Abstract
Although PTEN (phosphatase and tensin homologue deleted on chromosome 10) is one of the most commonly mutated tumour suppressors in human cancers, loss of PTEN expression in the absence of mutation appears to occur in an even greater number of tumours. PTEN is phosphorylated in vitro on Thr366 and Ser370 by GSK3 (glycogen synthase kinase 3) and CK2 (casein kinase 2) respectively, and specific inhibitors of these kinases block these phosphorylation events in cultured cells. Although mutation of these phosphorylation sites did not alter the phosphatase activity of PTEN in vitro or in cells, blocking phosphorylation of Thr366 by either mutation or GSK3 inhibition in glioblastoma cell lines led to a stabilization of the PTEN protein. Our data support a model in which the phosphorylation of Thr366 plays a role in destabilizing the PTEN protein.
Keywords: phosphatase, phosphoinositide, protein stability, PTEN (phosphatase and tensin homologue deleted on chromosome 10), tumour suppressor
Abbreviations: CK2, casein kinase 2; DMAT, 2-dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole; GSK3, glycogen synthase kinase 3; HEK-293 cell, human embroynic kidney cell; MDCK cell, Madin–Darby canine kidney cell; PKB, protein kinase B; PI3K, phosphoinositide 3-kinase; PTEN, phosphatase and tensin homologue deleted on chromosome 10; ROCK, Rho-associated kinase
INTRODUCTION
PTEN (phosphatase and tensin homologue deleted on chromosome 10) regulates many cellular processes, including cell proliferation, survival, growth and motility, principally by inhibiting PI3K (phosphoinositide 3-kinase)-dependent signalling via its PtdIns(3,4,5)P3 phosphatase activity [1,2]. It is one of the most commonly mutated tumour suppressors in human cancers, with genetic alterations occurring in a wide range of human tumour types, but at especially high frequency in endometrial carcinoma and glioblastoma [3]. However, evidence has indicated that loss of PTEN expression in the absence of biallelic mutation occurs even more frequently [4–7]. Although possible mechanisms causing the lack of expression of PTEN in tumours retaining at least one wild-type PTEN copy have been identified, such as promoter methylation [8–10], it appears that other, unknown, mechanisms may be acting in many tumours [9,10]. Understanding the mechanisms regulating PTEN expression seems to be particularly important, as, unlike many tumour suppressors, strong evidence indicates that partial loss of PTEN expression can enhance tumour development [11–13].
It is clear that PTEN stability can be regulated through the C-terminal tail, which is phosphorylated upon a cluster of serine and threonine residues, Ser380, Thr382, Thr383 and Ser385. This phosphorylation appears to stabilize the PTEN protein as well as to inhibit its biological activity [14,15]. Also, a protein named PICT1/GLTSCR2 (protein interacting with C-terminal tail 1/glioma tumour suppressor candidate region gene 2) has been described that binds to the C-terminal tail of PTEN, knockdown of which by RNAi (RNA interference) also leads to reduced PTEN protein stability [16]. Although PTEN ubiquitination and proteasomal degradation have been implicated previously [15,17,18], it has recently been shown that PTEN stability can be regulated through ubiquitination mediated by the NEDD4-1 ubiquitin ligase [19]. Although it seems likely that C-terminal cluster phosphorylation regulates PTEN stability through regulating a conformational change in the protein [20], and thus ubiquitination, further mechanistic details are not yet clear [21–24].
Two other phosphorylation sites within the PTEN C-terminal tail have been identified, Ser370 and Thr366 [23,25]. Ser370 was first identified as a phosphorylation site by metabolic labelling and mutational analysis and also by MS [23,25]. It can be phosphorylated efficiently in vitro by CK2 (casein kinase 2). Thr366 was identified as a phosphorylation site based upon the combined use of MS, mutational analysis and the use of phospho-threonine/proline-specific antibodies [25]. It appears to be phosphorylated efficiently in vitro and probably in cells by GSK3 (glycogen synthase 3) [25]. In the present study, we have raised phospho-specific antibodies to phospho-Ser370 and phospho-Thr366, and used these to analyse the phosphorylation of these sites by CK2 and GSK3 respectively. We show that, although the phosphorylation of these sites does not appear to alter PTEN activity in vitro or in cells, phosphorylation of Thr366 specifically can lead to destabilization of the PTEN protein.
EXPERIMENTAL
Cell culture
U87MG glioblastoma cells and NIH 3T3 fibroblasts were obtained from the ECACC (European Collection of Animal Cell Cultures) and maintained in the recommended media. Standard cell culture media, additives and sera were from Invitrogen/Gibco. Other chemicals were from Sigma. PTEN was expressed in U87MG cells using an adapted baculoviral delivery system. Adapted baculoviruses containing the PTEN cDNA downstream of a CMV (cytomegalovirus) promoter were prepared in SF9 cells, using standard protocols developed for recombinant protein expression in insect cells, and added to low-confluence U87MG cell cultures for 24 h at 5% (v/v) culture volume. The use of fluorescently marked proteins and functional studies show that this routinely led to relatively even expression of target proteins in well over 95% of the cultured U87MG cells as described previously [26]. In most experiments in U87MG cells, baculoviruses were used to express PTEN at similar levels to endogenous levels in other cultured cells (see for example, Supplementary Figure 1 at http://www.BiochemJ.org/bj/405/bj4050439add.htm), although, in protein stability experiments, levels were 5–10 times higher in order to help the detection of 35S-labelled PTEN in immunoprecipitates.
Antibodies and Western blotting
Phospho-specific antibodies against PTEN phospho-Thr366 and PTEN phospho-Ser370 were raised using the phosphopeptides TSVT*PDV and TPDVS*DNE respectively (where * indicates the phosphorylation site). These peptides, along with a PTEN N-terminal peptide MTAIIKEIVSRNKRRY, were synthesized by Dr Graham Bloomberg (Molecular Recognition Centre, University of Bristol, Bristol, U.K.) and were injected into sheep at Diagnostics Scotland (Edinburgh, U.K.). Sheep were also immunized with full-length hexahistidine-tagged PTEN protein expressed and purified from bacteria. Antibodies were purified from serum by affinity for the immunized peptide or protein. All blotting experiments using affinity-purified phospho-specific antibodies included co-incubation with the corresponding dephosphopeptide (at 10 μg/ml) to block non-phospho-specific immunoreactivity. Mouse monoclonal (clone A2B1) and rabbit polyclonal antibodies to PTEN were purchased from Santa Cruz Biotechnology and Biosource respectively. Polyclonal antibodies to phospho-PTEN (Ser380/Thr382/Thr383) and phospho-Ser473 Akt/PKB (protein kinase B) were purchased from Cell Signaling Technologies. A rabbit polyclonal antibody raised against PTEN phospho-Ser385 was purchased from Biosource. To prepare cellular samples for protein gel electrophoresis, the following cell lysis buffer was used: 25 mM Hepes (pH 7.4), 150 mM NaCl, 1% Triton X-100, 0.1% 2 mercaptoethanol, 1 mM EDTA, 1 mM EGTA, 10 mM β-glycerophosphate, 50 mM sodium fluoride, 10 μg/ml leupeptin, 100 μM PMSF and 1 mM benzamidine. Proteins were separated by PAGE using pre-cast 4–12% gradient gels and blotted on to PVDF membranes (Polyscreen; NEN/PerkinElmer). Most reagents for electrophoresis and blotting were purchased from Invitrogen, and standard manufacturers' protocols were followed. Immunoprecipitation of PTEN used the A2B1 monoclonal antibody from Santa Cruz Biotechnology, pre-conjugated with agarose beads. The assay of immunoprecipitated Akt/PKB activity followed methods published previously [26,27]. Quantification was performed using AIDA image analysis software. In the case of data in Supplementary Figures 2 and 3 (at http://www.BiochemJ.org/bj/405/bj4050439add.htm), the scanned and presented images derived from processed film were used. In the case of data from Figure 4 and Supplementary Figure 4 (at http://www.BiochemJ.org/bj/405/bj4050439add.htm), blots were analysed directly using a CCD camera.
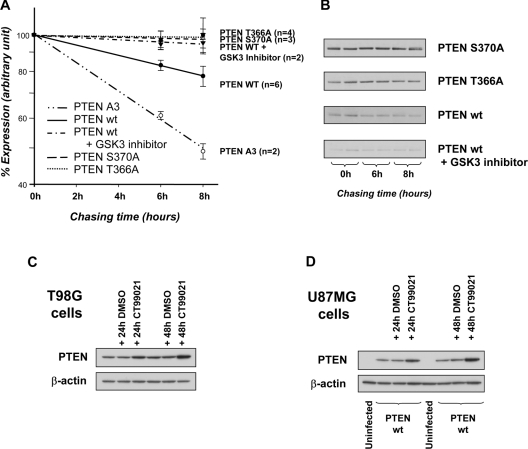
Figure 4. Thr366 phosphorylation regulates PTEN stability.
The stability of the PTEN proteins indicated was analysed by metabolic 35S amino-acid labelling of U87MG cells infected with viruses encoding wild-type (wt) PTEN, PTEN T366A, PTEN S370A or PTEN A3 (S380A T382A T383A) and chasing with unlabelled amino-acid-containing growth medium for the indicated times. The effects of the GSK3 inhibitor CT99021 on wild-type PTEN stability was also investigated (5 μM; present during both pulse and chase). (A) Results are shown on a log scale as the mean percentage expression from the indicated number of experiments (each performed in duplicate) ±S.E.M. from all data. A Student's t test indicated that results for all conditions were significantly different from wild-type at both 6 and 8 h (P <0.05), except PTEN 366A, which was significantly different at 6 h, but at 8 h gave P <0.1. (B) Immunoprecipitated PTEN proteins from one experiment detected using a phosphorimager. Relatively short chase times were used as this was found to reduce experimental variability. (C, D) The effect of the GSK3 inhibitor CT99021 on PTEN expression is shown in T98G cells (C) and U87MG cells (D). T98G cells expressing an endogenous PTEN protein or U87MG cells infected with viruses expressing wild-type PTEN were treated for 24 or 48 h with either DMSO vehicle or CT99021 before expression of PTEN was assessed by Western blotting for PTEN and β-actin. Quantification of these results is shown in Supplementary Figure 4 at http://www.BiochemJ.org/bj/405/bj4050439add.htm.
Purified recombinant proteins and in vitro assays
PTEN proteins were expressed in bacteria, purified, cleaved and isolated from the GST (glutathione S-transferase) tag as described previously [28]. Experiments addressing primed GSK3 phosphorylation required a 1 h phosphorylation at 30 °C with recombinant CK2 using 1 mM unlabelled ATP, followed by the addition of 100 μM CK2 inhibitor DMAT (2-dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole), 0.2 MBq [γ-32P]ATP and 1.0 unit/ml of GSK3β for a further 1 h at 30 °C. Stoichiometry calculations were based upon the known quantities of PTEN protein and the measured specific activities of the ATP stocks used. PTEN phosphatase assays against PtdIns(3,4,5)P3 and Ins(1,3,4,5)P4 followed methods described previously [29].
Analysis of cellular PTEN stability
U87MG cells were infected with baculoviruses encoding PTEN, then, 24 h after infection, cells were washed and incubated for 1 h in methionine/cysteine-free medium containing 10% (v/v) dialysed FCS (fetal calf serum). Cells were then labelled with [35S]methionine/cysteine (70 μCi/ml) for 2 h (Amersham Biosciences). After three washes with complete medium, cells were incubated in complete medium and chased for different times before lysis. PTEN proteins were isolated by immunoprecipitation and resolved by PAGE. The labelled PTEN present at each time point was quantified using a Fuji FLA-2000 phosphorimager and AIDA software.
RESULTS AND DISCUSSION
Phosphorylation of PTEN on Thr366 and Ser370
In order to investigate the phosphorylation of PTEN upon Thr366 and Ser370, phospho-specific antibodies against these sites were raised. The specificity of these antibodies was verified using bacterially expressed PTEN phosphorylated in vitro using GSK3 and CK2 and using PTEN protein mutated at each site, PTEN T366A and PTEN S370A (Figure 1). These antibodies did not recognize bacterially expressed PTEN, but gave a robust signal from PTEN phosphorylated in vitro. These antibodies also recognized PTEN in all cellular samples that we have investigated [U87 cells, NIH 3T3 cells, MDCK (Madin–Darby canine kidney) cells, HEK-293 (human embroynic kidney) cells, HeLa cells and rat liver tissue], indicating a degree of constitutive phosphorylation of both sites in these cell types. We also attempted to raise phospho-specific antibodies using phospho-Ser362 peptides. However, we did not observe any imunoreactivity using these antibodies, indicating either that Ser362 is not phosphorylated in cells or in vitro by GSK3, or that the immunizations failed to yield useful antibodies.
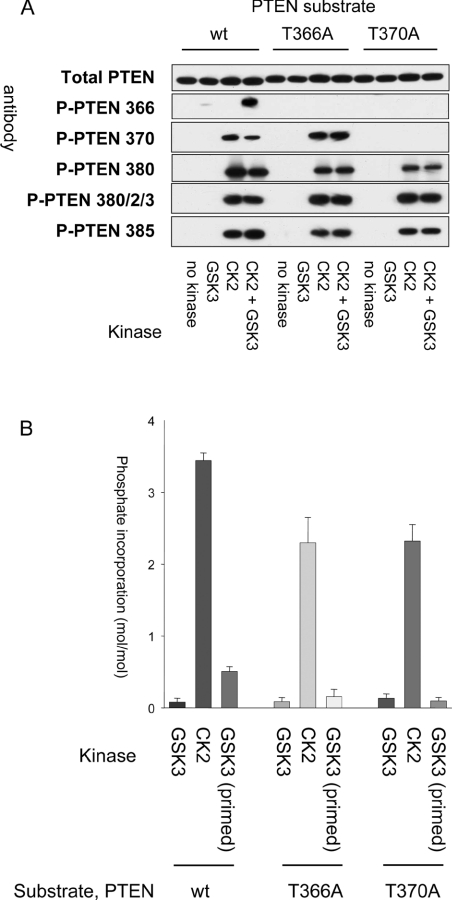
Figure 1. PTEN is phosphorylated in vitro by CK2 and GSK3.
(A) Purified recombinant PTEN, wild-type (wt), T366A mutant or S370A mutant, was phosphorylated in vitro using CK2, GSK3β or both kinases. PTEN protein was then analysed by Western blotting using the general (Total PTEN) and phospho-specific (P-PTEN) PTEN antibodies indicated. (B) Stoichiometry of phosphorylation by CK2 and GSK3. The stoichiometry of phosphate incorporation was calculated after phosphorylation in vitro of wild-type PTEN, PTEN T366A or PTEN S370A with CK2 or GSK3.
It has been proposed that Ser370 and Thr366 are phosphorylated by CK2 and GSK3 respectively, with CK2 phosphorylation acting as a priming event for subsequent GSK3 phosphorylation [25]. The enhancement of GSK3 phosphorylation by a priming phosphorylation event four residues C-terminal to the GSK3 substrate residue is well understood [30]. We first verified the phosphorylation of PTEN by CK2 and GSK3 in vitro using recombinant proteins. In agreement with previous studies, CK2 phosphorylated PTEN upon several sites, including Ser370 (Figure 1A). In order to assess whether the phosphorylation detected using phospho-specific antibodies represented a level of efficiency likely to occur in cells, we conducted more detailed experiments determining the stoichiometry of these phosphorylation events in vitro. This showed that CK2 incorporated between 3 and 4 mol phosphate/mol PTEN protein (Figure 1B), close to that described previously [15], although phosphorylation of Ser370 was less efficient than that of the cluster sites (residues 380–385) (Supplementary Figure 2 at http://www.BiochemJ.org/bj/405/bj4050439add.htm). GSK3 phosphorylated PTEN poorly without priming (mean stoichiometry of 0.11). However, after prior phosphorylation with CK2, GSK3 phosphorylated PTEN efficiently upon Thr366 (mean stoichiometry of 0.52; Figure 1B). GSK3 did not efficiently phosphorylate PTEN if either Thr366 or Ser370 was mutated. Comparison of cellular samples with PTEN phosphorylated to known stoichiometry in vitro allowed a crude estimate of the stoichiometry of phosphorylation of these sites in cells, suggesting a low level of phosphorylation of approx. 5–10% (Supplementary Figure 1 at http://www.BiochemJ.org/bj/405/bj4050439add.htm).
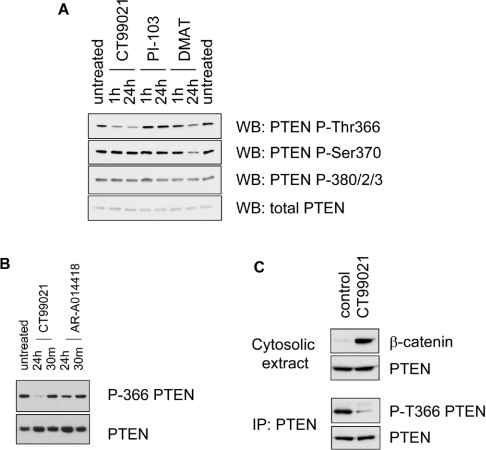
Investigating the phosphorylation of PTEN Thr366 by a panel of 37 protein kinases in vitro indicated that other protein kinases exist that can efficiently phosphorylate this site experimentally in addition to CK2 and GSK3 (results not shown). We therefore chose to address the effects on PTEN phosphorylation of a number of well-characterized small-molecule protein and lipid kinase inhibitors to determine the extent to which CK2 and GSK3 account for the phosphorylation of these sites in cells. PTEN phosphorylation upon both Thr366 and Ser370 in cells was inhibited by the CK2 inhibitor DMAT, whereas the GSK3 inhibitors CT99021 and AR-A014418 both inhibited the basal phosphorylation of PTEN on Thr366, but not Ser370 (Figure 2). The specificity of these inhibitors has been tested using large panels of protein kinases [31,32]. Strong suppression of phosphorylation by these inhibitors required long incubation times, of several hours or longer, and varied between cell types, being slower when PTEN was expressed in U87MG cells than with endogenous PTEN in NIH 3T3 cells (Figure 2, and results not shown). This suggests that under normal conditions dephosphorylation of these sites is slow. We also addressed other plausible or proposed mechanisms regulating PTEN phosphorylation, such as prolinedirected kinase phosphorylation of Thr366, and phosphorylation by ROCK (Rho-associated kinase)- and PI3K-dependent feedback phosphorylation [22,33]. In these experiments, the phosphorylation on Thr366 and Ser370 of wild-type PTEN expressed in U87MG cells was not affected by incubation with the DYRK (dual-specificity tyrosine-phosphorylated and -regulated kinase) inhibitor harmine (10 μM), the MEK (mitogen-activated protein kinase/extracellular-signal-reulated kinase kinase) inhibitor U0126 (5 μM), the CDK2 (cyclin-dependent kinase 2) inhibitor roscovitine (20 μM) or the ROCK inhibitor Y27632 (5 μM) (results not shown). The phosphorylation of PTEN on Thr366 and Ser370 was not greatly affected by the PI3K inhibitors LY294002 (50 μM), wortmannin (100 nM) or PI-103 (1 μM) (Figure 2A, and results not shown), although a reduction in the expression level of PTEN was often observed, consistent with the previously proposed PI3K feedback regulation of PTEN stability [22]. Significantly, the efficient inhibition of phosphorylation by specific GSK3 and CK2 inhibitors in cultured cells also suggests that other kinases are not responsible for the majority of the phosphorylation seen. These results strongly support the conclusion that in unstimulated cells PTEN is phosphorylated upon Thr366 and Ser370, principally by the protein kinases GSK3 and CK2 respectively, and that Ser370 phosphorylation acts to prime PTEN for phosphorylation upon Thr366 by GSK3.
Figure 2. Inhibition of GSK3 and CK2 reduces phosphorylation of PTEN on Thr366 and Ser370 respectively.
(A) NIH 3T3 cells were treated for the indicated times with the GSK3 inhibitor CT99021 (5 μM), the PI3K inhibitor PI-103 (1 μM) or the CK2 inhibitor DMAT (20 μM) and phosphorylation of PTEN was investigated by Western blotting using phospho-specific antibodies. Very similar results were obtained in U87MG cells expressing wild-type PTEN. (B) U87MG cells expressing PTEN were treated with the GSK3 inhibitors CT99021 (5 μM) or AR-A0144-18 (10 μM) for either 30 min (30 m) or 24 h before PTEN phosphorylation was investigated using PTEN phospho-Thr366-specific antibodies (P-366 PTEN). (C) MDCK cells were left untreated or treated with 2 μM CT99021 for 6 h. Cells were then subjected to hypotonic lysis to prepare a cytosolic extract, and the presence of PTEN and β-catenin in this extract and the phosphorylation of PTEN were investigated by immunprecipitation and Western blotting using phospho-Thr366-specific antibodies (P-T366 PTEN). The stabilization of cytosolic β-catenin was used to verify the efficacy of GSK3 inhibition. All results are representative of at least two separate experiments in the same cell type. Quantification of these results is shown in Supplementary Figure 3 at http://www.BiochemJ.org/bj/405/bj4050439add.htm.
Thr366 phosphorylation reduces PTEN stability in glioblastoma cell lines
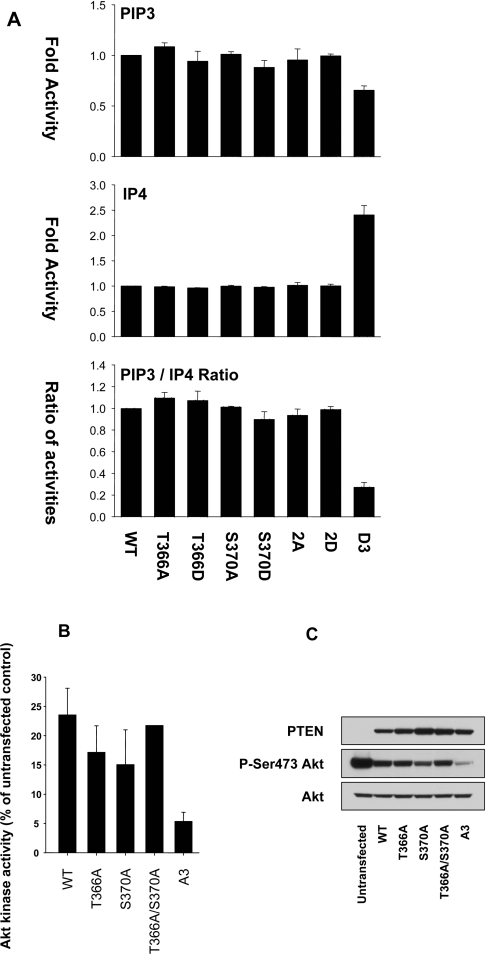
Phosphorylation of the C-terminal cluster sites of PTEN (residues 380–385) has been shown to lead to its reduced biological activity in the regulation of PI3K-dependent signalling, probably through an electrostatic shift in PTEN conformation causing reduced association with the plasma membrane and reduced metabolism of PtdIns(3,4,5)P3 [14,20,34,35]. We sought to investigate whether phosphorylation of Thr366 and Ser370 also affected the activity of PTEN, either in vitro or in cells. There was no significant effect of mutation of either phosphorylation site to alanine or aspartic acid on the in vitro phosphatase activity of these proteins against the lipid substrate PtdIns(3,4,5)P3, the soluble inositol phosphate Ins(1,3,4,5)P4 or the model peptide substrate poly(GluTyr) (Figure 3, and results not shown). Importantly, there was no indication of a shift in the ratio of activities against PtdIns(3,4,5)P3 and Ins(1,3,4,5)P4, a sensitive measure of C-terminal phosphorylation [29] (Figure 3).
Figure 3. The PTEN phosphorylation sites Thr366 and Ser370 do not affect phosphatase activity in vitro or in cells.
(A) Bacterially expressed PTEN proteins were assayed in vitro against phosphatidylcholine/PtdIns(3,4,5)P3 vesicles (100 ng of enzyme) and against the soluble substrate Ins(1,3,4,5)P4 (500 ng of enzyme). PTEN 2A and 2D refer to double mutants with either alanine or aspartic acid replacements respectively, of both Thr366 and Ser370. PTEN D3 refers to an aspartic acid triple replacement mutant of Ser380, Thr382 and Thr383. Results are shown for each substrate as the mean±S.E.M. activity from triplicate samples and also the ratio of these two activities, shown as the mean±S.E.M. ratio from these paired triplicate samples. (B, C) U87MG cells were infected with viruses encoding the indicated PTEN proteins. PTEN A3 refers to an alanine triple replacement mutant of Ser380, Thr382 and Thr383. At 24 h after infection, cells were lysed and the activity (B) and phosphorylation (C) of endogenous Akt/PKB was determined by immunoprecipitate kinase assay and Western blotting respectively. WT, wild-type.
We also addressed the cellular activity of these proteins by expressing them in the PTEN-null glioblastoma cell line U87MG and observing the effect on the activation state of the downstream PtdIns(3,4,5)P3-dependent kinase Akt/PKB. In these experiments, expression of wild-type PTEN reduced Akt/PKB activity, whereas PTEN A3 (an alanine triple replacement mutant of Ser380, Thr382 and Thr383) had a substantially greater effect than the wild-type enzyme. The effect of PTEN T366A, PTEN S370A or a double mutant was similar to that of the wild-type enzyme (Figure 3). These results suggest that phosphorylation of these latter sites may not directly regulate biological activity in the manner of phosphorylation on the cluster sites Ser380, Thr382 and Thr383.
During these studies in U87MG cells, it became evident that long-term treatment with GSK3 inhibitors frequently caused a clear increase in PTEN protein levels (see, for example, Figure 2B). Similarly, parallel samples using several preparations of expression vectors or viruses in mammalian cells encoding wild-type PTEN and PTEN T366A or S370A mutants invariably led to greater expression levels of the mutant proteins (see, for example, Figure 3). These results suggested that phosphorylation at Thr366 might regulate protein stability. To address this possibility, we investigated the effects of PTEN mutation and GSK3 inhibitors on the stability of PTEN as measured using metabolic amino acid labelling and pulse/chase analysis. In order to address the stability of different PTEN mutants and also increase the abundance of PTEN protein in these experiments, we continued to perform these experiments using PTEN proteins expressed in U87MG cells. These experiments showed that PTEN T366A and S370A are both more stable than the wild-type enzyme, and also that treatment of cells with the GSK3 inhibitor CT99021 caused an increase in the stability and expression of wild-type PTEN (Figures 4A, 4B and 4D). As established previously, mutation of three of the C-terminal cluster of phosphorylation sites to alanine (PTEN A3 or PTEN T380A, S382A and T383A) had the opposite effect, reducing the stability of the PTEN protein [14,15].
We performed experiments to address the regulation of PTEN by Thr366 phosphorylation in other cells types, first in another glioma cell line, T98G, which expresses an endogenous mutant PTEN protein that is catalytically inactive ([36], and results not shown). Prolonged treatment of T98G cells with the GSK3 inhibitor CT99021 led to a strong increase in PTEN expression (approx. 3-fold; Figure 4C). However, treatment of NIH 3T3 fibroblasts, HEK-293 cells and MDCK epithelial cells for 24 or 48 h with CT99021 had no effect on the expression of PTEN in these cells, despite reducing phosphorylation of Thr366 (Figure 2, and results not shown). This suggests that additional circumstances must be met before the effects of Thr366 phosphorylation on protein stability can be revealed, which, in our experiments, are only fulfilled in the glioma cell type U87MG and T98G. This observed effect did appear very potent, as blocking Thr366 phosphorylation led to an almost complete block in detectable PTEN turnover (Figure 4).
Our results establish a role for the phosphorylation of Thr366 in regulating the stability of the PTEN protein. Cellular PTEN abundance controls basal levels of PtdIns(3,4,5)P3 and downstream signalling, and even modest effects on PTEN expression have significant effects both on normal physiology and development and on tumour development in many tissues [11–13,37]. Thus a phosphorylation event that destabilizes the PTEN protein may have an important role in regulating PTEN expression levels in some normal and tumour cells and potentially allow the development of novel therapeutic strategies to stabilize this important tumour suppressor.
Online data
Acknowledgments
We thank the protein and antibody production teams of the Division of Signal Transduction Therapy (DSTT) in Dundee, co-ordinated by James Hastie and Hilary McLauchlan, for purified proteins, and Jenny Bain and Matt Elliott for provision of the kinase inhibitors. We thank the MRC (Medical Research Council), the Association for International Cancer Research, and the pharmaceutical companies of the DSTT consortium (Astra Zeneca, Boehringer Ingelheim, GlaxoSmithKline, Merck and Co., Merck KGaA and Pfizer) for financial support.
References
- 1.Sulis M. L., Parsons R. PTEN: from pathology to biology. Trends Cell Biol. 2003;13:478–483. doi: 10.1016/s0962-8924(03)00175-2. [DOI] [PubMed] [Google Scholar]
- 2.Maehama T., Taylor G. S., Dixon J. E. PTEN AND MYOTUBULARIN: Novel phosphoinositide phosphatases. Annu. Rev. Biochem. 2001;70:247–279. doi: 10.1146/annurev.biochem.70.1.247. [DOI] [PubMed] [Google Scholar]
- 3.Ali I. U., Schriml L. M., Dean M. Mutational spectra of PTEN/MMAC1 gene: a tumor suppressor with lipid phosphatase activity. J. Natl. Cancer Inst. 1999;91:1922–1932. doi: 10.1093/jnci/91.22.1922. [DOI] [PubMed] [Google Scholar]
- 4.Leslie N. R., Downes C. P. PTEN function: how normal cells control it and tumour cells lose it. Biochem. J. 2004;382:1–11. doi: 10.1042/BJ20040825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parsons R. Human cancer, PTEN and the PI-3 kinase pathway. Semin. Cell Dev. Biol. 2004;15:171–176. doi: 10.1016/j.semcdb.2003.12.021. [DOI] [PubMed] [Google Scholar]
- 6.Zhou X. P., Loukola A., Salovaara R., Nystrom-Lahti M., Peltomaki P., de la Chapelle A., Aaltonen L. A., Eng C. PTEN mutational spectra, expression levels, and subcellular localization in microsatellite stable and unstable colorectal cancers. Am. J. Pathol. 2002;161:439–447. doi: 10.1016/S0002-9440(10)64200-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sansal I., Sellers W. R. The biology and clinical relevance of the PTEN tumor suppressor pathway. J. Clin. Oncol. 2004;22:2954–2963. doi: 10.1200/JCO.2004.02.141. [DOI] [PubMed] [Google Scholar]
- 8.Whang Y. E., Wu X., Suzuki H., Reiter R. E., Tran C., Vessella R. L., Said J. W., Isaacs W. B., Sawyers C. L. Inactivation of the tumor suppressor PTEN/MMAC1 in advanced human prostate cancer through loss of expression. Proc. Natl. Acad. Sci. U.S.A. 1998;95:5246–5250. doi: 10.1073/pnas.95.9.5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salvesen H. B., Stefansson I., Kretzschmar E. I., Gruber P., MacDonald N. D., Ryan A., Jacobs I. J., Akslen L. A., Das S. Significance of PTEN alterations in endometrial carcinoma: a population-based study of mutations, promoter methylation and PTEN protein expression. Int. J. Oncol. 2004;25:1615–1623. [PubMed] [Google Scholar]
- 10.Khan S., Kumagai T., Vora J., Bose N., Sehgal I., Koeffler P. H., Bose S. PTEN promoter is methylated in a proportion of invasive breast cancers. Int. J. Cancer. 2004;112:407–410. doi: 10.1002/ijc.20447. [DOI] [PubMed] [Google Scholar]
- 11.Kwabi-Addo B., Giri D., Schmidt K., Podsypanina K., Parsons R., Greenberg N., Ittmann M. Haploinsufficiency of the Pten tumor suppressor gene promotes prostate cancer progression. Proc. Natl. Acad. Sci. U.S.A. 2001;98:11563–11568. doi: 10.1073/pnas.201167798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma L., Teruya-Feldstein J., Behrendt N., Chen Z., Noda T., Hino O., Cordon-Cardo C., Pandolfi P. P. Genetic analysis of Pten and Tsc2 functional interactions in the mouse reveals asymmetrical haploinsufficiency in tumor suppression. Genes Dev. 2005;19:1779–1786. doi: 10.1101/gad.1314405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trotman L. C., Niki M., Dotan Z. A., Koutcher J. A., Di Cristofano A., Xiao A., Khoo A. S., Roy-Burman P., Greenberg N. M., Van Dyke T., Cordon-Cardo C., Pandolfi P. P. Pten dose dictates cancer progression in the prostate. PLoS Biol. 2003;1:e59. doi: 10.1371/journal.pbio.0000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vazquez F., Ramaswamy S., Nakamura N., Sellers W. R. Phosphorylation of the PTEN tail regulates protein stability and function. Mol. Cell. Biol. 2000;20:5010–5018. doi: 10.1128/mcb.20.14.5010-5018.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torres J., Pulido R. The tumor suppressor PTEN is phosphorylated by the protein kinase CK2 at its C terminus: implications for PTEN stability to proteasomemediated degradation. J. Biol. Chem. 2001;276:993–998. doi: 10.1074/jbc.M009134200. [DOI] [PubMed] [Google Scholar]
- 16.Okahara F., Ikawa H., Kanaho Y., Maehama T. Regulation of PTEN phosphorylation and stability by a tumor suppressor candidate protein. J. Biol. Chem. 2004;279:45300–45303. doi: 10.1074/jbc.C400377200. [DOI] [PubMed] [Google Scholar]
- 17.Tolkacheva T., Boddapati M., Sanfiz A., Tsuchida K., Kimmelman A. C., Chan A. M. Regulation of PTEN binding to MAGI-2 by two putative phosphorylation sites at threonine 382 and 383. Cancer Res. 2001;61:4985–4989. [PubMed] [Google Scholar]
- 18.Wu W., Wang X., Zhang W., Reed W., Samet J. M., Whang Y. E., Ghio A. J. Zinc-induced PTEN protein degradation through the proteasome pathway in human airway epithelial cells. J. Biol. Chem. 2003;278:28258–28263. doi: 10.1074/jbc.M303318200. [DOI] [PubMed] [Google Scholar]
- 19.Wang X., Trotman L. C., Koppie T., Alimonti A., Chen Z., Gao Z., Wang J., Erdjument-Bromage H., Tempst P., Cordon-Cardo C., Pandolfi P. P., Jiang X. NEDD4 is a proto-oncogenic ubiquitin ligase for PTEN. Cell. 2007;128:129–139. doi: 10.1016/j.cell.2006.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vazquez F., Grossman S. R., Takahashi Y., Rokas M. V., Nakamura N., Sellers W. R. Phosphorylation of the PTEN tail acts as an inhibitory switch by preventing its recruitment into a protein complex. J. Biol. Chem. 2001;276:48627–48630. doi: 10.1074/jbc.C100556200. [DOI] [PubMed] [Google Scholar]
- 21.Hong K. W., Kim K. Y., Shin H. K., Lee J. H., Choi J. M., Kwak Y. G., Kim C. D., Lee W. S., Rhim B. Y. Cilostazol prevents tumor necrosis factor-α-induced cell death by suppression of phosphatase and tensin homolog deleted from chromosome 10 phosphorylation and activation of Akt/cyclic AMP response element-binding protein phosphorylation. J. Pharmacol. Exp. Ther. 2003;306:1182–1190. doi: 10.1124/jpet.103.052365. [DOI] [PubMed] [Google Scholar]
- 22.Birle D., Bottini N., Williams S., Huynh H., deBelle I., Adamson E., Mustelin T. Negative feedback regulation of the tumor suppressor PTEN by phosphoinositide-induced serine phosphorylation. J. Immunol. 2002;169:286–291. doi: 10.4049/jimmunol.169.1.286. [DOI] [PubMed] [Google Scholar]
- 23.Miller S. J., Lou D. Y., Seldin D. C., Lane W. S., Neel B. G. Direct identification of PTEN phosphorylation sites. FEBS Lett. 2002;528:145–153. doi: 10.1016/s0014-5793(02)03274-x. [DOI] [PubMed] [Google Scholar]
- 24.Okamura A., Iwata N., Tamekane A., Yakushijin K., Nishikawa S., Hamaguchi M., Fukui C., Yamamoto K., Matsui T. Casein kinase Iε down-regulates phospho-Akt via PTEN, following genotoxic stress-induced apoptosis in hematopoietic cells. Life Sci. 2006;78:1624–1629. doi: 10.1016/j.lfs.2005.07.041. [DOI] [PubMed] [Google Scholar]
- 25.Al-Khouri A. M., Ma Y., Togo S. H., Williams S., Mustelin T. Cooperative phosphorylation of the tumor suppressor phosphatase PTEN by casein kinases and glycogen synthase kinase 3β. J. Biol. Chem. 2005;280:35195–35202. doi: 10.1074/jbc.M503045200. [DOI] [PubMed] [Google Scholar]
- 26.Leslie N. R., Bennett D., Gray A., Pass I., Hoang-Xuan K., Downes C. P. Targeting mutants of PTEN reveal distinct subsets of tumour suppressor functions. Biochem. J. 2001;357:427–435. doi: 10.1042/0264-6021:3570427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alessi D. R., Andjelkovic M., Caudwell B., Cron P., Morrice N., Cohen P., Hemmings B. A. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 1996;15:6541–6551. [PMC free article] [PubMed] [Google Scholar]
- 28.McConnachie G., Pass I., Walker S. M., Downes C. P. Interfacial kinetic analysis of the tumour suppressor phosphatase, PTEN: evidence for activation by anionic phospholipids. Biochem. J. 2003;371:947–955. doi: 10.1042/BJ20021848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ning K., Miller L. C., Laidlaw H. A., Burgess L. A., Perera N. M., Downes C. P., Leslie N. R., Ashford M. L. A novel leptin signalling pathway via PTEN inhibition in hypothalamic cell lines and pancreatic β-cells. EMBO J. 2006;25:2377–2387. doi: 10.1038/sj.emboj.7601118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frame S., Cohen P. GSK3 takes centre stage more than 20 years after its discovery. Biochem. J. 2001;359:1–16. doi: 10.1042/0264-6021:3590001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pagano M. A., Andrzejewska M., Ruzzene M., Sarno S., Cesaro L., Bain J., Elliott M., Meggio F., Kazimierczuk Z., Pinna L. A. Optimization of protein kinase CK2 inhibitors derived from 4,5,6,7-tetrabromobenzimidazole. J. Med. Chem. 2004;47:6239–6247. doi: 10.1021/jm049854a. [DOI] [PubMed] [Google Scholar]
- 32.Murray J. T., Campbell D. G., Morrice N., Auld G. C., Shpiro N., Marquez R., Peggie M., Bain J., Bloomberg G. B., Grahammer F., et al. Exploitation of KESTREL to identify NDRG family members as physiological substrates for SGK1 and GSK3. Biochem. J. 2004;384:477–488. doi: 10.1042/BJ20041057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z., Dong X., Wang Z., Liu W., Deng N., Ding Y., Tang L., Hla T., Zeng R., Li L., Wu D. Regulation of PTEN by Rho small GTPases. Nat. Cell Biol. 2005;7:399–404. doi: 10.1038/ncb1236. [DOI] [PubMed] [Google Scholar]
- 34.Vazquez F., Matsuoka S., Sellers W. R., Yanagida T., Ueda M., Devreotes P. N. Tumor suppressor PTEN acts through dynamic interaction with the plasma membrane. Proc. Natl. Acad. Sci. U.S.A. 2006;103:3633–3638. doi: 10.1073/pnas.0510570103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Das S., Dixon J. E., Cho W. Membrane-binding and activation mechanism of PTEN. Proc. Natl. Acad. Sci. U.S.A. 2003;100:7491–7496. doi: 10.1073/pnas.0932835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steck P. A., Pershouse M. A., Jasser S. A., Yung W. K., Lin H., Ligon A. H., Langford L. A., Baumgard M. L., Hattier T., Davis T., et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat. Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 37.Di Cristofano A., Kotsi P., Peng Y. F., Cordon-Cardo C., Elkon K. B., Pandolfi P. P. Impaired Fas response and autoimmunity in Pten+/− mice. Science. 1999;285:2122–2125. doi: 10.1126/science.285.5436.2122. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.