Abstract
The mTOR (mammalian target of rapamycin) protein kinase is an important regulator of cell growth. Two complexes of mTOR have been identified: complex 1, consisting of mTOR–Raptor (regulatory associated protein of mTOR)–mLST8 (termed mTORC1), and complex 2, comprising mTOR–Rictor (rapamycininsensitive companion of mTOR)–mLST8–Sin1 (termed mTORC2). mTORC1 phosphorylates the p70 ribosomal S6K (S6 kinase) at its hydrophobic motif (Thr389), whereas mTORC2 phosphorylates PKB (protein kinase B) at its hydrophobic motif (Ser473). In the present study, we report that widely expressed isoforms of unstudied proteins termed Protor-1 (protein observed with Rictor-1) and Protor-2 interact with Rictor and are components of mTORC2. We demonstrate that immunoprecipitation of Protor-1 or Protor-2 results in the co-immunoprecipitation of other mTORC2 subunits, but not Raptor, a specific component of mTORC1. We show that detergents such as Triton X-100 or n-octylglucoside dissociate mTOR and mLST8 from a complex of Protor-1, Sin1 and Rictor. We also provide evidence that Rictor regulates the expression of Protor-1, and that Protor-1 is not required for the assembly of other mTORC2 subunits into a complex. Protor-1 is a novel Rictor-binding subunit of mTORC2, but further work is required to establish its role.
Keywords: cancer, mammalian target of rapamycin (mTOR), mTOR complex-2 (mTORC2), proline-rich repeat protein-5 (PRR-5), protein kinase, rapamycin-insensitive companion of mTOR (Rictor), protein observed with Rictor-1 (Protor)
Abbreviations: DTT, dithiothreitol; EST, expressed sequence tag; HEK-293 cells, human embryonic kidney cells; HRP, horseradish peroxidase; IGF-1, insulin-like growth factor-1; I.M.A.G.E., Integrated Molecular Analysis of Genomes and their Expression; mTOR, mammalian target of rapamycin; mTORC, mTOR complex; NCBI, National Center for Biotechnology Information; PDK1, phosphoinositide-dependent kinase 1; PI3K, phosphoinositide 3-kinase; PKB, protein kinase B; Raptor, regulatory associated protein of mTOR; Rictor, rapamycin-insensitive companion of mTOR; Protor, protein observed with Rictor; S6K, S6 kinase; siRNA, small interfering RNA; SPAK, Ste20/SPS1-related proline/alanine-rich kinase; TAP, tandem affinity purification
INTRODUCTION
mTOR (mammalian target of rapamycin) plays a vital role in coupling cell growth to signalling pathways, availability of nutrients and cellular energy supplies [1]. Recent studies in yeast and mammals indicate that mTOR exists in two different complexes which have been termed mTORC1 (mTOR complex 1) and mTORC2 (mTOR complex 2) (reviewed in [1,2]). mTORC1 consists of mTOR, Raptor (regulatory associated protein of mTOR) and mLST8 (previously known as GβL) and is acutely inhibited by the macrolide rapamycin [3–5]. mTORC1 is activated by growth factors via a PI3K (phosphoinositide 3-kinase)-regulated pathway, involving PKB (protein kinase B)-mediated phosphorylation of TCS2 (tuberous sclerosis complex 2) protein and activation of the Rheb GTPase (reviewed in [6]). The activity of mTORC1 is also stimulated by amino acids through a poorly characterized network, which might involve the Vps34 PI3K [7,8] and a Ste20-related protein kinase termed MAP4K-3 (mitogen-activated protein kinase kinase kinase kinase-3) [9]. A key cellular substrate for mTORC1 is the ribosomal p70 S6K (S6 kinase), which is phosphorylated by mTORC1 at its hydrophobic motif residue (Thr389) [4,5], thereby promoting S6K activation by PDK1 (phosphoinositide-dependent kinase 1) [10,11].
mTORC2 consists of mTOR, Rictor (rapamycin-insensitive companion of mTOR; also known as mAVO3), Sin1 (also known as mSin1 or MIP1) and mLST8 [12–17]. Unlike mTORC1, mTORC2 is insensitive to acute rapamycin treatment, although prolonged treatment disrupts mTORC2 assembly in certain cell lines [18]. mTORC2 is thought to be activated by PI3K through an unknown mechanism, but, unlike mTORC1, its activity is not regulated by amino acids [12,13,15,19]. mTORC2 phosphorylates PKB at its hydrophobic motif (Ser473) [19,20], which, together with phosphorylation of the T-loop of PKB (Thr308) by PDK1, is required for maximal activation of PKB [21]. In the present study, we identify a novel Rictor-binding component of mTORC2 as a previously unstudied protein that we have termed Protor-1 (protein observed with Rictor-1). Moreover, we demonstrate that an isoform of this protein, termed Protor-2, also interacts with mTORC2.
MATERIALS AND METHODS
Materials
Protein G–Sepharose, calmodulin–Sepharose 4B and glutathione–Sepharose were purchased from Amersham Bioscience; Colloidal Blue, protease-inhibitor cocktail tablets, precast SDS polyacrylamide Bis-Tris gels and oligofectamine were from Invitrogen; Tween 20, rabbit IgG–agarose and dimethyl pimelimidate were from Sigma, and CHAPS was from Calbiochem. The hexahistidine-tagged TEV (tobacco etch virus) protease was expressed in Escherichia coli by Gursant Kular (MRC Protein Phosphorylation Unit, School of Life Sciences, University of Dundee, Dundee, Scotland, U.K.), using a construct kindly provided by David Barford (Institute of Cancer Research, Chester Beatty Laboratories, London, U.K.), and was purified using nickel–agarose affinity chromatography and gel filtration.
Antibodies
The following antibodies were raised in sheep and affinity-purified on the appropriate antigen: anti-mLST8 was raised against the human full-length mLST8 protein expressed in E. coli (used for immunoblotting), anti-mTOR [residues 2–20 of human mTOR (LGTGPAAATTAATTSSNVS), and used for immunoblotting and immunoprecipitation in HEK-293 cells (human embryonic kidney cells)], anti-(Protor-1) was raised against the human full-length Protor-1 protein expressed in E. coli (used for immunoblotting and immunoprecipitation), anti-Raptor [residues 1–20 of human Raptor (MESEMLQSPLLGLGEEDEAD), and used for immunoblotting and immunoprecipitation], anti-Rictor [residues 6–20 of human Rictor (RGRSLKNLRVRGRND), and used for immunoblotting and immunoprecipitation], anti-Sin1 was raised against the human full-length Sin1 protein expressed in E. coli (used for immunoblotting and immunoprecipitation), and anti-SPAK (Ste20/SPS1-related proline/alanine-rich kinase) was raised against the human full-length SPAK protein expressed in E. coli (used for immunoblotting and immunoprecipitation). The anti-mTOR antibody used for immunoblotting of mouse mTOR in mouse fibroblasts was purchased from Santa Cruz Biotechnology (#sc-1549). The anti-(Filamin A) monoclonal antibody raised against a peptide near the N-terminus was purchased from Santa Cruz Biotechnology (#sc-17749), and was used for immunoblotting and immunoprecipitation. The monoclonal antibody recognizing the FLAG epitope tag was purchased from Sigma (#F1804), and secondary antibodies coupled with HRP (horseradish peroxidase) used for immunoblotting were obtained from Pierce.
General methods
Tissue culture, immunoblotting, restriction enzyme digests, DNA ligations and other recombinant DNA procedures were performed using standard protocols. DNA constructs used for transfection were purified from E. coli DH5α using Qiagen plasmid Mega or Maxi kits, according to the manufacturer's protocol, and transfection studies were carried out using polyethyleneimine, as described previously [22], unless otherwise stated. All DNA constructs were verified by DNA sequencing, which was performed by The Sequencing Service, School of Life Sciences, University of Dundee, Dundee, Scotland, U.K. using DYEnamic ET terminator chemistry (Amersham Biosciences) on Applied Biosystems automated DNA sequencers. Mouse embryonic fibroblasts were cultured with additional non-essential amino acids and 1% sodium pyruvate solution.
Buffers
The following buffers were used: CHAPS lysis buffer [50 mM Tris/HCl (pH 7.5), 1 mM EGTA, 1 mM EDTA, 0.3% (w/v) CHAPS, 1 mM sodium orthovanadate, 10 mM sodium β-glycerophosphate, 50 mM sodium fluoride, 5 mM sodium pyrophosphate, 0.27 M sucrose, 0.15 M NaCl, 0.1% (v/v) 2-mercaptoethanol and either 1 mM benzamidine and 0.1 mM PMSF or complete protease-inhibitor cocktail (one tablet/50 ml)], Triton lysis buffer [this was identical with CHAPS lysis buffer, except that 1% (v/v) Triton X-100 replaced the CHAPS detergent], buffer A [50 mM Tris/HCl (pH 7.5), 0.1mM EGTA and 1 mM DTT (dithiothreitol)], buffer B [50 mM Tris/HCl (pH 7.5), 0.15M NaCl, 0.27M sucrose, 0.3% (w/v) CHAPS and 1mM DTT], buffer C [50 mM Tris/HCl (pH 7.5), 0.15 M NaCl, 1 mM MgCl2, 1 mM imidazole, 2 mM CaCl2, 0.27 M sucrose and 1 mM DTT], buffer D [10 mM Tris/HCl (pH 7.5), 1 mM imidazole, 20 mM EGTA and 1 mM DTT] and TBS-Tween buffer [50 mM Tris/HCl (pH 7.5), 0.15M NaCl and 0.25% Tween 20].
Plasmids
All I.M.A.G.E. (Integrated Molecular Analysis of Genomes and their Expression) Consortium clones were purchased from Geneservice Ltd. A Sin1 cDNA (GenBank® accession number NM_001006617) I.M.A.G.E. Consortium clone 2823015 encoding for a splice variant termed mSin1.2 [15], lacking 36 amino acids between residues 320–356, was obtained. Standard PCR methods were used to create a clone that was identical in sequence with the longest Sin1 isoform (mSin1.1 [15]; GenBank® accession number NM_001006617), which was subcloned into Strataclone blunt PCR vectors (Stratagene) and subcloned further as a Bgl2/Not1 fragment into different expression vectors (pGEX6p, FLAG-pCMV and pEGB2T). A full-length clone of Protor-1α [NCBI (National Center for Biotechnology Information) Entrez Protein accession number NP_851850] I.M.A.G.E. Consortium clone 5013578 was obtained, amplified and subcloned into Strataclone blunt PCR vectors and then subcloned further into different expression vectors (pGEX6p, FLAG-pCMV and pEGB2T) as Bgl2/Not1 fragments. This cDNA was utilized to produce by PCR the Protor-1β (NCBI Entrez Protein accession number NP_056181) and Protor-1γ (NCBI Entrez Protein accession number NP_001017530) splice variants. A full length clone of Protor-2 (NCBI Entrez Protein accession number CAE45978) was obtained from RZPD German Resource Centre for Genome Research [EST (expressed sequence tag) DKFZp686N03132]. The open reading frame was amplified, the PCR product was subcloned into Strataclone blunt PCR vectors and then subcloned further into different expression vectors (pGEX6p, FLAG-pCMV and pEGB2T) as a BamH1/Not1 fragment. Full-length cDNA of human mTOR (NCBI Entrez Protein accession number NP_004949) was obtained from the I.M.A.G.E. Consortium (clone 40125717). N-terminal FLAG- and HA (haemagglutinin)-tagged forms of mTOR were created by PCR and subcloned as a Not1/Not1 fragment into vectors (FLAG-pCMV, HA-pCMV and pEGB2T). A pRL62-3 construct encoding for mouse Rictor (GenBank® accession number AY497009) was kindly provided by M. Hall and R. Loewith (Biozentrum, University of Basel, Basel, Switzerland). This construct was used as a PCR template to subclone Rictor into the pSC-A vector (Stratagene). Rictor was subsequently subcloned as a Bcl1/Bcl1 fragment into several expression vectors (FLAG-pCMV, HA-pCMV and pEGB2T, and to pEBGFP-C2-TAP [23]). A human mLST8-coding sequence (Genbank® accession number BC020499) was PCR-amplified from the I.M.A.G.E. Consortium EST clone 3903638, cloned into the pGEM-T Easy vector (Promega) and, subsequently, subcloned as a BamHI/NotI fragment into different expression vectors (pGEX6p, FLAG-pCMV5, HA-pCMV5 and pEBG2T). The Raptor cDNA (NCBI Entrez Protein accession number NP_065812) was amplified from placental mRNA (using Superscript III kit; Invitrogen) with the following primers 5′-AT-GCTGCAATCGCCTCTTCTGGGCCTG-3′ and 5′-CTATCTG-ACACGCTTCTCCACCGAGT-3′. The resulting PCR product was ligated into PCR 2.1 TOPO vector (Invitrogen), sequenced and subcloned as an EcoR1/EcoR1 or BamH1/Not1 fragment into different expression vectors (FLAG-pCMV5, HA-pCMV5 and pEBG2T).
Immunoprecipitation of endogenous Rictor
Rictor and pre-immune IgG antibodies were covalently coupled to Protein G–Sepharose at a ratio of 1 mg of antibody to 1 ml of resin using a dimethyl pimelimidate cross-linking procedure [24]. A total of 250 mg of HEK-293 cell suspension cell lysate was pre-cleared by incubation with 0.5 ml of Protein G–Sepharose for 30 min at 4°C. The supernatant was incubated with 0.5 ml of Rictor or pre-immune IgG antibodies conjugated with Protein G–Sepharose for 1.5 h at 4°C on a rolling shaker. The immunoprecipitates were washed four times with CHAPS lysis buffer and three times with buffer A. The resin was resuspended in 0.6 ml of 1/80-diluted NuPAGE® LDS sample buffer (Invitrogen) for 30 min at 25°C, the resin was removed by filtration through a 0.45 μm Spin-X filter and the eluate was concentrated by a Speed-Vac to approx. 20 μl. DTT was added to a concentration of 10 mM to the sample, which was heated for 1 min at 95°C and allowed to cool to 25°C. Iodoacetamide was added to a final concentration of 50 mM in order to alkylate cysteine residues. After incubation in the dark for 30 min at 25°C, samples were electrophoresed on a precast SDS/4–12% polyacrylamide gel, which was stained with Colloidal Blue and photographed. The bands labelled in Figure 1(A) were excised, washed and digested with trypsin, as described previously [25]. Peptides were analysed by combined MALDI–TOF, MALDI–TOF/TOF (matrix-assisted laser-desorption ionization–tandem time-of-flight) MS analysis on an Applied Biosystems 4700 TOF/TOF Proteomics Analyser using 5 mg/ml α-cyanocinnamic acid in 10 mM ammonium phosphate as the matrix, or by LC–MS on an Applied Biosystems 4000 Q-TRAP. The Celera Discovery System (Applied Biosystems) human database was searched using the Mascot search algorithm (http:www.matrixscience.com; [26]).
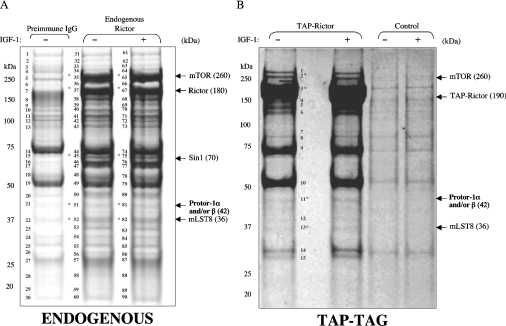
Figure 1. Identification of Protor-1.
(A) HEK-293 Cells were deprived of serum overnight and left untreated or stimulated with IGF-1 (50 ng/ml) for 30 min. HEK-293 cell extracts were subjected to immunoprecipitation with an anti-Rictor or pre-immune IgG antibody. The immunoprecipitates were electrophoresed on a polyacrylamide gel and the protein bands were visualized following Colloidal Blue staining. Each band visualized in the immunoprecipitate was given the indicated number, excised together with the equivalent region of the pre-immune immunoprecipitate, digested with trypsin and analysed by MS. The results obtained are shown in Supplementary Table 1 at http://www.BiochemJ.org/bj/405/bj4050513add.htm. The bands identified as Protor-1α and/or Protor-1β and other mTORC2 components are indicated with an arrow and were not detected within the control immunoprecipitate undertaken with pre-immune IgG antibody. The molecular mass (in kDa) of the indicated proteins is shown in parentheses, and asterisks denote mTORC2 components. (B) HEK-293 cells stably expressing TAP–Rictor were treated in the absence or presence of IGF-1 as in (A). TAP–Rictor was affinity-purified, electrophoresed on a polyacrylamide gel, and the protein bands were visualized following Colloidal Blue staining, labelled and analysed as in (A). The results are shown in Supplementary Table 2 at http://www.BiochemJ.org/bj/405/bj4050513add.htm.
Generation of stable cell line
HEK-293 cells were cultured in 10-cm-diameter dishes to 30–50% confluence and transfected with 2 μg of the pEGFP-TAP construct encoding human full-length Rictor using Fugene 6 reagent (Roche), according to the manufacturer's instructions. After 48 h, cells were split into five 10-cm-diameter dishes. G418 was added to the medium to a final concentration of 3 mg/ml after another 24 h allowing cells to recover, and then the medium was changed every 24 h containing fresh G418 for further antibiotic selection. After 14–20 days, individual surviving colonies expressing moderate levels of GFP (green flourescent protein) were selected and expanded. FACS analysis was also performed to ensure uniform expression of GFP in the selected cell lines. In addition, anti-Rictor immunoblotting analysis of lysed cells was performed to ensure that the expressed protein migrated at the expected molecular mass. For TAP (tandem affinity purification), we selected the stable cell lines that expressed low levels of GFP–TAP–Rictor to maximize the proportion of purified enzyme that is bound to an endogenous binding partner.
TAP
The purification method was adapted from the TAP protocol described previously [23,27]. For each TAP, we cultured ten 75 ml flasks of the confluent cell line, trypsinized and resuspended the cells in 500 ml of HEK-293 cell suspension culture medium (Pro-293 DMEM; Cambrez) with 3% (v/v) fetal bovine serum. When the cell density reached approx. 3×106 cells/ml, cells were harvested by centrifugation at 150 g for 5 min, washed twice with ice-cold PBS and lysed in 30 ml of ice-cold CHAPS lysis buffer. The lysates were centrifuged at 26000 g for 30 min at 4°C, and the supernatant was incubated with 0.5 ml of rabbit IgG–agarose beads for 1 h at 4°C. The IgG–agarose was washed three times with CHAPS lysis buffer, followed by two washes in buffer B, prior to incubation with 2 ml of buffer B containing 100 μg of TEV protease for 3.5 h at 4°C. The cleaved TAP-tagged Rictor protein was eluted with 7.5 ml of buffer C containing 7.5 μl of 1 M CaCl2 and was incubated with 0.25 ml of rabbit calmodulin–Sepharose equilibrated in buffer C for 1 h at 4°C. The calmodulin–Sepharose beads were washed twice with buffer C and then twice with buffer D. The samples were eluted from the beads in NuPAGE® LDS sample buffer, electrophoresed and analysed following digestion with trypsin and MS, as described above.
Immunoprecipitation of FLAG–Protor-1α for MS analysis
A total of 15 10-cm-diameter dishes of HEK-293 cells at 30–50% confluence were transiently transfected with 5 μg of FLAG–Protor-1α DNA per dish. At 36 h post-transfection, cells were lysed in 0.5 ml of CHAPS lysis buffer, clarified by centrifugation, and 60 μl of anti-FLAG antibody covalently attached to Protein G–Sepharose was incubated with 12 mg of cell lysate for 1 h at 4°C on a vibrating platform. The immunoprecipitates were washed four times with CHAPS lysis buffer and twice with buffer A. The resin was resuspended in 60 μl of 1/8-diluted NuPAGE® LDS sample buffer, and the eluted proteins were concentrated, electrophoresed on a polyacrylamide gel and analysed following digestion with trypsin and MS, as described above.
Immunoprecipitation of forms of FLAG–Protor for immunoblotting analysis
In order to generate samples for immunoblotting, 3 mg of cell lysate derived from transfected cells was incubated with 10 μl of anti-FLAG antibody covalently attached to protein G–Sepharose for 1 h at 4°C on a vibrating platform. The immunoprecipitates were washed six times with CHAPS lysis buffer in which 2-mercaptoethanol was omitted, followed by two washes with buffer A in which DTT was omitted. The immunoprecipitates were resuspended in 30 μl of NuPAGE® LDS sample buffer (not containing DTT) and filtered through a 0.45 μm Spin-X filter to remove the Sepharose resin. DTT was added to a concentration of 10 mM to the eluted samples. These were then subjected to electrophoresis and immunobloting analysis, as described below.
Immunoprecipitation of endogenous mTOR complexes for immunoblotting analysis
HEK-293 cells were lysed in CHAPS lysis buffer, and mouse tissues were homogenized in CHAPS lysis buffer. Debris were removed from cell lysates/tissue extracts by centrifugation at 12000 g for 20 min. A total of 1 mg of lysate or tissue extract was pre-cleared by incubating with 5 μl of Protein G–Sepharose. The lysates or tissue extracts were then incubated with 5 μl of Protein G–Sepharose conjugated with 5 μg of the indicated antibodies. Except in the case of Filamin-A, all of the other antibodies were covalently conjugated with Protein G–Sepharose. Immunoprecipitations were carried out for 1 h at 4°C on a vibrating platform. The immunoprecipitates were washed six times with CHAPS lysis buffer or Triton lysis buffer in which 2-mercaptoethanol was omitted, followed by two washes with buffer A in which DTT was omitted. The immunoprecipitates were resuspended in 30 μl of NuPAGE® LDS sample buffer (not containing DTT) and filtered through a 0.45 μm Spin-X filter. DTT was added to a concentration of 10 mM, and the samples were subjected to electrophoresis and immunoblot analysis, as described below.
siRNA (small interfering RNA) transfection
All siRNAs, including the control (non-targeting) sequence, were ordered from Dharmacon. The sequences of the strands of the siRNA that were used were: Protor-1 (sense, GGACAAGAUUCGCUUCUAUdTdT; and antisense, AUAGAAGCGAAUCUUGUCCdTdT); Sin1 {siGENOME™ SMARTpool siRNA (#M-014315-00-0050); Dharmacon [14]}; Rictor (sense, ACUUGUGAAGAAUCGUAUCdTdT; and antisense, GAUACGAUUCUUCACAAGUdTdT; [13]) and mTOR (sense, CCCUGCCUUUGUCAUGCCUdTdT; and antisense, AGGCAUGACAAAGGCAGGGdTdT; [5]). HeLa cells at 80% confluence were transfected with 65 nM siRNA using oligofectamine. After 24 h, the medium was replaced (without siRNA) and cells were cultured for a further 48 h before harvesting in CHAPS lysis buffer.
Immunoblotting
Total cell lysate (20 μg) or immunoprecipitated samples were heated at 70°C for 5 min in NuPAGE® LDS sample buffer, and proteins were subjected to PAGE and electrotransfer on to nitrocellulose membranes. Membranes were blocked for 1 h in TBS-Tween buffer containing 5% (w/v) skimmed milk. The membranes were probed with 1 μg/ml of the indicated antibodies in TBS-Tween containing 5% (w/v) skimmed milk for 16 h at 4°C. Detection was performed using HRP-conjugated secondary antibodies and enhanced chemiluminescence reagents.
RESULTS
Identification of Protor-1 as a Rictor-associated protein
In an attempt to characterize novel mTORC2-interacting proteins, we undertook an analysis of proteins that were co-immunoprecipitated with either endogenous Rictor (Figure 1A) or TAP-tagged Rictor that had been stably expressed in HEK-293 cells (Figure 1B). In both purifications, Rictor was a dominant Colloidal-Blue-staining band migrating at the expected molecular mass of approx. 180 kDa. As expected, the previously described mTORC2 components mTOR (approx. 260 kDa) [1], mLST8 (approx. 37 kDa) [3,28], as well as the recently discovered Sin1 (approx. 70 kDa) [14–16], were also associated with Rictor in these studies. Only one other protein of approx. 42 kDa of unknown function (NCBI Entrez Protein accession no. NP_851850), which we have named Protor-1 (protein observed with Rictor-1), was also present in both preparations of Rictor (Figure 1, and Supplementary Tables 1 and 2 at http://www.BiochemJ.org/bj/405/bj4050513add.htm). In one study, the gene encoding this protein was called PRR-5 (proline-rich repeat protein-5) [29]. We also observed that stimulation of HEK-293 cells with IGF-1 (insulin-like growth factor-1) did not alter the amounts of Protor-1 or mTOR, Sin1 and mLST8 associated with Rictor (Figure 1).
Protor-1 mRNA is reportedly expressed as three splice variants [29], which we have termed Protor-1α (residues 1–388; NCBI Entrez Protein accession number NP_851850), Protor-1β (residues 10–388; NCBI Entrez Protein accession number NP_056181) and Protor-1γ (residues 97–388; NCBI Entrez Protein accession number NP_001017530). Protor-1 possesses no recognizable functional domains and has not been studied previously at the protein level. The apparent molecular mass with which the Protor-1 protein migrated on an SDS/polyacrylamide gel (approx. 42 kDa; Figure 1) and MS analysis of the peptide sequences suggested that the splice variant of Protor associated with Rictor in the studies shown in Figure 1 was either Protor-1α or Protor-1β, which only differ by the presence of nine additional residues at the N-terminus (which we failed to detect in the MS analysis; Figure 1 and Supplementary Tables 1 and 2). Database analysis suggested that there is a second isoform of human Protor, encoded by an unstudied gene, that we have termed Protor-2 (NCBI Entrez Protein accession number CAE45978). Protor-2 encodes a protein of 368 residues displaying 39% identity in amino acid sequence with Protor-1α (Supplementary Figure 1 at http://www.BiochemJ.org/bj/405/bj4050513add.htm). The highest region of conservation between Protor-1 and Protor-2 encompasses residues 42–225 of Protor-1α (Supplementary Figure 1).
A number of other proteins were associated with immunoprecipitated endogenous Rictor (Figure 1A), which were not identified in the purification of the stably expressed TAP-tagged Rictor (Figure 1, and Supplementary Tables 1 and 2). Further investigation is required to determine whether any of these proteins interact with Rictor or whether these were non-specifically immunoprecipitated with the Rictor antibody used. Two of these proteins, MLEL1 and CDYL, which we have tested thus far, failed to interact when co-expressed with Rictor in HEK-293 cells (L.R. Pearce, unpublished work). We also found that three isoforms of 14-3-3 interacted with TAP–Rictor (Figure 1B, and Supplementary Table 2), but these adaptor proteins were not detected in the MS analysis of the endogenous Rictor immunoprecipitates (Figure 1A, and Supplementary Table 1). To our knowledge, neither Rictor nor other components of mTOR complexes have been identified in large-scale proteomic analysis of 14-3-3-interacting proteins.
Protor-1 interacts with mTORC2
We next expressed FLAG–Protor-1α in HEK-293 cells and investigated by MS which endogenously expressed proteins co-immunoprecipitated with it. Consistent with the notion that Protor-1 might comprise a component of mTORC2, mTOR, Rictor and Sin1 were readily detected in the FLAG–Protor-1α immunoprecipitates (Figure 2A). Although MS analysis did not detect mLST8 within the FLAG–Protor-1α immunoprecipitates, its presence was confirmed by immunoblotting analysis (Figure 2D). Raptor was not detected in immunoprecipitates of FLAG–Protor-1α by either MS (Figure 2A) or immunoblotting (Figure 2D). We also co-expressed FLAG–Protor-1α with GST–Rictor or six other GST fusion proteins in HEK-293 cells and observed that Protor-1α only interacted with Rictor, suggesting that the interaction was specific (Supplementary Figure 2 at http://www.BiochemJ.org/bj/405/bj4050513add.htm).
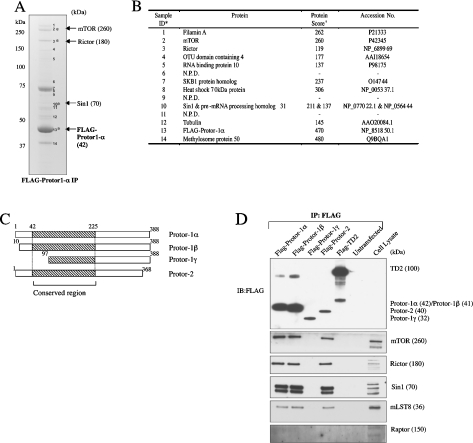
Figure 2. Protor-1 binds mTORC2.
(A) HEK-293 cells were transfected with a DNA construct encoding FLAG–Protor-1α. At 36 h post-transfection, cells were lysed, and the FLAG–Protor-1α was immunoprecipitated (IP) and electrophoresed on a polyacrylamide gel. The major protein bands were visualized following Colloidal Blue staining and labelled as indicated. The molecular mass (in kDa) of the indicated proteins is shown in parentheses, and asterisks denote mTORC2 components. (B) The labelled Colloidal-Blue-stained bands identified in (A) were excised from the gel, digested with trypsin and their identities determined by tryptic peptide mass-spectral fingerprint, as described in the Materials and methods section. Accession numbers are for the NCBI Entrez Protein database. †Mascot protein score, where a value >67 is considered significant (P<0.05). N.P.D., no significant protein identity determined. (C) Schematic representation of the Protor-1 isoforms and Protor-2. The conserved region is shaded and numbering of residues is based on the human sequence. For a sequence alignment of these proteins, see Supplementary Figure 1 at http://www.BiochemJ.org/bj/405/bj4050513add.htm. (D) As in (A), except that HEK-293 cells were transfected with the indicated forms of Protor and a control FLAG-epitope-tagged protein (TD2; Genbank® accession number NM_014779). Immunoprecipitates were immunoblotted (IB) with the indicated antibodies. Similar results were obtained in three separate experiments.
Forms of Protor that interact with mTORC2
We next investigated whether Protor-1 splice variants as well as Protor-2 were capable of interacting with endogenous mTORC2 components when overexpressed in HEK-293 cells. The results of these studies demonstrate that Protor-1β and Protor-2 bind to mTORC2 components to the same extent as Protor-1α (Figure 2D). Interestingly, Protor-1γ that lacks the N-terminal part of the region conserved between Protor-1 and Protor-2 (Supplementary Figure 1) failed to interact with any component of mTORC2, despite being expressed at the same level as Protor-2 in HEK-293 cells (Figure 2D).
Endogenous Protor-1 interacts with mTORC2 components
Using an antibody raised against Protor-1, immunoprecipitation of endogenous Protor-1 in HEK-293 cells resulted in the co-immunoprecipitation of endogenous mTOR, Rictor, Sin1 and mLST8 (Figure 3A). Furthermore, Protor-1 was clearly detected within immunoprecipitates of endogenous mTOR, Rictor and Sin1 (Figure 3A). Consistent with the view that Protor-1 associates with mTORC2, rather than mTORC1, Raptor was not detected within immunoprecipitates of endogenous Protor-1. As expected, Raptor was detected within immunoprecipitates of mTOR, but not within immunoprecipitates of Rictor or Sin1 (Figure 3A). We noticed that the cytoskeletal protein Filamin-A co-immunoprecipitated with FLAG–Protor-1α (Figure 2A). However, as we have observed Filamin-A to be a contaminant of other FLAG immunoprecipitations we have undertaken (D. R. Alessi, unpublished work), we were concerned that this was a non-specific interaction. Consistent with this, we failed to observe co-immunoprecipitation of endogenous forms of Filamin-A with either Protor-1α or other mTORC2 components (Figure 3A).
Figure 3. Endogenous Protor-1 binds to mTORC2.
(A) HEK-293 cell lysates were subjected to immunoprecipitation (IP) with the indicated antibodies raised against different mTORC2 components as well as the SPAK protein kinase as a control, which would not be expected to bind mTORC2 and Filamin-A detected within the FLAG–Protor-1α immunoprecipitate shown in Figure 2(A). Immunoprecipitates were immunoblotted with the indicated antibodies. Similar results were obtained in three separate experiments. The molecular mass (in kDa) of the indicated proteins is shown in parentheses. (B) As in (A), except that cells were lysed in a buffer containing either no detergents or the concentrations of the indicated detergents.
Effects of detergent on the association of Protor-1 with mTORC2 components
Previous studies have demonstrated that the integrity of mTORC2 is disrupted when cells are lysed with 1% (v/v) Triton X-100 detergent, but not with 0.3% (w/v) CHAPS [13]. In the previous experiments undertaken in the present study to demonstrate an association of Protor-1 with mTORC2, 0.3% (w/v) CHAPS detergent was employed. To investigate how detergents affected the association of Protor-1 with mTORC2, we lysed HEK-293 cells either in the absence of detergent or in the presence of one of four different detergents. None of the detergents employed affected the ability of Protor-1 to associate with Rictor and Sin1, suggesting that the interaction between these proteins is detergent-insensitive. Strikingly, when cells were lysed with Triton X-100 or n-octylglucoside, neither mTOR nor mLST8 associated with Protor-1 (Figure 3B). When cells were lysed with Tween 20, a similar association of Protor-1 with mTOR and mLST8 was observed as found with CHAPS (Figure 3B). The highest levels of mTOR and mLST8 associated with Protor-1 were seen when cells were lysed with no detergent.
Protor-1 interacts specifically with Rictor
In order to establish which components of mTORC2 Protor-1α interacts with, mTORC2 subunits and Protor-1 were co-expressed in HEK-293 cells. Immunoprecipitation of Protor-1α resulted in the co-immunoprecipitation of mTOR, Rictor and Sin1 as expected (Figure 4A). Omitting individual subunits suggested that Protor-1α is principally binding to mTORC2 by virtue of its capability to bind Rictor, as, in the absence of Rictor, Protor-1α failed to interact with co-expressed mTOR or Sin1. In the absence of mTOR, Protor-1 still associated with Rictor and Sin1 (Figure 4A, middle panel). In the absence of Sin1, Protor-1 still interacted with Rictor. Consistent with previous reports [14–16], mTOR failed to interact with Rictor in the absence of Sin1, as Sin1 is required for the stable binding of Rictor to mTOR (Figure 4A, middle panel). Further evidence that Protor-1 does not interact with Sin1 is that, in the absence of Rictor, immunoprecipitation of Sin1 did not result in the co-immunoprecipitation of Protor-1 (Figure 4A, lower panel).
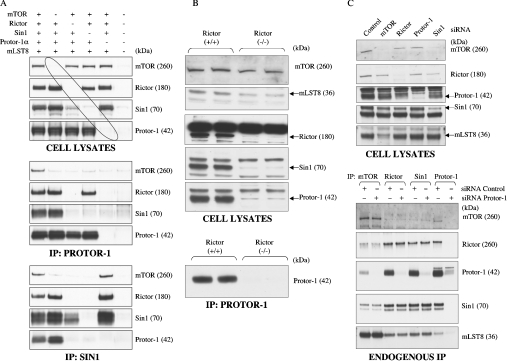
Figure 4. Rictor interacts with Protor-1 and regulates its expression.
(A) HEK-293 cells were co-transfected with the indicated DNA constructs encoding the indicated mTORC2 components. At 36 h post-transfection, cells were lysed and Protor-1 and Sin1 were immunoprecipitated (IP). The cell lysates (upper panel), the Protor-1 immunoprecipitates (middle panel) and Sin1 immunoprecipitates (lower panel) were immunoblotted with the indicated antibodies. Similar results were obtained in two separate experiments. The molecular mass (in kDa) of the indicated proteins is shown in parentheses. (B) Cell lysates derived from control wild-type Rictor+/+ and knockout Rictor−/− mouse embryonic fibroblast cells [30] were subjected to immunoblot analysis with the indicated antibodies. To confirm the marked reduction in Protor-1 expression, endogenous Protor-1 was immunoprecipitated (IP) and immunoblotted (lower panel). (C) HeLa cells were transfected with the indicated siRNAs (sequences as described in the Materials and methods section). At 72 h post-transfection, cells were lysed and cell lysates (upper panel) and the immunoprecipitates (lower panel) were immunoblotted with the indicated antibodies. Similar results were obtained in two separate experiments. The molecular mass (in kDa) of the indicated proteins is shown in parentheses.
Rictor regulates the expression of Protor-1 and Sin1
To investigate further the importance of Rictor in enabling Protor-1 to interact with mTORC2 components, we utilized Rictor−/− knockout mouse embryonic fibroblast cells that have been generated recently [30]. Strikingly, both Protor-1 and Sin1 expression was greatly decreased in the absence of Rictor (Figure 4B), suggesting that the stability and/or expression of Protor-1 and Sin1 are dependent upon the presence of Rictor.
Knockdown of Protor-1 does not affect the association of other mTORC2 components
To investigate the effect of knockdown of Protor-1 on the expression and ability of other mTORC2 subunits to interact with each other, HeLa cells were transfected with siRNA targeting individual mTORC2 components. siRNA treatments reduced the expression of each of the targeted subunits by 80–90% (Figure 4C, upper panel). Knockdown of Protor-1 did not affect the levels of mTOR, Sin1, mLST8 or Rictor in cell extracts. Moreover, immunoprecipitation of mTOR, Rictor or Sin1 in cells depleted of Protor-1 revealed that the depletion of Protor-1 did not affect the ability of the other mTORC2 components to form a complex (Figure 4C, lower panel). In contrast, knockdown of other mTORC2 components had significant effects on the expression of other subunits. For example, knockdown of mTOR reduced the expression of mLST8, lowering Rictor also decreased expression of Protor-1 and Sin1, and knockdown of Sin1 moderately reduced the expression of mTOR and Rictor.
DISCUSSION
In the present study, we have demonstrated in several ways that Protor-1 comprises a novel component of mTORC2. Our results suggest that Protor-1 binds specifically to the Rictor subunit of the complex, explaining why Protor-1 does not interact with the Raptor-containing mTORC1, which is devoid of Rictor. Based on our observations and the work undertaken by others, we summarize in Figure 5 the proposed subunit organization of mTORC2. Our present data also suggest that, in the presence of detergents such as Triton X-100 or n-octylglucoside, the interaction between Rictor and mTOR is disrupted, but the interactions between Protor-1, Rictor and Sin1 are maintained (Figure 3B). We find that the association of mTORC2 subunits is best preserved when cells are lysed with no detergent (Figure 3B). We would recommend in future studies when mTORC2 is isolated from cells or tissues that all detergents are omitted from the lysis procedure.
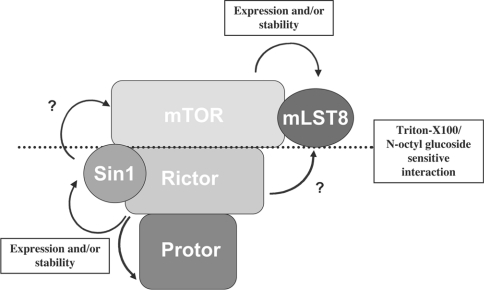
Figure 5. Model of the subunit organization of mTORC2.
The dotted line indicates the detergent-sensitive interaction between mTOR/mLST8 and Rictor/Sin1/Protor. ? indicates that direct interaction of these subunits is uncertain.
Our present findings also suggest that Rictor controls the expression and/or stability of Protor-1 as, in Rictor−/− cells, the expression of Protor-1 and Sin1 is markedly reduced (Figure 4B). Reduction of Sin1 protein levels in Rictor−/− cells has also been reported recently [14]. This observation emphasizes further the link between the Protor and Rictor proteins. Further work is required to investigate the mechanism by which Rictor controls the expression and/or stability of Protor-1. It is possible that Rictor ensures that expression of Protor-1 and Sin1 are maintained at similar stoichiometric levels to Rictor so that there is not an excess of these subunits. siRNA knockdown of mTOR, Rictor and Sin1 has moderate-to-significant effects on the expression of several other mTORC2 subunits (Figure 4C). Similar observations have been made for other protein kinases and their interacting subunits; for example, in cells lacking the LKB1 protein kinase expression of its regulatory STRAD (STE-related adaptor) subunit is also hugely decreased [31].
Protor-1 as well as Protor-2 possesses no recognizable functional domains or motifs within their sequence. Hence it is difficult to predict what their physiological role(s) might be, whether they possess enzymatic activity and how they might influence mTORC2. We have attempted siRNA experiments to reduce the expression of Protor-1, Sin1, Rictor and mTOR in HeLa cells to investigate how this affects Ser473 phosphorylation of PKB, the proposed target of mTORC2. Although we were able to reduce expression of these components by 80–90% (Figure 4C), this reduction of protein was not sufficient to significantly reduce phosphorylation of PKB at Ser473 (L. R. Pearce, unpublished work). Sabatini and co-workers [19] have elegantly demonstrated that it is essential to completely ablate expression of mTOR and Rictor in order to reduce phosphorylation of Ser473 of PKB, conditions that we have so far been unable to recapitulate in our laboratory. We are currently generating Protor knockout mice to address the physiological roles of this subunit in regulating mTORC2 function.
Sequence alignments of Protor-1 and Protor-2 and Protor-1 homologues found in other vertebrates (Supplementary Figure 3 at http://www.BiochemJ.org/bj/405/bj4050513add.htm) also highlight that the N-terminal region of Protor encompassing residues 42–225 of human Protor-1 is the most highly conserved and may therefore form a functional domain. Protor-1γ that lacks the first 96 residues, hence part of the conserved domain, was not capable of binding Rictor (Figure 2D). Further work is required to delineate the residues of Protor isoforms and Rictor that directly interact with each other, but it is likely that the Rictor-binding domain is within the conserved region on Protor. In addition, it remains to be investigated whether Protor-2 plays the same role as Protor-1. In our present MS studies, we failed to detect an association of Protor-2 with endogenous Rictor or TAP–Rictor; however, in the absence of a specific anti-(Protor-2) antibody, it is currently unknown whether Protor-2 is expressed in the HEK-293 cells utilized in our experiments.
Analysis of the tissue distribution patterns of mRNA expression of Protor-1 and Protor-2 in humans using the NCBI UniGene database (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=unigene) suggests that these proteins, similar to the other known components of mTOR complexes, are widely expressed in all tissues. Northern blot analysis of Protor-1 mRNA in 12 human tissues also supported this conclusion [29]. We have analysed the expression levels of the Protor-1 protein in nine different mouse tissue extracts and found that Protor-1 was broadly expressed, with highest levels in adipose, kidney, spleen and testis and lowest levels in skeletal muscle (Supplementary Figure 4 at http://www.BiochemJ.org/bj/405/bj4050513add.htm). In the only previous study in which the Protor-1 gene was investigated, it was reported that Protor-1 mRNA was overexpressed in 13 out of 16 colorectal tumours studied [29]. It was also reported that, in human metastatic breast cancer, Protor-1 mRNA was overexpressed in two out of eight tumours, whilst being down-regulated in five out the eight cancers [29]. No somatic mutations in the exons encoding for the Protor-1 gene in human breast or colorectal tumours have been identified yet.
Surprisingly, we have not been able to detect any obvious homologues of Protor-1 or Protor-2 in lower eukaryotic species, including yeast and Drosophila, frequently used for genetic analysis of mTOR pathways. In Saccharomyces cerevisiae, two TOR complexes termed TORC1 and TORC2, believed to comprise functional homologues of mTORC1 and mTORC2, have been characterized [3,32]. S. cerevisiae homologues of Rictor (AVO3), Sin1 (AVO1) and mLST8 (LST8) have been identified in TORC2 isolated from yeast extracts [3,32]. Two other S. cerevisiae proteins, namely AVO2 [3] and BIT61 [33], associate with purified TORC2. To our knowledge, no homologues of these proteins have yet been reported in mammals, and neither do AVO2 nor BIT61 possess evident identity with Protor-1/Protor-2. A clear homologue of Protor is found in frogs and fish (Supplementary Figure 3).
In conclusion, we have identified Protor-1 and potentially Protor-2 as novel Rictor-binding components of mTORC2. Further work is required to define the role that Protor plays in regulating the functions of mTORC2. In particular, it would be crucial to define whether Protor-1 is required to enable mTOR complexes to couple cell growth to the many signalling networks and environmental cues that mTOR complexes respond to [1].
Online data
Acknowledgments
We thank the Sequencing Service (School of Life Sciences, University of Dundee, Dundee, Scotland, U.K.) for DNA sequencing, the Post Genomics and Molecular Interactions Centre (School of Life Sciences, University of Dundee, Dundee, Scotland, U.K.) for MS facilities, and the protein production and antibody purification teams (Division of Signal Transduction Therapy, University of Dundee, Dundee, Scotland, U.K.), co-ordinated by Hilary McLauchlan and James Hastie, for generation and purification of antibodies. L. R. P. is supported by a U.K. MRC (Medical Research Council) studentship. We thank the Association for International Cancer Research, Diabetes UK, MRC, the Moffat Charitable Trust and the pharmaceutical companies supporting the Division of Signal Transduction Therapy Unit (AstraZeneca, Boehringer-Ingelheim, GlaxoSmithKline, Merck & Co. Inc, Merck KgaA and Pfizer) for financial support.
References
- 1.Wullschleger S., Loewith R., Hall M. N. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 2.Sarbassov D. D., Ali S. M., Sabatini D. M. Growing roles for the mTOR pathway. Curr. Opin. Cell. Biol. 2005;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 3.Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J. L., Bonenfant D., Oppliger W., Jenoe P., Hall M. N. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 4.Hara K., Maruki Y., Long X., Yoshino K., Oshiro N., Hidayat S., Tokunaga C., Avruch J., Yonezawa K. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–189. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- 5.Kim D. H., Sarbassov D. D., Ali S. M., King J. E., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. mTOR interacts with Raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 6.Li Y., Corradetti M. N., Inoki K., Guan K. L. TSC2: filling the GAP in the mTOR signaling pathway. Trends Biochem. Sci. 2004;29:32–38. doi: 10.1016/j.tibs.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 7.Byfield M. P., Murray J. T., Backer J. M. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J. Biol. Chem. 2005;280:33076–33082. doi: 10.1074/jbc.M507201200. [DOI] [PubMed] [Google Scholar]
- 8.Nobukuni T., Joaquin M., Roccio M., Dann S. G., Kim S. Y., Gulati P., Byfield M. P., Backer J. M., Natt F., Bos J. L., et al. Amino acids mediate mTOR/Raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc. Natl. Acad. Sci. U.S.A. 2005;102:14238–14243. doi: 10.1073/pnas.0506925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Findlay G. M., Yan L., Procter J., Mieulet V., Lamb R. F. A MAP4 kinase related to Ste20 is a nutrient-sensitive regulator of mTOR signalling. Biochem. J. 2007;403:13–20. doi: 10.1042/BJ20061881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biondi R. M., Kieloch A., Currie R. A., Deak M., Alessi D. R. The PIF-binding pocket in PDK1 is essential for activation of S6K and SGK, but not PKB. EMBO J. 2001;20:4380–4390. doi: 10.1093/emboj/20.16.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins B. J., Deak M., Arthur J. S., Armit L. J., Alessi D. R. In vivo role of the PIF-binding docking site of PDK1 defined by knock-in mutation. EMBO J. 2003;22:4202–4211. doi: 10.1093/emboj/cdg407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacinto E., Loewith R., Schmidt A., Lin S., Ruegg M. A., Hall A., Hall M. N. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 2004;6:1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- 13.Sarbassov D. D., Ali S. M., Kim D. H., Guertin D. A., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and Raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 14.Yang Q., Inoki K., Ikenoue T., Guan K. L. Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev. 2006;20:2820–2832. doi: 10.1101/gad.1461206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frias M. A., Thoreen C. C., Jaffe J. D., Schroder W., Sculley T., Carr S. A., Sabatini D. M. mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Curr. Biol. 2006;16:1865–1870. doi: 10.1016/j.cub.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 16.Jacinto E., Facchinetti V., Liu D., Soto N., Wei S., Jung S. Y., Huang Q., Qin J., Su B. SIN1/MIP1 maintains Rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127:125–137. doi: 10.1016/j.cell.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 17.Guertin D. A., Stevens D. M., Thoreen C. C., Burds A. A., Kalaany N. Y., Moffat J., Brown M., Fitzgerald K. J., Sabatini D. M. Ablation in mice of the mTORC components Raptor, Rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCα, but not S6K1. Dev. Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 18.Sarbassov D. D., Ali S. M., Sengupta S., Sheen J. H., Hsu P. P., Bagley A. F., Markhard A. L., Sabatini D. M. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol. Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 19.Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. Phosphorylation and regulation of Akt/PKB by the Rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 20.Hresko R. C., Mueckler M. mTOR.RICTOR is the Ser473 kinase for Akt/protein kinase B in 3T3-L1 adipocytes. J. Biol. Chem. 2005;280:40406–40416. doi: 10.1074/jbc.M508361200. [DOI] [PubMed] [Google Scholar]
- 21.Mora A., Komander D., Van Aalten D. M., Alessi D. R. PDK1, the master regulator of AGC kinase signal transduction. Semin. Cell. Dev. Biol. 2004;15:161–170. doi: 10.1016/j.semcdb.2003.12.022. [DOI] [PubMed] [Google Scholar]
- 22.Durocher Y., Perret S., Kamen A. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 2002;30:E9. doi: 10.1093/nar/30.2.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al-Hakim A. K., Goransson O., Deak M., Toth R., Campbell D. G., Morrice N. A., Prescott A. R., Alessi D. R. 14-13-3 cooperates with LKB1 to regulate the activity and localization of QSK and SIK. J. Cell Sci. 2005;118:5661–5673. doi: 10.1242/jcs.02670. [DOI] [PubMed] [Google Scholar]
- 24.Harlow E., Lane D. New York: Cold Spring Harbor Laboratory Publications; 1988. Antibodies: a Laboratory Manual. [Google Scholar]
- 25.Woods Y. L., Rena G., Morrice N., Barthel A., Becker W., Guo S., Unterman T. G., Cohen P. The kinase DYRK1A phosphorylates the transcription factor FKHR at Ser329 in vitro, a novel in vivo phosphorylation site. Biochem. J. 2001;355:597–607. doi: 10.1042/bj3550597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 27.Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 28.Kim D. H., Sarbassov D. D., Ali S. M., Latek R. R., Guntur K. V., Erdjument-Bromage H., Tempst P., Sabatini D. M. GβL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between Raptor and mTOR. Mol. Cell. 2003;11:895–904. doi: 10.1016/s1097-2765(03)00114-x. [DOI] [PubMed] [Google Scholar]
- 29.Johnstone C. N., Castellvi-Bel S., Chang L. M., Sung R. K., Bowser M. J., Pique J. M., Castells A., Rustgi A. K. PRR5 encodes a conserved proline-rich protein predominant in kidney: analysis of genomic organization, expression, and mutation status in breast and colorectal carcinomas. Genomics. 2005;85:338–351. doi: 10.1016/j.ygeno.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 30.Shiota C., Woo J. T., Lindner J., Shelton K. D., Magnuson M. A. Multiallelic disruption of the Rictor gene in mice reveals that mTOR complex 2 is essential for fetal growth and viability. Dev. Cell. 2006;11:583–589. doi: 10.1016/j.devcel.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 31.Hawley S. A., Boudeau J., Reid J. L., Mustard K. J., Udd L., Makela T. P., Alessi D. R., Hardie D. G. Complexes between the LKB1 tumor suppressor, STRADα/β and MO25α/β are upstream kinases in the AMP-activated protein kinase cascade. J. Biol. 2003;2:28. doi: 10.1186/1475-4924-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wullschleger S., Loewith R., Oppliger W., Hall M. N. Molecular organization of target of rapamycin complex 2. J. Biol. Chem. 2005;280:30697–30704. doi: 10.1074/jbc.M505553200. [DOI] [PubMed] [Google Scholar]
- 33.Wedaman K. P., Reinke A., Anderson S., Yates J., 3rd, McCaffery J. M., Powers T. Tor kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisiae. Mol. Biol. Cell. 2003;14:1204–1220. doi: 10.1091/mbc.E02-09-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.