mM cysteine. A control reaction of recombinant His–MENT and cathepsin L at 1:8 molar ratio was set up in parallel. Reactions were incubated at 37 °C for 30 min and then placed on ice before SDS/PAGE reducing buffer was added. A low-pH reducing buffer was added to the cysteine proteases [0.125 M Tris/HCl, pH 4.3, 4% (w/v) SDS, 20% (v/v) glycerol, 0.2 M dithiothreitol and 0.02% Bromophenol Blue]. Samples were then denatured at 100 °C for 5 min before being separated on reducing SDS/12.5% PAGE and being transferred to PVDF membrane (Millipore, North Ryde, NSW, Australia) by Western blotting. Membranes were then probed with mouse anti-His antibody (1:2000) (Qiagen) and goat anti-mouse-HRP (horseradish peroxidase) (Chemicon, North Ryde, NSW, Australia) (1:5000). Proteins were detected with chemiluminescent substrate (Pierce, Griffith, NSW, Australia) and visualized by exposure to Curix Ortho HT-G film (Agfa, Wayville, SA, Australia).
SI (stoichiometry of inhibition)
SI values were determined for the interaction of thrombin, plasmin and trypsin with His–centerin. His–centerin (0–1 μM) was incubated with a constant concentration of thrombin (200 nM) at 37 °C for 6 h and residual activity was assayed at 405 nm with the colorimetric substrate S2238 (Helena Laboratories, Beaumont, TX, U.S.A.) on standard microtitre plates. This was performed in 50 mM Tris (pH 7.4), 150 mM NaCl and 0.2% PEG [poly(ethylene glycol)] 3350. Inhibition of plasmin (200 nM) was titrated with increasing concentrations of His–centerin (0–1 μM) at 37 °C for 6 h and the residual activity was assayed at 405 nm with the colorimetric substrate S2251 (Helena Laboratories) on standard microtitre plates. This was performed in 20 mM Tris (pH 7.5), 150 mM NaCl and 0.01% Tween 80. Trypsin (200 nM) was incubated with increasing concentrations of His–centerin (0–800 nM) at 37 °C for 30 min and residual activity was assayed at 355 nm/460 nm with the fluorogenic substrate Z (benzyloxycarbonyl)-Phe-Arg-7-amido-4-methylcoumarin hydrochloride (Sigma) on fluorogenic microtitre plates (PerkinElmer). This was performed in 25 mM Tris (pH 8.0), 100 mM NaCl, 10 mM CaCl2 and 0.2% PEG 3350. Following linear regression analysis of the plot of residual enzyme activity against serpin concentration, the SI was determined by extrapolating the protease/serpin ratio to the point where protease activity is zero. The SI value represents the average of two to three separate experiments. Some reactions were performed in the presence of 1–5 units/ml unfractionated heparin or 20 μg/ml chondroitin sulfate.
Association rate constant: discontinuous method
A discontinuous method [7] was used to determine the rate of inhibition (ka) of thrombin, plasmin and trypsin by His–centerin. The pseudo-first-order rate constant with thrombin (2 nM) and His–centerin (60–160 nM) was determined by incubation for different periods of time (0–2 h), followed by measurement of residual thrombin activity at 405 nm with the colorimetric substrate S2238 (Helena Laboratories) on standard microtitre plates. The pseudo-first-order rate constant with plasmin (2.5 nM) and His–centerin (20–62.5 nM) was determined by incubation for different periods of time (0–1 h) followed by measurement of residual plasmin activity at 405 nm with the colorimetric substrate S2251 (Helena Laboratories) on standard microtitre plates. The pseudo-first-order rate constant with trypsin (2 nM) and His–centerin (10–16 nM) was determined by incubation for different periods of time (0–3 min) followed by measurement of residual trypsin activity at 355 nm/460 nm with the fluorogenic substrate Z-Phe-Arg-7-amido-4-methylcoumarin hydrochloride (Sigma) on fluorogenic microtitre plates. The pseudo-first-order constant, kobs, was determined from the slope of a semi-logarithmic plot of the residual protease activity against time. The kobs values were then plotted against serpin concentration and the slope of the line of best fit gave an estimate of the second-order rate constant, ka. The ka values represent the average of two to three separate experiments. Some reactions were performed in the presence of 1–5 units/ml unfractionated heparin or 20 μg/ml chondroitin sulfate.
Time course of inhibition of trypsin, plasmin and thrombin
His–centerin (20 nmol) was incubated with 2 nM trypsin (10:1, serpin/protease molar ratio), over a time course of 240, 180, 120, 60, 30, 10, 5 and 0 min, followed by measurement of residual trypsin activity at 355 nm/460 nm with the fluorogenic substrate Z-Phe-Arg-7-amido-4-methylcoumarin hydrochloride (Sigma) on fluorogenic microtitre plates. His–centerin (40 nmol) was incubated with 2 nM thrombin (20:1, serpin/protease molar ratio), over a time course of 240, 180, 120, 60, 30, 10, 5 and 0 min, followed by measurement of residual thrombin activity at 405 nm with the colorimetric substrate S2238 (Helena Laboratories) on standard microtitre plates. His–centerin (40 nmol) was incubated with 2 nM plasmin (20:1, serpin/protease molar ratio), over a time course of 240, 180, 120, 60, 30, 10, 5 and 0 min, followed by measurement of residual plasmin activity at 405 nm with the colorimetric substrate S2251 (Helena Laboratories) on standard microtitre plates.
RESULTS
Expression and purification of His–centerin
Centerin cDNA was amplified from reverse transcribed RNA isolated from Raji cells. The 5′-end of cDNA corresponded to residue 33, 18 residues N-terminal to the predicted first α-helix. Nucleotide sequencing of the centerin cDNA isolated from Raji cells agreed with the published sequence [1], except for Ala348 →Val. This variation is conserved in other members of the serpin family and is therefore unlikely to affect the structural or functional properties of the protein. The centerin cDNA was inserted into the vector pET His and expressed in BL21(DE3) pLysS cells.
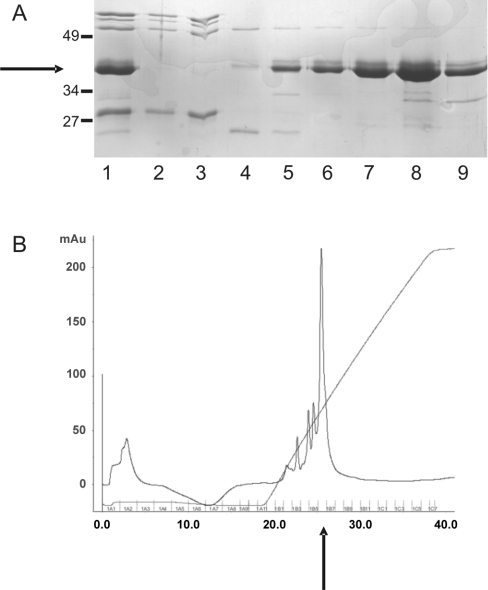
Crude bacterial extract was chromatographed on a nickel-HisTrap column and eluted with an imidazole step gradient yielding a partially pure preparation of His–centerin (Figure 1A, lane 1). Pooled fractions from the nickel-HisTrap column eluate were further purified by mono S ion-exchange chromatography. This produced protein of approx. 90% purity (Figures 1A, lane 4–9, and 1B). A final yield of 4 mg of pure recombinant His–centerin was generated from 5 litres of culture. The identity of His–centerin was confirmed by Western-blot analysis using an antibody to the His tag (results not shown) and MS analysis showed the recombinant protein had a molecular mass of 43.9 kDa as predicted from the amino acid sequence.
Figure 1. Purification of His–centerin.
Bacterial lysate containing recombinant His–centerin was chromatographed on a nickel-HisTrap column and the pooled eluate (A; lane 1) was then applied to a mono-S cation-exchange column. (A) The Figure shows flow through (lanes 2 and 3) and fractions eluted with a 0.125–1 M NaCl gradient (lanes 4–9) (Coomassie-stained SDS/PAGE gel). Recombinant His–centerin is arrowed on left. Molecular mass standards are shown to the left of the Figure. Sizes are in kDa. (B) The Figure shows a protein elution profile [A280 (absorbance)] from mono-S cation-exchange chromatography step. The major protein peak (arrowed) corresponds to fraction 8 (A).
Biophysical characterization of His–centerin
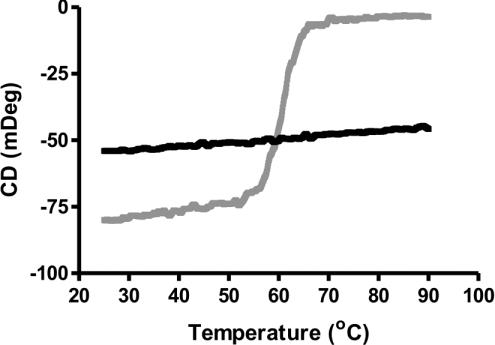
To assess whether His–centerin has the biophysical characteristics of an inhibitory serpin, thermal denaturation studies were performed. Figure 2 (grey line) shows thermal denaturation of His–centerin demonstrating a sigmoidal unfolding pattern, and a melting temperature of 61.5 °C, typical of a serpin in its native conformation [8]. RCL-cleaved His–centerin analysed by thermal denaturation showed a loss of the sigmoidal unfolding transition, which is indicative of RCL insertion into the serpin A-β-sheet (Figure 2, black line) [9]. These results demonstrated that recombinant His–centerin was able to undergo the stressed to relaxed conformational transition, which is an essential feature of the serpin inhibitory mechanism [9].
Figure 2. CD analysis of native and RCL-cleaved His–centerin.
Thermal unfolding profile of native His–centerin (grey line) and RCL-cleaved His–centerin (black line) was monitored using far-UV CD at 222 nm with a heating rate of 1 °C/min.
GAG/DNA binding to His–centerin
The calculated pI of centerin is 9.62, giving it a strong net positive charge at physiological pH. We therefore investigated the ability of His–centerin to bind to negatively charged entities such as DNA and GAGs.
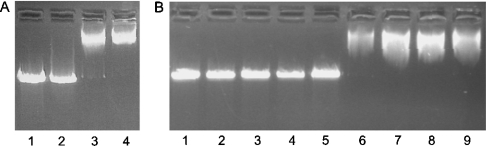
Increasing concentrations of His–centerin were incubated with linearized double-stranded DNA and then electrophoresed on an agarose gel (Figure 3A). Lanes 2–4 show that 10 and 20 μg of His–centerin significantly retarded the migration of DNA into the gel compared with the buffer control (Figure 3A, lane 1). The strength of the His–centerin–DNA interaction was investigated by performing the gel retardation assay in the presence of increasing ionic strength. Figure 3(B) (lanes 6–9) show that retardation of migration of the DNA by His–centerin occurred in NaCl concentrations up to 300 mM, suggesting a high-affinity interaction.
Figure 3. DNA binding to His–centerin.
(A) The Figure shows a gel retardation assay in which 1.5 μg of linearized plasmid DNA was incubated with 0 μg (lane 1), 2 μg (lane 2), 10 μg (lane 3) and 20 μg (lane 4) of His–centerin for 20 min prior to 1% agarose gel electrophoresis and visualization using ethidium bromide. (B) The Figure shows the effects of salt concentration on binding of His–centerin to DNA. Lane 1 shows 1.5 μg of DNA alone. Lanes 2–5 contain 1.5 μg of DNA incubated with increasing concentrations of NaCl (lane 2, 160 mM; lane 3, 200 mM; lane 4, 250 mM; and lane 5, 300 mM). Lanes 6–9 show 1.5 μg of DNA incubated with 5 μg of His–centerin in the presence of increasing concentrations of NaCl (lane 6, 160 mM; lane 7, 200 mM; lane 8, 250 mM; and lane 9, 300 mM).
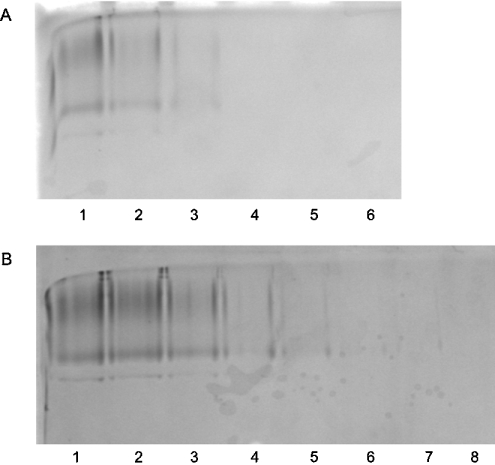
The ability of His–centerin to bind GAGs was also investigated using a gel retardation assay on acid-native PAGE. Figure 4(A) shows the effect of incubating His–centerin with increasing concentrations of unfractionated heparin from 0 to 50 units/ml (lanes 1–6). At the lowest concentration of heparin (lane 2) there was already some loss of intensity of the His–centerin band. At heparin concentrations from 10 units/ml (lane 4) there was complete loss of His–centerin from the gel, indicating that a His–centerin–heparin complex forms, but fails to enter the gel. Direct binding of His–centerin to heparin was demonstrated by applying the protein to a heparin–agarose column and elution with a linear sodium chloride gradient. The serpin was eluted at 518 mM NaCl. His–centerin binding to the GAG, chondroitin sulfate, was also investigated by gel retardation on acid-native PAGE (Figure 4B) with similar results. As the concentration of chondroitin sulfate was increased from 0 to 10 μg/ml there was a progressive decrease in intensity of the His–centerin band (Figure 4B, lanes 2–8). These results are again consistent with the formation of a His—centerin–chondroitin sulfate complex.
Figure 4. Unfractionated heparin/chondroitin sulfate binding to His–centerin.
Gel retardation assays using acid-native PAGE are shown. (A) His–centerin (10 μg) was incubated with 0, 2.5, 5, 10, 20 and 50 units/ml (lanes 1–6 respectively) unfractionated heparin. (B) His–centerin (10 μg) was incubated with 0, 2, 4, 6, 8, 10, 12 and 14 μg/ml (lanes 1–8 respectively) chondroitin sulfate.
Inhibitory activity of His–centerin
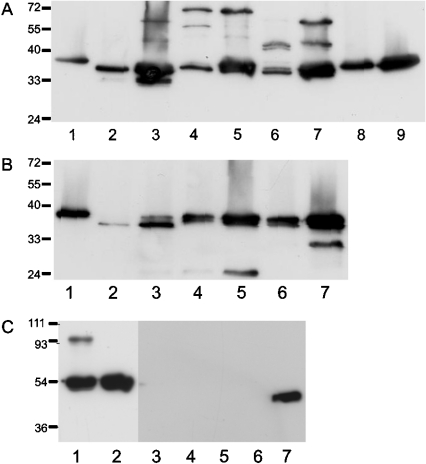
The physiological target protease of centerin in the germinal centre is unknown, although the predicted P1–P1′ residues suggest that it is a trypsin-like enzyme. To confirm this prediction, His–centerin was incubated with a panel of trypsin-like proteases including trypsin, thrombin, plasmin, urokinase and Factor Xa, together with other serine proteases including chymotrypsin, neutrophil elastase and cathepsin G and the cysteine proteases cathepsin L and cathepsin V. The formation of a covalent complex between His–centerin and protease was detected by a band shift on SDS/12.5% PAGE approximating to the molecular mass of His–centerin plus the protease. Figure 5(A) shows that His–centerin formed a complex with trypsin (lane 3), plasmin (lanes 4 and 5) and thrombin (lanes 6 and 7) at 1:1 and/or 5:1 molar ratios. Incubation of His–centerin at equimolar concentrations with chymotrypsin (Figure 5B, lanes 2 and 3) resulted in complete degradation of the serpin, while at a ratio of 5:1 (serpin/enzyme), there was a decrease in apparent molecular mass of the serpin, suggestive of RCL cleavage. Similarly, incubation of His–centerin with either cathepsin G or neutrophil elastase at 1:1 or 5:1 molar ratio (Figure 5B, lanes 4–7) resulted in a small decrease in the molecular mass of the serpin compared with control (Figure 5B, lane 1), consistent with RCL cleavage. Incubation of His–centerin with Factor Xa (Figure 5A, lanes 8 and 9) appeared to have no effect on the serpin. Incubation of His–centerin with the cysteine proteases, cathepsin L or V (Figure 5C, lanes 3–6), resulted in complete degradation of the serpin.
Figure 5. Complex formation between His–centerin and a panel of proteases.
His–centerin was incubated with a panel of proteases at either 1:1 or 5:1 molar ratio (centerin/enzyme) for 30 min at 37 °C and then electrophoresed on SDS/PAGE and Western blotted. His–centerin was detected using anti-His monoclonal antibody and goat anti-mouse HRP-conjugated secondary antibody. (A) The Figure shows the results of incubations of His–centerin with trypsin-like proteases: lane 1, His–centerin with no protease; lane 2, trypsin 1:1; lane 3, trypsin 5:1; lane 4, thrombin 1:1; lane 5, thrombin 5:1; lane 6, plasmin 1:1; lane 7, plasmin 5:1; lane 8, Factor Xa 1:1; lane 9, Factor Xa 5:1. (B) The Figure shows the results of incubations of His–centerin with other serine proteases: lane 1, His–centerin with no protease; lane 2, chymotrypsin 1:1; lane 3, chymotrypsin 5:1; lane 4, cathepsin G 1:1; lane 5, cathepsin G 5:1; lane 6, elastase 1:1; lane 7, elastase 5:1. (C) The Figure shows results obtained using cysteine proteases. Lanes 1 and 2 show a positive control using the serpin MENT, which forms an SDS-stable complex under the conditions of this assay: lane 1, MENT-cathepsin L; lane 2, MENT alone. Lanes 3–7 show the results of incubations of His–centerin with cathepsins L and V at either 1:1 or 5:1 molar ratio (serpin/protease). Lane 3, cathepsin L 1:1; lane 4, cathepsin L 5:1; lane 5, cathepsin V 1:1; lane 6, cathepsin V 5:1; lane 7, His–centerin with no protease.
Inhibition kinetics of His–centerin
As His–centerin formed SDS/PAGE stable complexes with trypsin, thrombin and plasmin, we proceeded to perform kinetic analysis of the interaction. Measurement of the SI of His–centerin demonstrated detectable inhibitory activity against thrombin and plasmin with values of 2.57 and 1.72 respectively (Table 1). The stability of the inhibitory complex of centerin with thrombin or plasmin was assessed over 3 h by measuring residual proteolytic activity. This demonstrated progressive loss of activity (Figure 6). The association rate constant (ka) of His–centerin with these proteases was measured under pseudo-first-order conditions using a discontinuous method and the results are shown in Table 1. The ka values for the interaction of His–centerin with thrombin and plasmin were 1.17×103 and 1.92×103 M−1·s−1 respectively.
Table 1. Comparison of inhibitory kinetics of His–centerin with thrombin, plasmin and trypsin, with or without GAGs.
UH, unfractionated heparin; CS, chondroitin sulfate.
| Protease | Condition | n | SI | ka (M−1·s−1) | ka×SI |
|---|---|---|---|---|---|
| Thrombin | Control | 3 | 2.57±0.04 | 1.17±0.094×103 | 3.00×103 |
| UH (1 unit/ml) | 3 | 3.83±0.01 | 5.21±0.460×103 | 2.0×104 | |
| UH (2.5 units/ml) | 3 | 3.74±0.10 | 1.14±0.175×104 | 4.26×104 | |
| UH (5 units/ml) | 3 | 3.55±0.14 | 1.45±0.027×104 | 5.15×104 | |
| CS (20 μg/ml) | 3 | 3.15±0.15 | 2.36±0.072×103 | 7.43×103 | |
| Plasmin | Control | 3 | 1.72±0.05 | 1.92±0.18×103 | 3.30×103 |
| UH (10 units/ml) | 2 | 2.87±0.21 | 1.54±0.05×103 | 4.42×103 | |
| Trypsin | Control | 3 | 2.12±0.09 | 1.90±0.06×105 | 4.02×105 |
| UH (10 units/ml) | 2 | 3.23±0.16 | 1.76±0.14×105 | 5.68×105 |
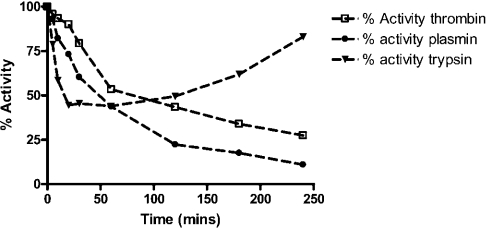
Figure 6. Time course of trypsin, thrombin and plasmin inhibition by centerin.
His–centerin (20 nmol) was incubated with trypsin (molar ratio of 10:1, centerin/trypsin) and residual trypsin (▼) activity was measured at intervals, as shown, over 240 min. His–centerin (40 nmol) was incubated with plasmin or thrombin (molar ratio of 20:1, centerin/plasmin or centerin/thrombin) and residual plasmin (●) or thrombin (□) activity was measured at intervals, as shown, over 240 min.
When the SI for trypsin was measured, we observed that there was an initial rapid loss of proteolytic activity, followed by its reappearance (Figure 6). Using a shortened incubation time (30 min) the SI was calculated to be 2.12 and the ka value was 1.9×105 M−1·s−1 (Table 1).
Since the ka value for the association of His–centerin with thrombin and plasmin was slow, we investigated the possibility that the interactions were accelerated by heparin. The effect of GAGs on the kinetics of the inhibitory interactions of His–centerin with thrombin and plasmin are shown in Table 1. In the presence of 2.5 units/ml unfractionated heparin the ka value for the His–centerin–thrombin interaction was accelerated 10-fold (1.17×103 to 1.14×104 M−1·s−1). Unfractionated heparin had no affect on the inhibition of plasmin or trypsin. In the presence of 20 μg/ml chondroitin sulfate the ka value for His–centerin–thrombin interaction was approximately doubled (1.17×103 to 2.36×103 M−1·s−1).
DISCUSSION
We have presented the results of biochemical and biophysical studies using an N-terminally 6×His-tagged bacterial recombinant His–centerin. To ensure that the recombinant protein was authentically folded, we performed biophysical analysis that demonstrated that it undergoes the stressed to relaxed conformational change upon RCL cleavage, a prerequisite for protease inhibitory activity [10,11]. Complex formation assays on SDS/PAGE demonstrated that His–centerin bound thrombin, plasmin and trypsin. Kinetic analysis showed that His–centerin was an inefficient inhibitor of plasmin and thrombin, but was a rapid inhibitor of trypsin, with a physiologically relevant association rate constant of 1.9×105 M−1·s−1. Examination of the complex formation assay with His–centerin and trypsin demonstrated that as well as complex formation there was also significant RCL cleavage occurring. It was also noted that prolonged incubation of His–centerin with trypsin resulted in significant regeneration of enzyme activity. The most likely explanation for these observations is that His–centerin forms an inhibitory complex with trypsin leading to RCL cleavage and loop insertion, but without significant deformation of the protease; over time, hydrolysis of the RCL is completed, giving rise to release of the active enzyme and cleaved inhibitor. This has previously been observed with other serpins and strongly suggests that trypsin is not the physiological target for centerin [12,13]. The slow rate of inhibition of thrombin and plasmin also suggests that they are not the physiological target proteases.
Centerin is unusual among serpins because of its high pI (pI 9.62). This suggested the possibility of binding to negatively charged entities such as GAGs and DNA and we were able to demonstrate this directly using retardation assays in electrophoretic gels. However, when DNA was added to protease inhibition assays it did not enhance the rate (results not shown). Similarly, the addition of unfractionated heparin had no effect on the rate of inhibition of plasmin and trypsin, but there was a modest increase in the rate of inhibition of thrombin (ka value increased 10-fold). It is known that thrombin possesses a heparin-binding exosite [14], while trypsin and plasmin do not. When taken together this suggests that the acceleration of thrombin inhibition by His–centerin in the presence of heparin is most likely related to a template effect rather than conformation change in the serpin.
The significance of centerin's high pI and its binding to ligands such as DNA and heparin is unclear. The only other member of the superfamily with a similarly high pI is the intracellular serpin, MENT, which binds DNA and plays a role in packing of heterochromatin [15]. It has been observed that the centerin transcript gives rise to three splice variants in addition to the full-length form. Two of the observed splice variants are predicted to be intracellular and would therefore have the potential for interaction with either DNA or RNA [1,2]. The binding of His–centerin to GAGs may well be of significance for the secreted forms of the protein. Even if these ligands have no effect on the rate of inhibition of target proteases, it is likely that the avid binding of centerin to matrix-associated GAGs would lead to pericellular localization of the serpin with implications for local control of proteolysis.
Our results demonstrate that centerin has the potential to be a rapid and effective protease inhibitor in vivo. Its ability to bind to either DNA or GAGs raises the possibility of localization of that activity, either within or around the cell, depending on whether it is secreted or not. It is known that germinal centre B-cells are mobile, migrating into and out of the follicle, while undergoing somatic hypermutation and maturation [16]. It is likely that centerin is involved in controlling pericellular proteolytic activity and thereby regulates germinal centre B-cell movement. Possible target proteases for centerin would include members of the membrane-anchored serine protease family, which have trypsin-like activity [17]. This could explain why centerin expression confers a favourable prognosis in large-cell lymphomas, as protease inhibition would tend to immobilize malignant cells, possibly making them more susceptible to control by local negative regulatory factors such as cytokines, cell–cell contacts and apoptotic stimuli. Further progress in understanding the biology of centerin in germinal centre will depend on identification of its target protease, and this work is ongoing in our laboratory.
References
- 1.Frazer J. K., Jackson D. G., Gaillard J. P., Lutter M., Liu Y. J., Banchereau J., Capra J. D., Pascua V. Identification of centerin: a novel human germinal centre B cell-restricted serpin. Eur. J. Immunol. 2000;30:3039–3048. doi: 10.1002/1521-4141(200010)30:10<3039::AID-IMMU3039>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 2.Pan Z., Shen Y., Du C., Zhou G., Rosenwald A., Staudt L., Greiner T., McKeithan T., Chan W. Two newly characterised germinal centre B-cell-associated genes, GCET1 and GCET2, have differential expression in normal and neoplastic B cells. Am. J. Pathol. 2003;163:135–144. doi: 10.1016/S0002-9440(10)63637-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenwald A., Wright G., Chan W., Connors J., Campo E., Fisher R. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N. Eng. J. Med. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 4.Forsyth S., Horvath A., Coughlin P. A review and comparison of the murine alpha(1)-antitrypsin and alpha(1)-antichymotrypsin multigene clusters with the human clade A serpins. Genomics. 2003;81:336–345. doi: 10.1016/s0888-7543(02)00041-1. [DOI] [PubMed] [Google Scholar]
- 5.Zhou A., Faint R., Charlton P., Dafforn T. R., Carrell R. W., Lomas D. A. Polymerization of plasminogen activator inhibitor-1. J. Biol. Chem. 2001;276:9115–9122. doi: 10.1074/jbc.M010631200. [DOI] [PubMed] [Google Scholar]
- 6.Irving J. A., Shushanov S. S., Pike R. N., Popova E. Y., Bromme D., Coetzer T. H., Bottomley S. P., Boulynko I. A., Grigoryev S. A., Whisstock J. C. Inhibitory activity of a heterochromatin-associated serpin (MENT) against papain-like cysteine proteinases affects chromatin structure and blocks cell proliferation. J. Biol. Chem. 2002;277:13192–13201. doi: 10.1074/jbc.M108460200. [DOI] [PubMed] [Google Scholar]
- 7.Bjork I., Olson S. T., Shore J. D. Kinetic characterization of heparin-catalyzed and uncatalyzed inhibition of blood coagulation proteinases by antithrombin. Methods Enzymol. 1993;222:525–559. doi: 10.1016/0076-6879(93)22033-c. [DOI] [PubMed] [Google Scholar]
- 8.Gooptu B., Hazes B., Chang W. S., Dafforn T. R., Carrell R. W., Read R. J., Lomas D. A. Inactive conformation of the serpin alpha(1)-antichymotrypsin indicates two-stage insertion of the reactive loop: implications for inhibitory function and conformational disease. Proc. Natl. Acad. Sci. U.S.A. 2000;97:67–72. doi: 10.1073/pnas.97.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whisstock J. C., Skinner R., Carrell R. W., Lesk A. M. Conformational changes in serpins: I. The native and cleaved conformations of alpha(1)-antitrypsin. J. Mol. Biol. 2000;295:651–665. doi: 10.1006/jmbi.1999.3375. [DOI] [PubMed] [Google Scholar]
- 10.Silverman G. A., Bird P. I., Carrell R. W., Church F. C., Coughlin P. B., Gettins P. G., Irving J. A., Lomas D. A., Luke C. J., Moyer R. W., et al. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J. Biol. Chem. 2001;276:33293–33296. doi: 10.1074/jbc.R100016200. [DOI] [PubMed] [Google Scholar]
- 11.Gettins P. G. Serpin structure, mechanism, and function. Chem. Rev. 2002;102:4751–4804. doi: 10.1021/cr010170+. [DOI] [PubMed] [Google Scholar]
- 12.Barker-Carlson K., Lawrence D. A., Schwartz B. S. Acyl-enzyme complexes between tissue-type plasminogen activator and neuroserpin are short-lived in vitro. J. Biol. Chem. 2002;277:46852–46857. doi: 10.1074/jbc.M207740200. [DOI] [PubMed] [Google Scholar]
- 13.Shieh B. H., Potempa J., Travis J. The use of α2-antiplasmin as a model for the demonstration of complex reversibility in serpins. J. Biol. Chem. 1989;264:13420–13423. [PubMed] [Google Scholar]
- 14.Carter W. J., Cama E., Huntington J. A. Crystal structure of thrombin bound to heparin. J. Biol. Chem. 2005;280:2745–2749. doi: 10.1074/jbc.M411606200. [DOI] [PubMed] [Google Scholar]
- 15.Grigoryev S. A., Bednar J., Woodcock C. L. MENT, a heterochromatin protein that mediates higher order chromatin folding, is a new serpin family member. J. Biol. Chem. 1999;274:5626–5636. doi: 10.1074/jbc.274.9.5626. [DOI] [PubMed] [Google Scholar]
- 16.Tarlinton D. Germinal centres: form and function. Curr. Opin. Immunol. 1998;10:245–251. doi: 10.1016/s0952-7915(98)80161-1. [DOI] [PubMed] [Google Scholar]
- 17.Szabo R., Wu Q., Dickson R. B., Netzel-Arnett S., Antalis T. M., Bugge T. H. Type II transmembrane serine proteases. Thromb. Haemostasis. 2003;90:185–193. doi: 10.1160/TH03-02-0071. [DOI] [PubMed] [Google Scholar]