Abstract
The ETS transcription factor Ets-1 (E26 transformation-specific-1) plays a critical role in many physiological processes including angiogenesis, haematopoietic development and tumour progression. Its activity can be regulated by post-translational modifications, such as phosphorylation. Recently, we showed that Ets-1 is a target for SUMO (small ubiquitin-like modifier) modification and that PIASy [protein inhibitor of activated STAT (signal transducer and activator of transcription) Y], a specific SUMO-E3 ligase for Ets-1, represses Ets-1-dependent transcription. In the present study, we demonstrated that Ets-1 is degraded by the proteasome and that overexpression of PIASy increased the stability of endogenous and ectopically expressed Ets-1 protein by preventing proteasomal degradation. Moreover, knockdown of the endogenous PIASy expression by RNA interference reduced the protein level of endogenous Ets-1. The proteasome inhibitor MG132 reversed this effect. Deletion analysis showed that the TAD (transcriptional activation domain), which has been identified as the interaction domain with PIASy, was also required for Ets-1 ubiquitination and proteasomal degradation. However, the Ets-1 stabilization by PIASy was not due to reduced ubiquitination of Ets-1. Our results suggested that PIASy controls Ets-1 function, at least in part, by inhibiting Ets-1 protein turnover via the ubiquitin–proteasome system.
Keywords: E26 transformation-specific-1 (Ets-1), protein inhibitor of activated STAT (protein inhibitor of activated STAT) Y (PIASy), small ubiquitin-like modifier (SUMO), ubiquitin (Ub)
Abbreviations: AR, androgen receptor; CHX, cycloheximide; Ets-1, E26 transformation-specific-1; (E)GFP, (enhanced) green fluorescent protein; His6, hexahistidine; IκBα, inhibitory κBα (also called inhibitor of nuclear factor κBα); NIH, National Insttiutes of Health; Ni-NTA, Ni2+ -nitrilotriacetate; NLS, nuclear localization signal; PIAS, protein inhibitor of activated STAT (signal transducer and activator of transcription); RNAi, RNA interference; RT, reverse transcription; siRNA, small interfering RNA; Sox9, sex-determining-region-Y-related high-mobility-group box 9; SUMO, small ubiquitin-like modifier; TAD, transcriptional activation domain; WT, wild-type
INTRODUCTION
The Ets (E26 transformation-specific) family of transcription factors plays a critical role in a variety of physiological processes, including cell proliferation, differentiation, development and apoptosis [1]. Family members share a highly conserved DNA-binding domain of 80–90 amino acids, called the ‘Ets domain’, which recognizes a GGAA/T motif, designated the EBS (Ets-binding site) [2–4].
The proto-oncogene Ets-1 is the founding member of the Ets family of proteins [2,5]. Ets-1 is expressed in endothelial cells, vascular smooth-muscle cells, lymphoid cells and epithelial cells, and is involved in the regulation of angiogenesis and the differentiation of lymphoid lineages. Ets-1 has been associated with tumour progression in various carcinomas; high expression levels of the Ets-1 gene have been reported in many types of human cancers, including epithelial ovarian tumours and breast cancers [6–8]. Ets-1 regulates the expression of a number of genes involved in extracellular matrix remodelling; overexpression of Ets-1 is thought to be associated with the invasive phenotype of metastatic cancer cells [9,10].
SUMO (small ubiquitin-like modifier) modification (SUMOylation) is a form of post-translational modification involved in cell-cycle progression and transcriptional regulation [11,12]. SUMOylation of target proteins occurs in a manner analogous to ubiquitination and requires a heterodimeric E1-activating enzyme, Aos1/Uba2, an E2-conjugating enzyme, Ubc9, and an E3 ligase, such as PIAS (protein inhibitor of activated STAT), RanBP2 (RAN binding protein 2) or a polycomb group protein Pc2 [12]. Several ETS transcription factors are reported to undergo SUMOylation. Recently we have shown that SUMOylation regulates Ets-1 activity [13]; SUMOylation of Ets-1 occured at Lys15 and Lys227, both of which lie within previously identified synergy control motifs. Replacement of these lysine residues with arginine or overexpression of SENP1 (SUMO1/sentrin-specific peptidase 1), a deSUMOylation enzyme, enhanced the transactivational ability of Ets-1 [13]. Furthermore, we identified PIASy as a novel interaction partner and a specific SUMO-E3 ligase of Ets-1; PIASy represses Ets-1-dependent transcription and its repression is independent of the SUMOylation status of Ets-1 [13].
Ubiquitination is an important post-translational modification involved in cellular processes such as cell-cycle regulation and apoptosis [14]. Previously, ubiquitination of transcription factors, their co-regulators and ubiquitin-dependent proteolysis have been proposed to contribute to the regulation of transcriptional control [15,16]. Previously we observed that Ets-1 is a short-lived protein and, when co-expressed with PIASy, Ets-1 protein levels become elevated (T. Nishida, M. Terashima and K. Fukami, unpublished work). In the present study, we demonstrate that Ets-1 is degraded via the ubiquitin–proteasome system and that PIASy stabilizes Ets-1 protein levels by a mechanism that inhibits the proteasomal degradation of Ets-1. These findings indicate that PIASy plays an important role in the controlling of Ets-1 protein levels in addition to its role in Ets-1 SUMOylation.
EXPERIMENTAL
Plasmid constructs
pFLAG-Ets-1WT (where WT is wild-type), pFLAG-Ets-12KR (where 2KR is a simultaneous point mutation of Lys15 and Lys227 to arginine), pcDNA3.1/Myc-PIASyWT and pcDNA3.1/Myc-PIASyC342A (where C342A is a Cys342→Ala mutation) have been described previously [13]. Simultaneous point mutations of Lys377, Arg378 and Lys379 to alanine (AAA) and the deletion mutants of Ets-1 were created by PCR using specific primers, and both mutants were cloned into pFLAG-CMV-2 and p3×FLAG-CMV-7.1 (Sigma) (CMV is cytomegalovirus). pEGFP-PIASy and pEGFP-PIAS1 have been described previously [13,17]. A His6 (hexahistidine)-tagged ubiquitin expression plasmid was kindly provided by Dr Takayuki Ohshima (Faculty of Pharmaceutical Sciences at Kagawa Campus, Tokushima Bunri University, Sanuki City, Kagawa, Japan). A ubiquitin K48R mutant was amplified by PCR with specific primers and cloned into pcDNA3.1/His C (Invitrogen).
Cell culture and transfection
COS-7 and U2OS cells were grown in DMEM (Dulbecco's modified Eagles' medium) supplemented with 10% (v/v) FBS (fetal bovine serum). All cells were transiently transfected using Lipofectamine™ 2000 (Invitrogen) according to the manufacturer's protocol.
RNAi (RNA interference) in COS-7 cells was performed using Validated Stealth™ RNAi duplex for PIAS4 (Invitrogen), or by Stealth™ RNAi negative control duplex (Invitrogen). Cells were transfected using Lipofectamine™ 2000 according to the manufacturer's protocol.
Co-immunoprecipitation analysis
Transfected cells were washed with ice-cold PBS and then lysed in RIPA buffer (50 mM Tris/HCl, pH 7.4, containing 150 mM NaCl, 1% sodium deoxycholate, 1% Triton X-100, 1 mM dithiothreitol and a protease-inhibitor mixture). The cell lysates were sonicated (TOMY Sonicator UR-20P; Tomy Seiko, Tokyo, Japan; three 10 s bursts; 40% output power at 4 °C) and cell debris was removed by centrifugation (12 000 g for 10 min at 4 °C). The lysates were precleared by incubation with Protein A–Sepharose beads (Amersham) for 1 h at 4 °C and then immunoprecipitated with a polyclonal anti-FLAG antibody (Sigma) followed by incubation with Protein A beads. After extensive washing, the precipitates were analysed by SDS/PAGE followed by immunoblotting with an anti-FLAG antibody (M2; Sigma) or an anti-(T7 epitope tag) antibody (Novagen; where T7 is MASMTGGQQMG).
Immunofluorescence
U2OS cells were grown on coverslips and transfected with the indicated expression plasmids. At 24 h after transfection, cells were rinsed in PBS, fixed in 4% (w/v) paraformaldehyde and permeabilized with 0.2% Triton X-100. FLAG-tagged Ets-1 proteins were detected with a polyclonal anti-FLAG antibody (Sigma) and an Alexa Fluor® 568-conjugated goat anti-rabbit secondary antibody (Molecular Probes Inc.). The localization of GFP (green fluorescent protein) fusion proteins were directly observed by fluorescence microscopy.
Analysis of protein stability
COS-7 cells were transfected with pFLAG-Ets-1 (either WT or 2KR) and pcDNA3.1/Myc-PIASy. After 24 h, the transfected cells were treated with 20 μM MG132 (a proteasome inhibitor) or vehicle (DMSO) for 1 h. After 30 min, 100 μg/ml CHX (cycloheximide, a protein-biosynthesis inhibitor) was added to the medium. The cells were harvested at different time points after CHX treatment (0, 0.5, 1, 2, 4 and 8 h), lysed with 2×SDS sample buffer and subjected to immunoblotting with an anti-FLAG antibody (M2; Sigma), an anti-Myc antibody (9E10; Invitrogen) or an anti-β-actin antibody (Sigma). Endogenous Ets-1 protein was detected with an anti-Ets-1 antibody (C-20; Santa Cruz Biotechnology). Immunoblots were scanned and band density was determined by NIH (National Institutes of Health) ImageJ software for semi-quantitative analysis.
Total RNA extraction and semi-quantitative RT (reverse transcription)-PCR
Total RNA from cultured cells was purified using an RNeasy kit (Qiagen) according to the manufacturer's instructions, and 1 μg of the total RNA was converted to cDNA using the oligo(dT)12-18 primer (Amersham) and Superscript III reverse transcriptase (Invitrogen). Each single-stranded cDNA was then diluted and subjected to PCR amplification using Ex Taq DNA polymerase (Takara). The PCR conditions were as follows: initial denaturation at 94 °C for 2 min followed by 25 cycles of 94 °C for 30 s, 56 °C for 30 s and 72 °C for 30 s. The primer sequences used were: Ets-1: 5′-AGCCCGGCTCCCCGCGTTCCGCCTACT-3′ and 5′-TAGCCCCCTAACCAGCTGGAGAAGTCG-3′; β-actin: 5′-TTGTTACCAACTGGGACGACATGG-3′ and 5′-GATCTTGATCTTCATGGTGCTAGG-3′. The PCR products were resolved by electrophoresis in a 1.5%-(w/v)-agarose gel containing ethidium bromide and bands were visualized under UV light.
In vivo ubiquitination assay
COS-7 cells were transfected (in 100-mm-diameter dishes) with plasmids encoding either WT or mutant: p3×FLAG-Ets-1 (0.5 μg), pcDNA3.1/Myc-PIASy (5.0 μg) and His6-tagged ubiquitin (2.0 μg). At 24 h after the transfection, the cells were treated with MG132 (20 μM) or CHX (100 μg/ml) for 5 h. Next, the cells were lysed in buffer A (6 M guanidinium chloride, 0.1 M Na2HPO4/NaH2PO4, pH 8.0, and 10 mM imidazole). Lysates were affinity-purified with Ni-NTA (Ni2+-nitrilotriacetate)–agarose beads (Qiagen) and analysed by immunoblotting with an anti-FLAG antibody (M2) or anti-ubiquitin antibody (P4D1; Santa Cruz Biotechnology).
RESULTS
PIASy protects Ets-1 from proteasomal degradation
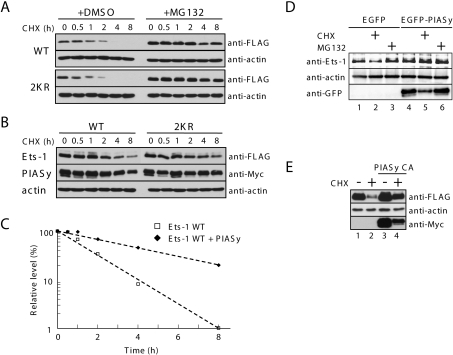
Previously, Ets-1 was shown to be rapidly degraded via the proteasome pathway [18]. To confirm this observation, COS-7 cells were transfected with a WT Ets-1 expression plasmid and treated with either vehicle (DMSO) or the proteasome inhibitor MG132, followed 30 min later by treatment with the protein-biosynthesis inhibitor CHX (Figure 1A, upper panel). As expected, WT Ets-1 protein levels were barely detectable after 4 h of CHX treatment. Furthermore, pretreatment with MG132 prevented clearance of the majority of WT Ets-1, demonstrating that degradation of Ets-1 in COS-7 cells requires the proteasome pathway. Levels of the SUMOylation-defective Ets-1 mutant, 2KR, were similar to WT Ets-1 in both the vehicle- and MG132-treated COS-7 cells following the addition of CHX (Figure 1A, lower panel), suggesting that SUMOylation of Ets-1 does not influence Ets-1 protein stability.
Figure 1. PIASy overexpression stabilizes Ets-1 against proteasomal degradation.
(A) COS-7 cells were transiently transfected with FLAG–Ets-1 (either WT or the 2KR mutant). At 24 h after transfection, cells were treated with vehicle (DMSO) as a control or MG132 (20 μM). After 30 min, CHX (100 μg/ml) was added to the medium. At the indicated time points after the addition of CHX, cell lysates were prepared and FLAG–Ets-1 and β-actin protein levels were detected by immunoblotting with anti-FLAG and β-actin monoclonal antibodies respectively. β-actin was used as a loading control. (B) COS-7 cells were transiently co-transfected with FLAG–Ets-1 (either WT or the 2KR mutant) and Myc–PIASy expression plasmids. After 24 h, CHX (100 μg/ml) was added to the medium. Cell lysates were collected at the indicated time and subjected to immunoblotting with the indicated antibodies, as described for (A). (C) Immunoblot band densities were determined using ImageJ (NIH) and Ets-1 levels were normalized to the loading control, β-actin. The percentage level of Ets-1 was calculated relative to zero time (100%). (D) COS-7 cells were transiently transfected with EGFP (enhanced green fluorescent protein)–PIASy or EGFP control vector. At 24 h after transfection, cells were treated with vehicle (DMSO) as a control, CHX (100 μg/ml) or MG132 (20 μM). After 5 h, cell lysates were prepared and subjected to immunoblotting with the indicated antibodies. (E) COS-7 cells were transiently co-transfected with FLAG–Ets-1 and Myc–PIASy C342A mutant. At 24 h after transfection, cells were treated with CHX (100 μg/ml) or left untreated. After 5 h, cell lysates were prepared and subjected to immunoblotting with the indicated antibodies.
Previously, we observed that co-expression of PIASy and Ets-1 leads to an increase in Ets-1 protein levels (T. Nishida, M. Terashima and K. Fukami, unpublished work). Therefore we examined whether Ets-1 was stabilized by overexpression of PIASy. We assessed the stability of ectopically expressed Ets-1 in COS-7 cells co-transfected with PIASy following treatment with CHX. Overexpression of PIASy markedly reduced the clearance of WT Ets-1 in cells treated with CHX (Figures 1B and 1C) and the Ets-1 mutant, 2KR, was equally stabilized by PIASy. As with ectopically expressed Ets-1, the levels of endogenously expressed Ets-1 in COS-7 cells were higher when PIASy was overexpressed, in both CHX-treated and untreated cells (Figure 1D). Furthermore, the C342A mutant PIASy retained the ability to stabilize Ets-1 protein levels (Figure 1E), indicating that the SUMO-E3 ligase activity of PIASy is not required for Ets-1 stabilization. These results suggest that PIASy overexpression protects Ets-1 from proteasomal degradation independently of Ets-1 SUMOylation.
PIASy specifically stabilizes Ets-1
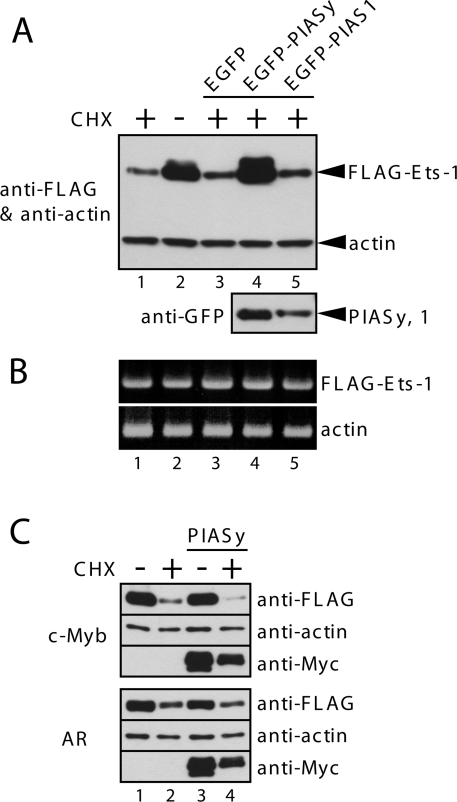
PIASy specifically interacts with Ets-1, and enhances its SUMOylation. To examine whether this Ets-1-protein-stabilization effect is specific to PIASy, we investigated whether another PIAS protein, PIAS1, could protect Ets-1 from proteasomal degradation. However, after 5 h of CHX treatment, PIAS1 overexpression did not protect Ets-1 from degradation (Figure 2A). To exclude any transcriptional effect on Ets-1, the level of Ets-1 mRNA was determined by semi-quantitative RT-PCR and products were visualized by agarose-gel electrophoresis (Figure 2B). The levels of Ets-1 mRNA in cells transfected with the Ets-1 plasmid alone, or with a combination of Ets-1 and PIASy expression plasmids, were similar, suggesting that the stabilization effect of PIASy on Ets-1 protein is probably due to a post-transcriptional event. To further examine the specificity of the effect of PIASy on the Ets-1 stability, we used two known PIASy-interacting proteins, c-Myb and the AR (androgen receptor), in stability assays. As shown in Figure 2(C), the expression levels of both proteins were reduced by CHX treatment (lane 2), most probably due to the proteasome-mediated degradation of these proteins, which has been reported previously [19,20]. However, the degradation of c-Myb and AR in CHX-treated cells was unaffected by co-expression with PIASy (lane 4), suggesting that PIASy specifically protects Ets-1 from proteasome-mediated degradation.
Figure 2. PIASy specifically stabilizes Ets-1.
(A) COS-7 cells were transiently co-transfected with FLAG–Ets-1 and Myc–PIASy C342A mutant. At 24 h after transfection, cells were treated with CHX (100 μg/ml) or left untreated. After 5 h, cell lysates were prepared and subjected to immunoblotting with the indicated antibodies. (A) COS-7 cells were transiently co-transfected with FLAG–Ets-1 and EGFP–PIASy or EGFP–PIAS1. CHX treatment and immunoblotting were performed as described for Figure 1(E). (B) Cells were co-transfected and treated as described for (A). Total cellular RNA was extracted and subjected to semi-quantitative RT-PCR. β-Actin was used as a control. (C) COS-7 cells were transiently co-transfected with FLAG–c-Myb or FLAG–AR and Myc–PIASy. CHX treatment and immunoblotting were performed as described for Figure 1(E).
siRNA (small interfering RNA)-mediated knockdown of PIASy reduces Ets-1 protein levels
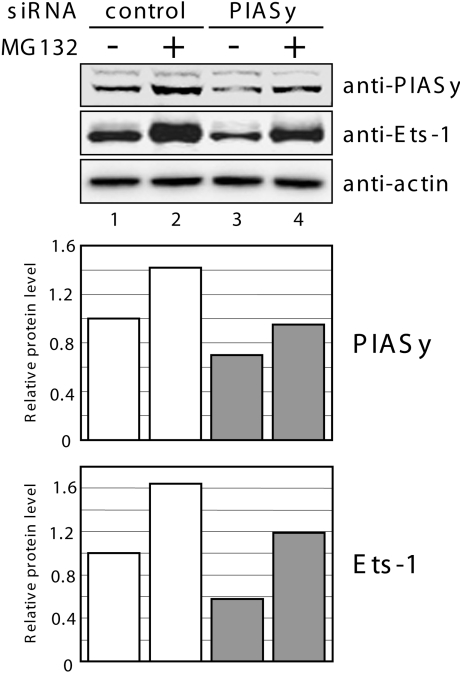
To further confirm the importance of PIASy in controlling Ets-1 protein levels, we postulated whether down-regulation of endogenous PIASy expression using an RNAi technique would destabilize endogenous Ets-1 protein. COS-7 cells were transfected with an siRNA duplex designed to silence PIASy expression. At 48 h after transfection, cell lysates were analysed with PIASy- and Ets-1-specific antibodies (Figure 3). In cells transfected with PIASy siRNA, endogenous PIASy protein expression was reduced compared with cells transfected with control siRNA (lane 3). Importantly, knockdown of PIASy protein expression resulted in a significant decrease of Ets-1 protein levels. Moreover, this Ets-1 protein reduction was rescued after treatment with the proteasome inhibitor MG132 (lane 4), suggesting that endogenous PIASy protects endogenous Ets-1 protein, at least in part, from proteasomal degradation. These observations confirmed that basal PIASy levels are important in controlling basal Ets-1 levels.
Figure 3. Suppression of PIASy expression down-regulates Ets-1 expression.
COS-7 cells were transiently transfected with either control siRNA or PIASy siRNA. At 48 h after transfection, cells were treated with vehicle (DMSO) as a control or MG132 (20 μM). After 5 h, cell lysates were prepared and subjected to immunoblotting with PIASy-, Ets-1- and β-actin-specific antibodies (top panel). Immunoblot band densities were determined using ImageJ (NIH), and PIASy and Ets-1 levels were normalized to the loading control, β-actin (middle and bottom panels respectively). The value for the control siRNA transfected cells in the absence of MG132 was assigned an arbitrary value of 1.0.
Mapping of the Ets-1 region responsible for the interaction with PIASy
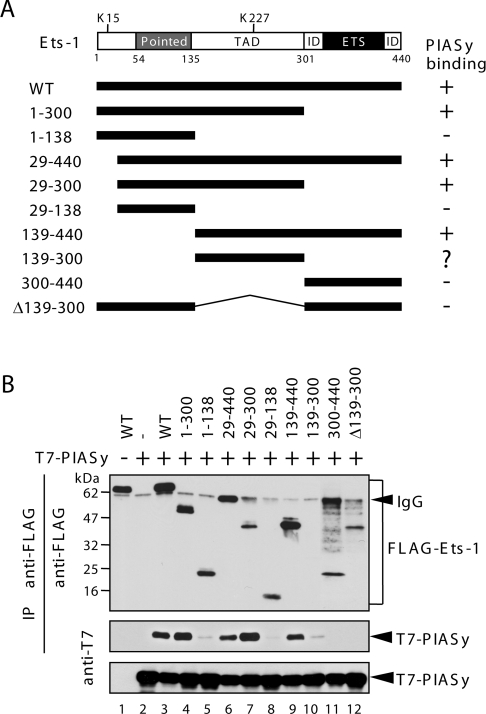
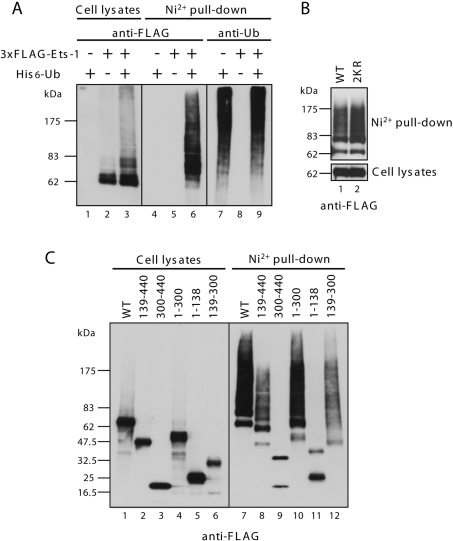
Previously, co-immunoprecipitation analysis revealed that PIASy interacts with Ets-1 [13]. To identify the region of Ets-1 that interacts with PIASy, we generated deletion mutants of Ets-1 (Figure 4A) and tested their ability to interact with PIASy in co-immunoprecipitation assays. Whole-cell lysates were prepared and co-immunoprecipitation assays were performed with an anti-FLAG antibody, followed by immunoblotting with an anti-T7 antibody. As shown in Figure 4(B) (middle panel), the interaction with PIASy was maintained when the C-terminal ETS domain was removed (lane 4; 1–300). Similarly, deletion of the N-terminal activation domain and the ‘pointed’ domain did not affect the interaction (lane 9; 139–440). By contrast, deletion of the internal TAD (transcriptional activation domain) completely abolished the binding (lanes 5, 11 and 12; 1–138, 300–440, Δ139–300). These data, which are summarized in Figure 4(A), indicate that the region containing the internal TAD (residues 139–300) is critical for the interaction with PIASy.
Figure 4. Mapping of Ets-1 for the PIASy interaction domain.
(A) Schematic diagrams of the Ets-1 constructs used. The two SUMOylation sites (Lys15 and Lys227) are indicated. Pointed, Pointed domain; ID, inhibitory domain; ETS, ETS domain. Numbers refer to amino acid residues. The interaction of Ets-1 mutants with PIASy following immunoprecipitation (IP) is summarized. (B) Lysates from COS-7 cells were co-transfected with FLAG–Ets-1 mutants and T7–PIASy were immunoprecipitated with an anti-FLAG antibody. The immunoprecipitates were then analysed by immunoblotting with an anti-FLAG antibody (top panel) or an anti-T7 antibody (middle panel). The middle panel shows the Ets-1-bound PIASy. The levels of transfected T7–PIASy protein in the total cell lysates are shown in the bottom panel.
The levels of immunoprecipitated full-length Ets-1 and deletion mutants were comparable, except for the 139–300 mutant (Figure 4B, upper panel), which was barely detectable by immunoblotting (lane 10). We reasoned that PIASy may destabilize this mutant. To test this hypothesis, we compared the levels of 139–300 mutant or WT Ets-1 when co-expressed with PIASy and found that PIASy efficiently decreased expression of the 139–300 mutant protein, but had no effect on the expression of WT Ets-1(Figure 4A: WT and 139–300). These results suggest that residues 139–300 of Ets-1 mediate the interaction with PIASy; however, this fragment alone might not be stable enough to interact specifically with PIASy.
The TAD and the C-terminal region, including the Ets domain, are required for PIASy-mediated stabilization
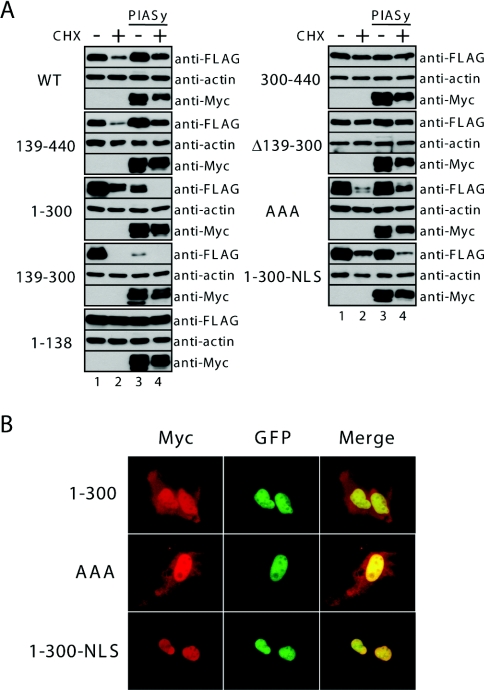
We investigated which region(s) of Ets-1 is/are required for its degradation and for PIASy-mediated stabilization. To achieve this, we performed CHX-chase experiments in COS-7 cells that transiently expressed Ets-1 deletion mutants with or without Myc–PIASy (Figure 5A). No alteration in proteasomal degradation or PIASy-mediated stabilization were observed for the N-terminal truncation mutant 139–440. Interestingly, PIASy did not stabilize the C-terminal deletion mutant 1–300 or the N-and C-terminal deletion mutant 139–300; rather, PIASy enhanced the degradation of these mutants following CHX treatment as described above. Furthermore, the deletion mutants 1–138, Δ139–300 and 300–440 were not rapidly degraded by the proteasome. These results indicate that the C-terminal region (residues 301–440) is necessary, but not sufficient, for PIASy-mediated stabilization, and that the region containing residues 139–300 is necessary for proteasomal degradation of Ets-1.
Figure 5. Ets-1 region responsible for stabilization by PIASy.
(A) COS-7 cells were transiently co-transected with FLAG–Ets-1 mutants and Myc–PIASy. At 24 h after transfection, CHX (100 μg/ml) was added to the medium and, after 5 h, cell lysates were prepared and subject to immunoblotting with the indicated antibodies. The results presented are representative of three independent experiments. (B) Subcellular localization of Ets-1 mutants. U2OS cells were grown on coverslips and transiently co-transfected with the indicated FLAG–Ets-1 mutants and EGFP–PIASy. At 24 h after transfection, the cells were incubated with an anti-FLAG antibody followed by an Alexa Fluor 568-conjugated secondary antibody, and visualized under a fluorescence microscope. The co-localization of FLAG–Ets-1 and EGFP–PIASy is shown in the merged images.
The C-terminal domain (residues 301–440) contains the Ets domain, which is a DNA-binding domain, and the NLS (nuclear localization signal) is located within this domain [21]. Indeed, both the NLS mutant (AAA) and the C-terminal deletion mutant 1–300, which lack both the Ets domain and therefore the NLS, become dispersed throughout the cell (Figure 5B). Although the AAA Ets-1 mutant was not efficiently stabilized by PIASy following CHX treatment, when compared with WT Ets-1, it was not destabilized by PIASy co-expression. We therefore examined whether enforced nuclear accumulation of a mutant that lacks the NLS leads to stabilization by PIASy after CHX treatment. For this purpose, we added the simian-virus-40 NLS to the C-terminus of the 1–300 mutant. As expected, the 1–300 NLS mutant accumulated in the nucleus (Figure 5B), suggesting that the NLS of this mutant recovered the stability of the 1–300 mutant. The efficient nuclear accumulation of Ets-1 appears to be important for the protection from PIASy-induced instability. However, PIASy did not stabilize 1–300 NLS efficiently when compared with WT Ets-1. These results suggest that the C-terminal region of Ets-1 plays a critical role in stabilization by PIASy, in addition to its role in nuclear localization.
Ets-1 is polyubiquitinated in cells
The fact that Ets-1 is largely degraded by the proteasome suggests that it may be polyubiquitinated, a post-translational modification that marks target proteins for proteasomal degradation [14]. In order to further characterize the molecular mechanism that underlies the stabilization of Ets-1 by PIASy, we tested whether Ets-1 could be polyubiquitinated. COS-7 cells were transfected with expression plasmids encoding 3×FLAG–Ets-1 and/or His6-ubiquitin. The transfected cells were lysed after MG132 treatment and the ubiquitinated proteins were isolated by binding to Ni-NTA beads. Immunoblots of the eluates were probed with an anti-FLAG or anti-ubiquitin antibody. As shown in Figure 6(A), high-molecular-mass bands representing polyubiquitinated Ets-1 were detected in cells that were co-transfected with 3×FLAG–Ets-1 and His6-ubiquitin (lane 6). The two SUMOylation sites of Ets-1 were not required for ubiquitination (Figure 6B). To determine which region of Ets-1 becomes ubiquitinated, a series of deletion mutants were used to perform in vivo ubiquitination assays. As expected, mutants lacking the transactivation region (residues 139–300), which abrogated the proteasome-dependent degradation in transfected cells, were not polyubiquitinated (Figure 6C, lanes 9 and 11).
Figure 6. Ets-1 is ubiquitinated in vivo.
(A) COS-7 cells were transiently co-transfected with 3×FLAG–Ets-1 and His6-ubiquitin as indicated. At 24 h after transfection, the cells were treated with MG132 for 4 h and lysed in buffer containing 6 M guanidinium chloride. His6-ubiquitinated species were purified with Ni-NTA–agarose beads and analysed by immunoblotting with an anti-FLAG antibody and an anti-ubiquitin antibody. Total cell lysates were subjected to immunoblotting and incubated with an anti-FLAG antibody. (B and C) Experiments analogous to those in (A) were performed using the indicated Ets-1 mutants.
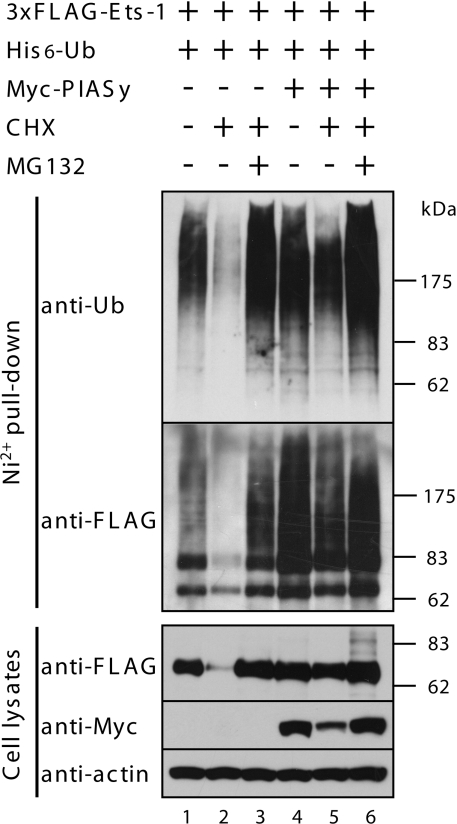
PIASy mediates the accumulation of polyubiquitinated Ets-1
To test whether PIASy had an effect on ubiquitination of Ets-1, COS-7 cells were transfected with expression plasmids encoding 3×FLAG–Ets-1, His6-ubiquitin and either vector or Myc–PIASy. Cells were then incubated in the presence or absence of CHX for 5 h, harvested and the ubiquitination state of Ets-1 was analysed. As shown in Figure 7, under control conditions, vector-transfected cells contained polyubiquitinated Ets-1 (lane 1); however, this polyubiquitination was reduced following treatment with CHX for 5 h (lane 2) by proteasomal degradation. Interestingly, co-expression of PIASy led to the accumulation of polyubiquitinated forms of Ets-1, regardless of CHX treatment (lanes 4 and 5). When c-Myb and AR were used as substrates for the ubiquitination assays, they were efficiently polyubiquitinated, and the polyubiquitination forms of these proteins were removed by proteasomal degradation after CHX treatment, regardless of the co-expression of PIASy (results not shown). These results suggest that overexpression of PIASy leads to the accumulation of polyubiquitinated forms of Ets-1.
Figure 7. PIASy protects polyubiquitinated Ets-1 proteins from proteasomal proteolysis.
COS-7 cells were transiently co-transfected with 3×FLAG–Ets-1, His6-ubiquitin and Myc–PIASy. Cells were treated with MG132 (20 μM) and CHX (100 μg/ml) for 5 h before lysis. Immunoblotting was performed as described for Figure 6(A).
DISCUSSION
In the present study we demonstrated that Ets-1 is unstable in transfected cells and that its degradation is independent of SUMOylation, but dependent on the ubiquitin–proteasome pathway. These results are in good agreement with those recently reported by Ji et al. [22]. In their paper they mainly addressed Ets-1 SUMOylation, but also dealt with ubiquitination. Their results concerning Ets-1 SUMOylation were almost identical with our published results [13], except for some differences caused by using different materials. In addition to these similar results, our published results [13] have shown the significant repressive activity of PIASy towards Ets-1-dependent transcription. Interestingly, this activity was partly dependent on its SUMO-E3 ligase activity, but independent of the SUMOylation status of Ets-1. In respect to Ets-1 ubiquitination, the results of Ji et al. [22] showed that the majority of Ets-1 polyubiquitin chains are Lys48-linked: an observation confirmed in our laboratory (result not shown). More importantly, Ji et al. [22] showed that Ets-1 SUMOylation does not affect stability and ubiquitination. SUMOylation can affect ubiquitination and the following proteasome-mediated degradation in some target proteins [23,24]. In the case of IκBα (inhibitory κBα), SUMOylation and ubiquitination occurred antagonistically at a same lysine residue. SUMOylated IκBα cannot be ubiquitinated and is resistant to proteasome-mediated degradation [23]. Modification of the transcription factor c-Myb with at least two molecules of SUMO-1 substantially increased its stability. Unlike IκBα, modification of SUMO-1 and ubiquitin do not compete for the same lysine residue [24]. However, mutations of Ets-1 SUMOylation sites did not alter stability and ubiquitination. Moreover, both non-sumoylated and sumoylated forms of Ets-1 show similar proteolytic processing rates [22]. Although our present results overlap with the findings of Ji et al. [22], we also showed that PIASy overexpression enhances Ets-1 stability by a mechanism that prevents its proteasome-mediated degradation. Conversely, siRNA-mediated knockdown of PIASy caused further destabilization of the Ets-1 protein. These findings suggested that PIASy expression levels control Ets-1 protein levels physiologically. PIASy interacts with the TAD of Ets-1, which is thought to be critical for the proteasomal degradation of Ets-1. In the present study we could not ascertain whether the direct interaction with PIASy was required for stabilization of Ets-1, as deletion of the TAD led to stable expression in the absence of PIASy. Since deletion of this domain led to a reduction in ubiquitinated forms of Ets-1, it is possible that both the ubiquitination sites and/or the ubiquitin ligase recognition region lie within the transactivation region: PIASy does not directly reduce Ets-1 ubiquitination. It is plausible that PIASy might inhibit the regulatory particles of the proteasome that recognize polyubiquitin chains. Clearly, this requires further investigation.
PIAS1 is another member of the PIAS family, which has been reported to regulate the stability of p73, a p53-related nuclear transcription factor [25]. PIAS1 enhances the SUMOylation of the p73α and ΔNp73α isoforms of p73, but not the C-terminally truncated isoforms, p73β and γ – yet this stabilization effect of PIAS1 did not require the SUMOylation of p73 [25]. Recent reports show that PIAS1 enhances Sox9 (sex-determining-region-Y-related high-mobility-group box 9) protein levels, by preventing its degradation via the proteasome pathway [26]. In a manner similar to the stabilization of Ets-1 by PIASy, the stabilization of Sox9 by PIAS1 seems to occur at the post-transcriptional level, although the mechanisms have not been characterized.
Importantly, increased levels of transcription factors such as p73 and Sox9 might result in increased transcriptional activity. It remains unclear whether ubiquitination directly regulates Ets-1 transcriptional activity; PIASy has been shown to repress Ets-1-dependent transcriptional activity in a dose-dependent manner [13]. This cannot be explained by a change in Ets-1 protein levels, because protein expression levels of transcription factors usually reflect their transcriptional activities. It appears that PIASy has a dual role in repressing transcriptional activity and enhancing protein levels of Ets-1. One potential explanation for these apparently contradictory findings is that ubiquitin-dependent proteasomal degradation is required for efficient transactivation of the Ets-1-dependent transcription. Several reports have proposed that proteasome-mediated degradation of transcription factors may have a requisite role in the mechanism of transcriptional activation [15,16]. PIASy might inhibit the proteasomal degradation process for specific transcription factors, but not the process of ubiquitination. As a result, ubiquitination forms are accumulated and it may increase the protein levels.
In conclusion, we have demonstrated that Ets-1 undergoes two distinct post-translational modifications: SUMOylation and ubiquitination. PIASy attenuates Ets-1-mediated transcription, at least in part, by preventing Ets-1 protein turnover via the ubiquitin–proteasome system in addition to functioning as a SUMO-E3 ligase. Our present results therefore suggest that PIASy plays multiple roles in regulating Ets-1 activity.
Acknowledgments
We thank Dr Takayuki Ohshima for his generosity in providing plasmids. This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to T.N.).
References
- 1.Graves B. J., Petersen J. M. Specificity within the Ets family of transcription factors. Adv. Cancer Res. 1998;75:1–55. doi: 10.1016/s0065-230x(08)60738-1. [DOI] [PubMed] [Google Scholar]
- 2.Seth A., Ascione R., Fisher R. J., Mavrothalassitis G. J., Bhat N. K., Papas T. S. The Ets gene family. Cell Growth Differ. 1992;3:327–334. [PubMed] [Google Scholar]
- 3.Janknecht R., Nordheim A. Gene regulation by Ets proteins. Biochim. Biophys. Acta. 1993;1155:346–356. doi: 10.1016/0304-419x(93)90014-4. [DOI] [PubMed] [Google Scholar]
- 4.Wasylyk B., Hahn S. L., Giovane A. The Ets family of transcription factors. Eur. J. Biochem. 1993;211:7–18. doi: 10.1007/978-3-642-78757-7_2. [DOI] [PubMed] [Google Scholar]
- 5.Sharrocks A. D., Brown A. L., Ling Y., Yates P. R. The ETS-domain transcription factor family. Int. J. Biochem. Cell Biol. 1997;29:1371–1387. doi: 10.1016/s1357-2725(97)00086-1. [DOI] [PubMed] [Google Scholar]
- 6.Span P. N., Manders P., Heuvel J. J., Thomas C. M., Bosch R. R., Beex L. V., Sweep C. G. Expression of the transcription factor Ets-1 is an independent prognostic marker for relapse-free survival in breast cancer. Oncogene. 2002;21:8506–8509. doi: 10.1038/sj.onc.1206040. [DOI] [PubMed] [Google Scholar]
- 7.Buggy Y., Maguire T. M., McGreal G., McDermott E., Hill A. D., O'Higgins N., Duffy M. J. Overexpression of the Ets-1 transcription factor in human breast cancer. Br. J. Cancer. 2004;91:1308–1315. doi: 10.1038/sj.bjc.6602128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katayama S., Nakayama T., Ito M., Naito S., Sekine I. Expression of the Ets-1 proto-oncogene in human breast carcinoma: differential expression with histological grading and growth pattern. Histol. Histopathol. 2005;20:119–126. doi: 10.14670/HH-20.119. [DOI] [PubMed] [Google Scholar]
- 9.Sementchenko V. I., Watson D. K. Ets target genes: past, present and future. Oncogene. 2000;19:6533–6548. doi: 10.1038/sj.onc.1204034. [DOI] [PubMed] [Google Scholar]
- 10.Dittmer J. The biology of the Ets1 proto-oncogene. Mol. Cancer. 2003;2:29–49. doi: 10.1186/1476-4598-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 2004;18:2046–2059. doi: 10.1101/gad.1214604. [DOI] [PubMed] [Google Scholar]
- 12.Hay R. T. SUMO: a history of modification. Mol. Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 13.Nishida T., Terashima M., Fukami K. PIASy-mediated repression of the Ets-1 is independent of its sumoylation. Biochem. Biophys. Res. Commun. 2006;345:1536–1546. doi: 10.1016/j.bbrc.2006.05.065. [DOI] [PubMed] [Google Scholar]
- 14.Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 15.Lipford J. R., Deshaies R. J. Diverse roles for ubiquitin-dependent proteolysis in transcriptional activation. Nat. Cell Biol. 2003;5:845–850. doi: 10.1038/ncb1003-845. [DOI] [PubMed] [Google Scholar]
- 16.Muratani M., Tansey W. P. How the ubiquitin–proteasome system controls transcription. Nat. Rev. Mol. Cell Biol. 2003;4:192–201. doi: 10.1038/nrm1049. [DOI] [PubMed] [Google Scholar]
- 17.Nishida T., Yasuda H. PIAS1 and PIASxα function as SUMO-E3 ligases toward androgen receptor and repress androgen receptor-dependent transcription. J. Biol. Chem. 2002;277:41311–41317. doi: 10.1074/jbc.M206741200. [DOI] [PubMed] [Google Scholar]
- 18.Vetter M., Blumenthal S. G., Lindemann R. K., Manns J., Wesselborg S., Thomssen C., Dittmer J. Ets-1 is an effector of protein kinase Cα in cancer cells. Oncogene. 2005;24:650–661. doi: 10.1038/sj.onc.1208234. [DOI] [PubMed] [Google Scholar]
- 19.Feikova S., Wolff L., Bies J. Constitutive ubiquitination and degradation of c-myb by the 26S proteasome during proliferation and differentiation of myeloid cells. Neoplasma. 2000;47:212–218. [PubMed] [Google Scholar]
- 20.Lin H. K., Wang L., Hu Y. C., Altuwaijri S., Chang C. Phosphorylationdependent ubiquitylation and degradation of androgen receptor by Akt require Mdm2 E3 ligase. EMBO J. 2002;21:4037–4048. doi: 10.1093/emboj/cdf406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boulukos K. E., Pognonec P., Rabault B., Begue A., Ghysdael J. Definition of an Ets1 protein domain required for nuclear localization in cells and DNA-binding activity in vitro. Mol. Cell. Biol. 1989;9:5718–5721. doi: 10.1128/mcb.9.12.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ji Z., Degerny C., Vintonenko N., Deheuninck J., Foveau B., Leroy C., Coll J., Tulasne D., Baert J. L., Fafeur V. Regulation of the Ets-1 transcription factor by sumoylation and ubiquitinylation. Oncogene. 2007;26:395–406. doi: 10.1038/sj.onc.1209789. [DOI] [PubMed] [Google Scholar]
- 23.Desterro J. M., Rodriguez M. S., Hay R. T. SUMO-1 modification of IκBα inhibits NF-κB activation. Mol. Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 24.Bies J., Markus J., Wolff L. Covalent attachment of the SUMO-1 protein to the negative regulatory domain of the c-Myb transcription factor modifies its stability and transactivation capacity. J. Biol. Chem. 2002;277:8999–9009. doi: 10.1074/jbc.M110453200. [DOI] [PubMed] [Google Scholar]
- 25.Munarriz E., Barcaroli D., Stephanou A., Townsend P. A., Maisse C., Terrinoni A., Neale M. H., Martin S. J., Latchman D. S., Knight R. A., et al. PIAS-1 is a checkpoint regulator which affects exit from G1 and G2 by sumoylation of p73. Mol. Cell. Biol. 2004;24:10593–10610. doi: 10.1128/MCB.24.24.10593-10610.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hattori T., Eberspaecher H., Lu J., Zhang R., Nishida T., Kahyo T., Yasuda H., de Crombrugghe B. Interactions between PIAS proteins and SOX9 result in an increase in the cellular concentrations of SOX9. J. Biol. Chem. 2006;281:14417–14428. doi: 10.1074/jbc.M511330200. [DOI] [PubMed] [Google Scholar]