Abstract
A thiamine biosynthesis gene, thi3, from maize Zea mays has been identified through cloning and sequencing of cDNA and heterologous overexpression of the encoded protein, THI3, in Escherichia coli. The recombinant THI3 protein was purified to homogeneity and shown to possess two essentially different enzymatic activities of HMP(-P) [4-amino-5-hydroxymethyl-2-methylpyrimidine (phosphate)] kinase and TMP (thiamine monophosphate) synthase. Both activities were characterized in terms of basic kinetic constants, with interesting findings that TMP synthase is uncompetitively inhibited by excess of one of the substrates [HMP-PP (HMP diphosphate)] and ATP. A bioinformatic analysis of the THI3 sequence suggested that these activities were located in two distinct, N-terminal kinase and C-terminal synthase, domains. Models of the overall folds of THI3 domains and the arrangements of active centre residues were obtained with the SWISS-MODEL protein modelling server, on the basis of the known three-dimensional structures of Salmonella enterica serotype Typhimurium HMP(-P) kinase and Bacillus subtilis TMP synthase. The essential roles of Gln98 and Met134 residues for HMP kinase activity and of Ser444 for TMP synthase activity were experimentally confirmed by site-directed mutagenesis.
Keywords: cDNA cloning, enzyme kinetics, protein overexpression, site-directed mutagenesis, structural modelling, thiamine phosphate diphosphorylase
Abbreviations: CBD, chitin-binding domain; HET, 4-methyl-5-(2-hydroxyethyl)thiazole; HET-P, HET phosphate; HMP, 4-amino-5-hydroxymethyl-2-methylpyrimidine; HMP-P, HMP phosphate; HMP-PP, HMP diphosphate; IPTG, isopropyl-β-D-thiogalactoside; RACE, rapid amplification of cDNA ends; RP, reverse-phase; RT, reverse transcriptase; TBA, tetrabutylammonium hydrogensulfate; TDP, thiamine diphosphate; TMP, thiamine monophosphate
INTRODUCTION
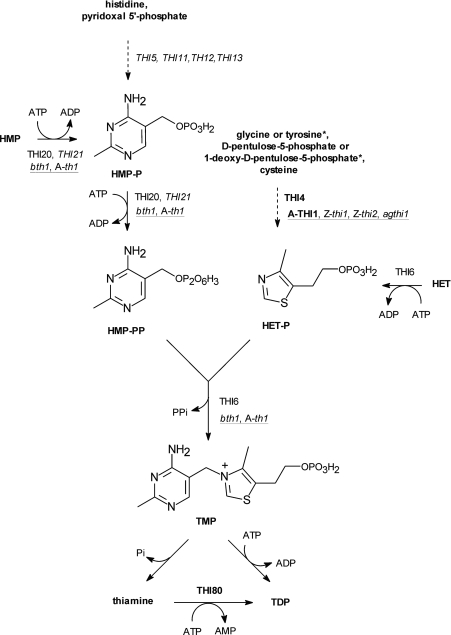
TDP (thiamine diphosphate) plays an essential role in all organisms as a cofactor of several universal enzymes involved in carbohydrate and amino acid metabolism, including pyruvate decarboxylase, pyruvate-, α-ketoglutarate- and branched fatty acid-dehydrogenases, transketolase and α-acetolactate synthase [1]. TDP is synthesized from exogenous thiamine taken with food (as vitamin B1) by higher animals, taken up by microbial cells from the environment [2,3] or from endogenous TMP (thiamine monophosphate) which is synthesized by many prokaryotic and eukaryotic micro-organisms as well as higher plants. The last step of de novo TMP biosynthesis involves a condensation of two precursors, HMP-PP (where HMP is 4-amino-5-hydroxymethyl-2-methylpyrimidine and PP is diphosphate) and HET-P [where HET is 4-methyl-5-(2-hydroxyethyl)thiazole and P is phosphate] which are separately synthesized on the routes that significantly differ between various groups of thiamine-synthesizing organisms. Many micro-organisms can additionally salvage HMP-PP and HET-P by phosphorylation of HMP and HET taken up from the medium (Scheme 1).
Scheme 1. Proposed scheme for thiamine and TDP biosynthesis in Eukaryota.
The biosynthesis pathways may differ in some details between yeasts and higher plants, as exemplified by different substrates (*) used by plants for the synthesis of the thiazole moiety HET-P. The sequenced genes in bold and purified proteins (italics) unequivocally assigned to thiamine biosynthesis (normal font) are shown. The symbols of proteins whose three-dimensional structures were reported are in higher plants (underlined) and bakers yeast. A, A. thaliana; Z, Z. mays; b, B. napus; ag, A. glutinosa.
The pathways of bacterial thiamine biosynthesis are well recognized (reviewed in [4]). Many of the enzymes which catalyse the specific steps of thiamine synthesis in model bacteria species such as Escherichia coli, Bacillus subtilis and Salmonella Typhimurium have been obtained as recombinant proteins and their three-dimensional structures have previously been determined (reviewed in [5]). In sharp contrast, our knowledge of this process in eukaryotic cells is at a very preliminary stage, in terms of both the identification of individual biosynthetic reactions and the structural biology of the enzymes involved. In Saccharomyces cerevisiae, at least fifteen TDP biosynthesis genes have been identified (reviewed in [6]) but products of only four genes, THI80, THI6, THI4 and THI20, are known as purified, natural or recombinant proteins [7–10]. Kinetic characteristics are available for the THI80 protein with thiamine diphosphokinase (EC 2.7.6.2) activity [11] and for the THI6 bifunctional enzyme with TMP synthase (TMP diphosphorylase, EC 2.5.1.3) and HET kinase (EC 2.7.1.50) activities [8]. Three-dimensional structures were reported for thiamine diphosphokinase [7], and THI4 [9] which is involved in thiazole synthesis [12] and possibly plays some role in yeast tolerance against DNA damage [13]. In higher plants, a limited number of thiamine biosynthesis genes have been identified and sequenced, including several orthologues of the yeast THI4 thiazole-synthesizing gene, which are found in Zea mays [14], Arabidopsis thaliana [15] and Alnus glutinosa [16], and a family of orthologues of the Brassica napus bth1 gene [17] which probably codes for a bifunctional enzyme with TMP synthase and HMP(-P) kinase (EC 2.7.1.49/EC 2.7.4.7) activities. The protein product of the bth1 gene has not been isolated, although it has been implicated in thiamine biosynthesis from the functional complementation of thiamine-requiring mutants in some bacteria [17]. Recently, a three-dimensional structure of the A. thaliana orthologue of yeast THI4 protein was reported [18] but its enzymatic activity remains to be defined.
In the present study, we cloned and sequenced the cDNA of a Z. mays gene designated thi3 which is orthologous to the B. napus bth1 gene. A product of this gene was isolated as a recombinant active enzyme and its dual TMP synthase and HMP(-P) kinase activity was investigated and kinetically characterized. Models for the structure of enzyme active centres are presented. This is the second plant thiamine-synthesizing enzyme which has been characterized on the purified protein level and the first one whose enzymatic properties have been quantitatively analysed.
EXPERIMENTAL
Cloning and sequencing of thi3 cDNA from Z. mays leaves
The 10-day-old leaves were ground in liquid nitrogen to a fine powder using a mortar and pestle followed by NucleoSpin filter homogenization (Macherey-Nagel). Total RNA from 100 mg of disrupted tissue was purified using the NucleoSpin RNA II Kit (Macherey-Nagel). The quantity and integrity of the total RNA was checked by formaldehyde agarose gel electrophoresis.
To identify the thi3 cDNA, a set of six degenerate primers on the basis of the known eukaryotic and prokaryotic conservative sequences of BTH1-related proteins (sequences derived from the EMBL database) were designed. The cDNA was synthesized using Superscript III reverse transcriptase (Invitrogen) with poly-(dT)20 primer. From nine reactions performed in the experiment, the MO1-F and MO2-R primer pair (Table 1) gave a dominant PCR product of approx. 1000 bp. The product was purified from the agarose gel, cloned into the pCR4-TOPO vector and sequenced. All PCR reactions were performed with Perpetual Opti Taq Polymerase (Eurx, Gdansk, Poland).
Table 1. Primers used in the present study.
| Name of primer | Sequence | Description |
|---|---|---|
| MO1-F | TCGTGAAGACTGGGATGCT(TC)CC | Degenerate, forward primer |
| MO2-R | GTAGTC(CT)GCACCATCTTTCCA | Degenerate, reverse primer |
| ZTH51 | GCTTCTTTCACATTTGGAGTGAC | 5′-RACE RT primer |
| ZTH52 | GTAGCGAGAGTAGATGGTCC | First 5′-RACE primer |
| ZTH53 | GCTGGTGGACACCATGACC | ‘Nested’ 5′-RACE primer |
| ZTH31 | GCGTCCATGTTGGCCAATC | First 3′-RACE primer |
| ZTH32 | CATACCAGCATGGGAAGTAC | ‘Nested’ 3′-RACE primer |
| Adaptor | GACTCGAGTCGACATCG | Adaptor 3′- and 5′-RACE primer |
| ZTH1-F | GGTGGTCATATGACGTCCGTACCGCTAC | Forward primer for PCR of thi3 cloned into pTYB1 vector |
| ZTH1-R | GGTGGTTGCTCTTCCGCAAGACCTTGAGTGAGCATTCTTC | Reverse primer for PCR of thi3 cloned into pTYB1 vector |
| ZTH11-F | GGTGGTTGCTCTTCCAACGCCACCCTTGGCTTCCGCA | Forward primer for PCR of thi3 cloned into pTYB11 vector |
| ZTH11-R | GGTGGTCTCGAGCTAAGACCTTGAGTGAGCATTC | Reverse primer for PCR of thi3 cloned into pTYB11 vector |
| MutQ98L | GCTGTCACCGCGCTGAACACCGTCGG | Mutagenetic primer, replaces Gln98 with leucine |
| MutQ373L | GGCGCTACCATTGTCCTACTGAGAGAAAAAGACGCC | Mutagenetic primer, replaces Gln373 with leucine |
| MutM134K | GGTGAAAACAGGGAAGCTCCCTTCAGCTGG | Mutagenetic primer, replaces Met134 with lysine |
| MutT472D | GTGTCTTCCCGACCACGGACAAGGCAAACAATCCTACC | Mutagenetic primer, replaces Thr472 with aspartic acid |
| MutS444A | GGAAAAATCATCGGCGTCGCATGTAAGACCCTGG | Mutagenetic primer, replaces Ser444 with alanine |
The 5′- and 3′-RACE (rapid amplification of cDNA ends) were performed according to Frohman [19]. On the basis of the partial sequence of thi3, a set of 5′-RACE reverse primers and 3′-RACE forward primers were designed (Table 1). The synthesis of cDNA from 4 μg of the total RNA was performed using the ZTH51 primer in 5′-RACE and oligo-(dT)17-adaptor primers in 3′-RACE. After the 5′-RACE RT (reverse transcriptase) reaction, primers, nucleotides and salts were removed by ultrafiltration with Microcon-100 columns (Millipore), diluted using sterile water. The purified cDNA was tailed using terminal transferase (Promega) in the presence of 0.25 mM dATP at 37 °C for 10 min. The second strand of cDNA was synthesized using oligo-(dT)17-adaptor primer. After addition of adaptor and ZTH52 primers, the first 5′-RACE reaction was performed, followed by ‘semi-nested’ PCR using the ZTH53 primer and adaptor primer. The discrete product was purified and cloned into the pCR4-TOPO sequencing vector.
The 3′-RACE product was amplified after addition of ZTH31 primer and adaptor primer to cDNA. Then the ZTH32 primer and adaptor primer were used in ‘semi-nested’ PCR. The specific product was purified and cloned into the pCR4-TOPO vector. All DNAs were analysed by sequencing (Sequencing Laboratory, Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Warsaw, Poland).
Expression and purification of THI3 protein in E. coli
The overexpression and purification of THI3 protein containing intein [with the CBD (chitin-binding domain)] attached at either the N- or C-terminus of the protein was performed according to the published method in [20]. For this purpose, the thi3 cDNA was cloned into the pTYB1 vector (New England Biolabs) utilizing the NdeI and SapI restriction sites, and into the pTYB11 vector utilizing the SapI and XhoI restriction sites, resulting in the pMO1 and pMO2 plasmids respectively. The N-tagged version of recombinant THI3 protein lacked the N-terminal 21-amino-acid sequence. Recombinant proteins containing the CBD tag were overexpressed after induction with 0.3 mM IPTG (isopropyl-β-D-thiogalactoside) in ER2566 E. coli cells at 16 °C for 16 h. The cells were suspended in 20 mM Tris/HCl buffer (pH 8.5) containing 500 mM NaCl and sonicated. After centrifugation at 40000 g for 20 min, the recombinant proteins containing the CBD tag were purified according to the IMPACT-CN expression system (New England Biolabs) from the soluble fraction using 50 mM dithiothreitol to induce tag cleavage.
The protein samples were dialysed against 20 mM Tris/HCl buffer (pH 7.5) containing 150 mM NaCl, and subjected to fine purification on a Superdex 200 HR 10/50 high performance size-exclusion column (Amersham) eluted with the same buffer. The purified protein samples were stored in aliquots at −20 °C. Under these storage conditions, the enzyme was stable for at least 3 months.
Protein samples from various preparation steps were analysed by SDS/PAGE in the Laemmli system [21] using a 10% resolution gel. The protein bands were either visualized by Coomassie Brilliant Blue R-250 staining or subjected to Western blotting on nitrocellulose membranes, using primary polyclonal anti-CBD antibodies (New England Biolabs) and a horseradish peroxidase conjugated secondary anti-rabbit antibody (Promega).
Site-directed mutagenesis
The QuikChange® multi-site-directed mutagenesis kit (Stratagene) with the pTyb11 bacterial expression plasmid incorporating the thi3 insert as a template were used for mutagenesis. Initially, two double mutants were produced: Q98L/Q373L and M134K/T472D. The single mutants Q98L, M134K, Q373L and T472D were constructed by digestion of the double mutants and the wild-type Tyb11-thi3 by SacI and XhoI restriction enzymes and appropriate ligation. Additionally, a single mutant in which Ser444 was replaced by an alanine residue was designed. Table 1 contains the primers used in site-directed mutagenesis. All seven mutagenesis constructs were sequenced.
Synthesis of HMP and its phosphate and diphosphate esters
HMP synthesis was performed through the intermediary syntheses of 4-amino-5-aminomethyl-2-methylpyrimidine [22] and 4-amino-5-bromomethyl-2-methylpyrimidine [23]. HMP-P and HMP-PP were obtained from HMP [24].
Enzyme assays
For the TMP synthase assay, the reaction mixture contained, unless otherwise specified, 15 μM HMP-PP, 15 μM HET-P and 10 mM MgCl2 in 50 mM Tris/HCl buffer (pH 8.0) and 5 μg of the pure enzyme in a final volume of 100 μl. After incubation at 37 °C for 60 min, the reaction was stopped with 16 μl of 10% metaphosphoric acid and the samples were centrifuged at 10000 g for 10 min, diluted 1:2 times with 0.1 M potassium phosphate and analysed for TMP content by RP-HPLC (reverse-phase HPLC) with post-column derivatization and fluorimetric detection [25]. A Merck LiChrosphere 100 RP-18 (5 μm) column (250 mm×4 mm) was used, with a two-solvent system including 0.15 M ammonium citrate (pH 4.2) (solvent 1) and 0.1 M formic acid containing 0.4% diethylamine (pH 3.2) (solvent 2). The gradient elution of 0–90% 2 in 16 min at a flow rate of 1 ml·min−1 was applied. The fluorogenic derivatization was performed with 0.0025% sodium hexacyanoferrate(III) in 2.25% NaOH, pumped through a peristaltic pump with a flow rate of 0.8 ml·min−1. The fluorescence emission at 430 nm excited at 365 nm was monitored.
The production of TMP from HMP or HMP-P due to the consecutive action of HMP(-P) kinase and TMP synthase were determined in analogous samples, with HMP-PP replaced by these substrates (15 μM) and with the 10 mM ATP supplement.
For the direct HMP kinase assay, the reaction mixtures contained 20 μM HMP, 1 mM ATP, 10 mM MgCl2, 50 mM Tris/HCl buffer (pH 8.0) and 5 μg of enzyme in a final volume of 100 μl. After 60 min incubation at 37 °C, the reaction was stopped and the sample, diluted 1:2 with 0.1 M potassium phosphate, was analysed for HMP-P content by RP-HPLC on a Supelcosil LC-18 column (250 mm×4.6 mm), eluted with a ternary solvent system composed of 4 mM TBA (tetrabutylammonium hydrogensulfate) in water (solvent A), 4 mM TBA in methanol (solvent B) and 4 mM TBA in 50 mM potassium phosphate buffer (pH 6.6) (solvent C). The following time program was applied: at t=0 min: 10% B, 30% C; at t=14 min: 50% B, 50% C; at t=16 min: 10% B, 80% C; at t=18 min: 10% B, 30% C; at t=35 min: 10% B, 30% C where the combination of solvents A, B and C equal 100%. The absorbance of the eluate at 245 nm was monitored.
Kinetic data analysis
The plots of the initial reaction velocity against substrate concentrations were analysed for various kinetic parameters as outlined by Bisswanger [26]. For hyperbolic kinetics, the apparent Km and Vmax parameters were estimated by fitting the classic Michaelis–Menten equation (untransformed) with the use of the GraphPad Prism software. In cases where the reaction velocity reached a maximum and decreased thereafter, indicative of inhibition by excess substrate, the following equation was fitted:
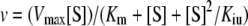 |
where v is the reaction velocity, [S] is the substrate concentration and Kiu is the apparent substrate (uncompetitive) inhibition constant. Kiu was first estimated from the abscissa intercept of the high-concentration part of the Dixon plot, 1/v against [S]. The value obtained was used as a constant during the fitting of the above equation by GraphPad Prism to estimate the remaining Km and Vmax parameters. The uncompetitive inhibition of TMP synthase by ATP was analysed from the plots of the reaction velocities against HET-P concentrations at various ATP concentrations which should follow the equation:
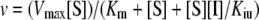 |
where [I] is the inhibitor concentration, and Kiu is the apparent uncompetitive inhibition constant. The Hanes–Woolf transformations, [S]/v against [S], were used to classify the type of inhibition, and their slopes, determined by linear regression, were re-plotted against [I] to estimate Kiu as the abscissa intercept of this secondary plot. In addition, all three parameters, Kiu, Km and Vmax, of the above equation were also estimated by direct non-linear regression fits to the untransformed data.
Bioinformatic analyses
Searching for protein sequence similarities was performed with the BLAST (v. 2.2.16) program [27] available on the server of National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). The multiple sequence alignments were generated with the use of ClustalW (v. 1.82) [28] from the ExPASy Proteomics Server of Swiss Institute of Bioinformatics (http://www.expasy.org) and visualized with JalView 2.3 [29] multiple alignment editor (http://www.jalview.org). Putative chloroplast transit peptides were found by the ChloroP (v. 1.1) program [30] at the server of Center for Biological Sequence Analysis of the Technical University of Denmark (http://www.cbs.dtu.dk/services/ChloroP). The THI3 protein sequence was also analysed using the Conserved Domains Search service [31] on the NCBI server. Models of three-dimensional structures of THI3 protein domains, on the basis of the known crystal structures of Salmonella Typhimurium HMP-P kinase (PDB code 1JXH, [32]) and B. subtilis TMP synthase (PDB code 2TPS, [33]), were generated using the SWISS-MODEL automated homology modelling server [34] of the Swiss Institute of Bioinformatics (http://swissmodel.expasy.org). The structures were visualized using the Swiss-PdbViewer and PyMOL programs available from the ExPASy Server.
RESULTS
Cloning and sequence analysis of maize thi3 cDNA
The amplification with a pair of degenerate MO1-F (forward) and MO2-R (reverse) primers (Table 1) resulted in a discrete, specific RT-PCR product, which was cloned into a sequencing vector. After E. coli transformation, ten clones were randomly chosen for sequencing. All clones contained the same 997-nucleotide DNA fragment. On the basis of this sequence, 5′-RACE and 3′-RACE were performed. In both reactions, nested PCR gave dominant products, which were cloned into a sequencing vector. For each RACE, four randomly chosen colonies were picked and plasmid purified from each clone was sequenced. All 3′-RACE sequences were identical. From clones received after 5′-RACE we chose one with the longest region upstream of the putative ATG start codon. This thi3 cDNA clone (GenBank® Accession No. AM167973) had a total length of 1947 bp and contained a 48 bp untranslated 5′ region, a 1653 bp-long open reading frame and a 246 bp untranslated 3′ region with the stop codon and a polyA tail. A putative open reading frame-encoded protein, THI3 (GenBank® Accession No. CAJ45026), consisted of 551 amino acid residues and its predicted molecular mass and isoelectric point were 57.8 kDa and 8.51 respectively.
Purification and basic molecular characterization of recombinant maize THI3 protein overproduced in E. coli
Of several versions of the THI3 recombinant protein tested, the protein lacking the N-terminal 21 amino acid fragment, and expressed in E. coli with the 55-kDa intein attached to the N-terminus as a tag protein, gave a satisfactory level of expression, especially when bacterial cultures were propagated at 16 °C. Under these conditions, the majority of the recombined fusion THI3 protein was found in the soluble fraction.
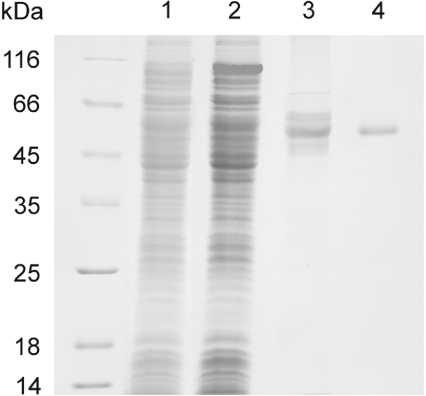
SDS/PAGE of a typical THI3 purification run is presented in Figure 1. After affinity chromatography on a chitin-containing column followed by Superdex-200 high-performance gel-filtration, a homogeneous protein was obtained in a typical yield of approx. 3 mg from 4 litres of bacterial culture. The apparent protein monomer molecular mass was 55 kDa as determined by SDS/PAGE. Upon Superdex gel-filtration, the protein eluted in a volume corresponding to an apparent molecular mass of 95 kDa (results not shown), suggesting that under non-denaturing conditions the protein existed as a dimer.
Figure 1. SDS/PAGE patterns from a representative purification of maize THI3 protein overproduced in E. coli.
The electrophoresis was carried out in the Laemmli system [21] using a 10% resolution gel. Protein bands were visualized by Coomassie Brilliant Blue R-250 staining. Left lane: molecular mass standards; lane 1: transformed cell lysate before IPTG induction; lane 2: transformed cell lysate after IPTG induction for 16 h at 16 °C; lane 3: sample after chitin-affinity chromatography; lane 4: sample after Superdex-200 gel filtration.
Characterization of enzymatic activities of THI3 protein
As shown in Table 2, the recombinant maize THI3 protein could synthesize TMP from HMP-PP and HET-P substrates. This TMP synthase activity required the presence of magnesium ions in the reaction mixture. For this reaction, HMP-PP could not be substituted by HMP or HMP-P, nor HET-P by HET. The TMP synthase activity was inhibited by ATP, but in the presence of ATP, TMP could be formed from HET-P plus HMP or HMP-P, clearly indicating that the enzyme possessed additional HMP- and HMP-P kinase activities. At the equivalent ATP concentration, the rate of TMP formation from HMP(-P) plus HET-P substrates, i.e. due to the sequential action of HMP(-P) kinase and TMP synthase, was approx. 30% of that from HMP-PP plus HET-P. On the other hand, no HET kinase activity could be assigned to THI3 as TMP was not formed from unphosphorylated thiazole in combination with any pyrimidine in the presence of ATP.
Table 2. Enzymatic activity of maize THI3 protein: the rates of TMP formation from various combinations of substrates and cofactors.
Concentrations in the reaction mixture were 15 μM HMP, HMP-P, HMP-PP, HET-P or HET, 10 mM ATP and 10 mM MgCl2. Samples containing 5 μg of protein/100 μl in 50 mM Tris/HCl buffer (pH 8.0) were incubated for 60 min at 37 °C and then analysed for TMP content by RP-HPLC with fluorogenic post-column derivatization. Values are the means±S.D. from at least three independent kinetic experiments.
| Activity tested | Substrates and cofactors | Specific activity (nmol of TMP/min per mg of protein) |
|---|---|---|
| TMP synthase | HMP-PP, HET-P | 0.52±0.05 |
| HMP-PP, HET-P, Mg2+ | 2.46±0.12 | |
| HMP-PP, HET-P, ATP | 0.05±0.02 | |
| HMP-PP, HET-P, Mg2+, ATP | 0.83±0.11 | |
| HMP kinase/TMP synthase | HMP, HET-P, ATP, Mg2+ | 0.22±0.03 |
| HMP-P kinase/TMP synthase | HMP-P, HET-P, ATP, Mg2+ | 0.24±0.02 |
| HET kinase/TMP synthase | HMP-PP, HET, ATP, Mg2+ | 0 |
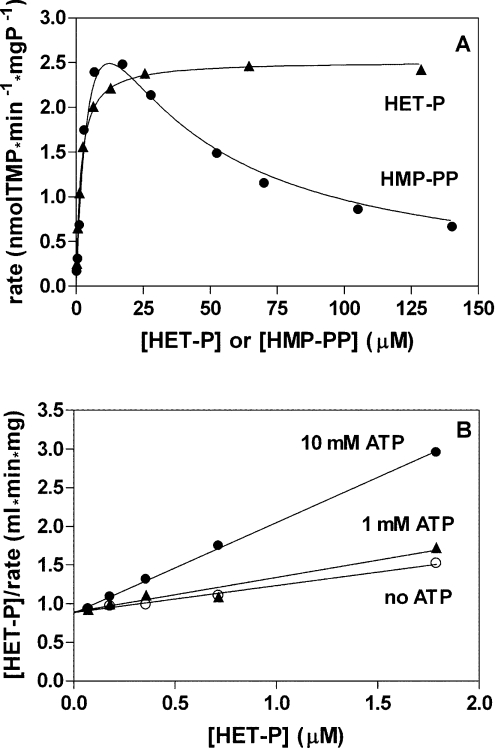
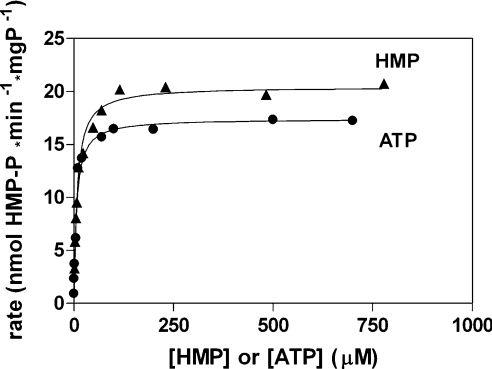
The conventional kinetic characteristics of maize TMP synthase is illustrated in Figure 2. The dependence of initial reaction rate on HET-P concentration was hyperbolic but the dependence on HMP-PP concentration showed a pattern indicating a strong inhibition by excess substrate (Figure 2A). The rate was maximal at 15–20 μM HMP-PP and then gradually dropped. The inhibition of TMP synthase by ATP was analysed against various HET-P concentrations at a constant HMP-PP concentration. An example analysis using Hanes–Woolf plots is shown in Figure 2(B). These plots, prepared for various ATP concentrations, had a common intercept with the rate axis, suggesting that the inhibition is uncompetitive [26]. The kinetic characteristics of HMP kinase activity was obtained through direct determinations of HMP phosphorylation. The dependences on HMP or ATP concentration, when the other substrate had a fixed concentration, were hyperbolic (Figure 3). The values of estimated kinetic constants of maize HMP kinase and TMP synthase are collected in Table 3.
Figure 2. Kinetic characteristics of TMP synthase activity of THI3 protein.
(A) Dependences of TMP-formation rate on concentrations of substrates. The line drawn through data points for the HMP-PP concentration dependence was generated with Kiu=24 μM found from its Dixon plot, and Km=6.35 μM and Vmax=5.05 nmol of TMP/min per mg of protein were estimated by fitting the equation for the uncompetitive inhibition by excess substrate. For the HET-P concentration dependence, the classic hyperbola with Km=1.74 μM and Vmax=2.52 nmol of TMP/min per mg of protein was fitted. (B) Hanes–Woolf plots of the dependences of TMP-formation rates on HET-P concentration in the presence of ATP. From the Hanes–Woolf plot obtained in the absence of ATP, the Km and Vmax values for HET-P were estimated as 2.55 μM and 2.88 nmol of TMP/min per mg of protein respectively, and the Kiu for ATP (4.3 mM in this example) was obtained from the abscissa intercept of the secondary plot of the slope against the ATP concentration.
Figure 3. Kinetic characteristics of HMP kinase activity of THI3.
THI3 has a dependence of HMP-P formation rate on concentrations of HMP and ATP substrates. The production of HMP-P was directly quantified by RP-HPLC with spectrophotometric detection. Ordinary hyperbolic fits are shown with Km and Vmax values of 7.46 μM and 20.5 nmol HMP-P per min per mg of protein respectively, for HMP concentration dependence, and of 6.27 μM and 17.4 nmol of HMP-P/min per mg of protein respectively, for the dependence on ATP concentration.
Table 3. Kinetic constants of TMP synthase and HMP kinase activities of THI3 protein.
The apparent kinetic parameters were determined at the second substrate concentration of 15 μM (HMP-PP), 1.5 μM (HET-P), 0.5 mM (HMP) or 5 mM (ATP). Km, Michaelis constant; KiuHMP-PP, substrate (HMP-PP) uncompetitive inhibition constants for TMP synthase; KiuATP, the constant for uncompetitive inhibition of TMP synthase by ATP (determined against variable concentration of HET-P as the substrate); Vmax, maximal velocity recalculated per 1 mg of protein. The applied kinetic equations with these parameters are specified in the Experimental section. Values are means ± S.D.; the number of independent kinetic experiments are given in parentheses.
| Activity | Parameter | Parameter value |
|---|---|---|
| TMP synthase | KmHMP-PP (μM) | 12.8±6.9 (4) |
| KiuHMP-PP (μM) | 14.8±6.3 (4) | |
| VmaxHMP-PP (nmol of TMP/min per mg of protein) | 8.5±3.9 (4) | |
| KmHET-P (μM) | 1.79±0.63 (8) | |
| VmaxHET-P (nmol of TMP/min per mg of protein) | 2.66±0.22 (8) | |
| KiuATP (mM) | 3.4±0.9 (3) | |
| HMP kinase | KmHMP (μM) | 6.58±1.55 (4) |
| VmaxHMP (nmol of TMP/min per mg of protein) | 20.1±1.90 (4) | |
| KmATP (μM) | 7.24±1.80 (7) | |
| VmaxATP (nmol of TMP/min per mg of protein) | 17.7±0.34 (7) |
Site-directed mutagenesis of THI3 at the TMP synthase and HMP(-P) kinase active centres
The active sites of THI3 were probed by substitution of several amino acid residues which were selected on the basis of the previously published data on bacterial TMP synthases and HMP kinases, as well as on the model of THI3 structure (see Discussion). The rates of TMP synthase and HMP kinase reactions in the extracts from cells transformed with mutant plasmids are presented in Table 4. Substitutions of Q98L and M134K were detrimental to HMP kinase activity whereas TMP synthase activity was fully preserved. A substitution of S444A represents the opposite situation of a complete elimination of TMP synthase activity without any significant effect on the HMP kinase activity. Two further single mutants, Q373L and T472D, exhibited full TMP synthase activity and moderately reduced (by 30 and 50% respectively) HMP kinase activity. A double mutant, Q98L/Q373L, differed from the Q98L mutant only in a detectable but still small (approx. 10%) HMP kinase activity. In contrast, another mutant with a double amino acid substitution, one in the TMP synthase and the other in the HMP(-P) kinase active centres, M134K/T472D, showed anomalous behaviour, with HMP kinase activity being too high, approx. 25% of that of the wild-type protein.
Table 4. Enzymatic activity of THI3 mutants.
Activity was determined in the lysates from E. coli cells transformed with plasmids containing the THI3 sequence with indicated amino acid substitutions. The expression levels of all mutants were similar to that of wild-type protein as judged from immunochemical analysis using CBD-specific antibodies (results not shown). The activities were expressed relatively to those determined in the wild-type protein samples. Values given are ranges from at least three independent experiments.
| Relative activity (%) | ||
|---|---|---|
| Mutant | TMP synthase | HMP kinase |
| Q98L | 104–124 | 0–2 |
| M134K | 97–111 | 0–5 |
| Q373L | 96–113 | 64–75 |
| S444A | 0–1 | 80–91 |
| T472D | 101–126 | 46–54 |
| Q98L/Q373L | 93–105 | 7–13 |
| M134K/T472D | 83–97 | 19–32 |
DISCUSSION
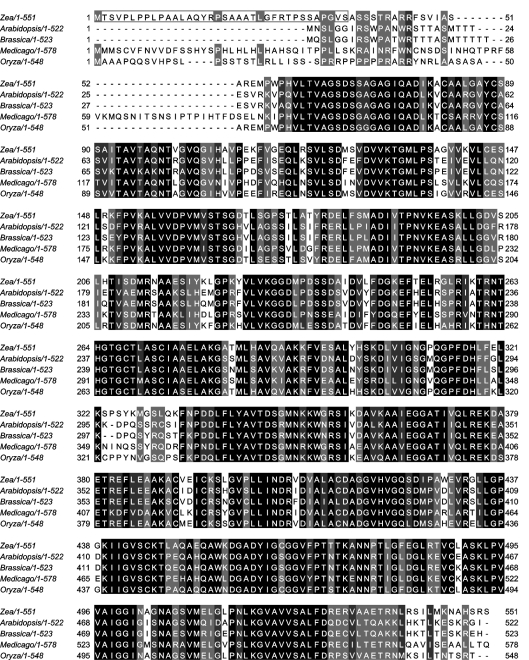
The Z. mays gene which was identified in the present study through cloning of its cDNA and characterization of its product in the form of pure, heterologously expressed recombinant protein is hereby designated thi3 as the third discovered gene of this species involved in thiamine biosynthesis. Searching through available protein databases revealed several orthologues of Z. mays THI3 in four higher plant species, including Oryza sativa, Medicago truncatula, A. thaliana and B. napus. A multiple sequence alignment of plant THI3 orthologues (one from each species) is shown in Figure 4. Amino acid identity with Z. mays THI3 varies from 77% for O. sativa ABA96049 protein to 63% for M. truncata ABE87727 protein. In the A. thaliana genome, three homologous sequences, AAM 91567 (Figure 4), BT020417 and AF000657 were identified. The first two differ by only one amino acid substitution; the third shows one short insertion and one deletion and may be the product of alternative splicing of the same gene. This gene exists in a single copy and maps to the locus AGT1G22950 on chromosome 1. Similarly, four homologous sequences, ABA96049 (Figure 4), EAZ19897, BAF29362 and ABG21903, were found in the rice genome. The first two probably originate by alternative splicing from one gene at the locus Os12g0192500 on chromosome 12, and two others are their truncated versions.
Figure 4. Multiple alignment of the amino acid sequences of THI3 and its orthologues occurring in various plant species.
The accession codes of the sequences compared were emb/CAJ45026 (Z. mays, the present study), gb/AAM91567 (A. thaliana), gb/AAC31298 (B. napus, BTH1), gb/ABE87727 (M. truncatula) and gb/ABA96049 (O. sativa). The alignment was performed using the ClustalW program and the result visualized in JalView. Black, dark grey and light grey highlights mark the positions in the primary structures where all 5, 4 out of 5 or 3 out of 5 residues are identical respectively. The box at the N-terminus of Z. mays THI3 protein includes the putative 36-amino-acid residue chloroplast transit peptide predicted by the ChloroP program [25].
Like all other members of this plant protein family, THI3 contains a putative N-terminal chloroplast transit peptide [35]. All THI3 orthologues were described as having two putative conserved domains: an N-terminal domain with high sequence similarity to many bacterial HMP(-P) kinases, and a C-terminal domain homologous with bacterial TMP synthases. As analysed with the use of NCBI CDD Search Server, THI3 shows an analogous two-domain structure, with the N-terminal kinase domain comprising residues 59–302 and the C-terminal synthase domain including residues 335–546.
In spite of the presence of putative HMP(-P) kinase and TMP synthase-encoding domains in cDNAs of several plant species, their protein products have not yet been obtained in isolated, pure form for which the suggested enzymatic activities could be directly demonstrated. However, the B. napus bthi1 cDNA was isolated on the basis of its ability to complement an E. coli mutant deficient in TMP synthase and was additionally shown to complement some HMP-kinase-deficient mutants [17]. Our maize THI3 protein, obtained by heterologous expression in E. coli, is the first pure bifunctional plant enzyme with confirmed HMP(-P) kinase and TMP synthase activity.
The recombinant THI3 protein was homogeneous as determined by SDS/PAGE and apparently existed in solution as a dimer, similarly to bacterial HMP kinase and TMP synthase, which form dimers in crystals [32,33]. The enzymatic properties of recombinant THI3 were readily detectable, although the determined specific activities (Table 3) were much lower than those reported for corresponding bacterial enzymes. For example, the specific activity of THI3 HMP kinase for HMP as the substrate (20 nmol/min per mg) was one-eighth of the lowest value reported for a bacterial HMP kinase, ThiD, from E. coli [36]. Similarly, the specific activities of bacterial TMP synthases, ranging from 65 nmol/min per mg for the enzyme isolated from E. coli [37] to 700 nmol/min per mg for ThiC from B. subtilis [38], exceed at least eight times the value for the plant bifunctional enzyme (8.5 nmol/min per mg). Whether these comparisons indicate an intrinsically lower turnover of the bifunctional plant enzyme or for some conformational imperfections of its recombinant form remains to be established. The kinetic characterization of maize THI3 protein revealed two interesting properties of the TMP synthase activity of this enzyme: the substrate inhibition by excess HMP-PP and uncompetitive inhibition by ATP. The uncompetitive inhibition by ATP, ADP and inorganic diphosphate was also reported for E. coli TMP synthase [37].
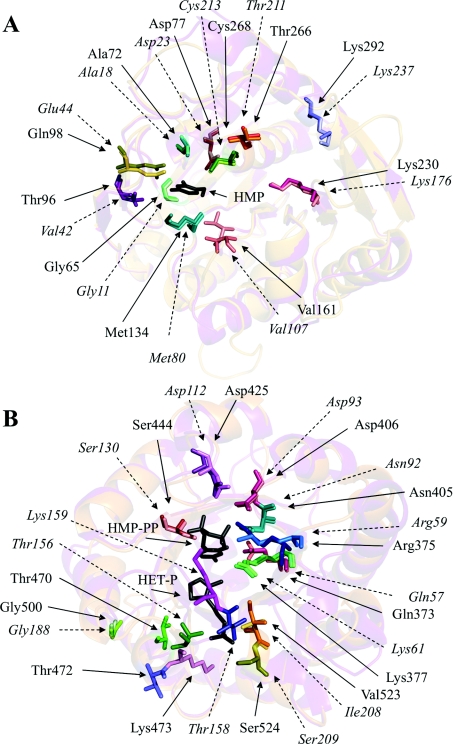
In an attempt to identify specific amino acid residues which form the active centres on the THI3 molecule we took advantage of a significant sequence similarity of THI3 with Salmonella Typhimurium HMP(-P) kinase (GenBank® Accession No. P55882) and B. subtilis TMP synthase (GenBank® Accession No. P39594) whose crystal structures have been reported [32,33]. As presented in Table 5, most of the amino acid residues that occur in the active sites of bacterial enzymes are conserved in the maize enzyme except that Thr96 and Gln98 replaced valine and glutamate respectively, in the HMP(-P) kinase domain and Val523 replaced isoleucine in the TMP synthase domain. The sequence identity of Salmonella Typhimurium HMP(-P) kinase and B. subtilis TMP synthase with the maize THI3 protein, approx. 39% as estimated by BLAST, was sufficient to use the three-dimensional structures of those bacterial enzymes as templates for modelling the THI3 structure. For this purpose, we used the SWISS-MODEL automatic homology-based modelling server of the Swiss Institute of Bioinformatics. The predicted overall structures of THI3 domains were very similar to those of the reference enzymes (Figure 5). Thus the model of the HMP kinase domain revealed a ribokinase-like fold with a core structure composed of a central eight strand β-sheet covered by two layers of five and three α-helices [32]. The modelled TMP synthase domain had the triosephosphate isomerase fold, containing a core barrel composed of eight α-helices and eight parallel β-strands [33,39]. More importantly, the predicted arrangements of active-site residues within the three-dimensional protein structure basically match those in the reference bacterial proteins. In the HMP kinase domain of THI3 protein (Figure 5A), eight residues, Gly65, Ala72, Asp77, Thr96, Gln98, Met134, Val161 and Cys268, may be involved in HMP binding. Gln98 probably forms a direct hydrogen bond to the 4-amino group of HMP and the other residues may interact with the pyrimidine ring indirectly, through fixed water molecules, or by making some hydrophobic contacts [32]. Three residues, Lys230, Thr266 and Lys292 may contribute to a putative ATP-binding site. At least three residues in the TMP synthase domain of THI3 (Figure 5B), Thr470, Thr472 and Lys473, are shifted from the locations which the corresponding residues of B. subtilis protein occupy to make direct hydrogen bonds with a thiazole phosphate group (Thr156 and Thr158) and van der Waals contacts with thiazole ethyl chain as well as electrostatic interactions with the HMP-PP diphosphate group (Lys159) [39]. A new location of Lys473 may provide favourable interactions with the thiazole phosphate group. Positions of other active centre residues of plant and bacterial proteins nearly match. Residues Ser524, Gly500 and Val523 may constitute the phosphate-binding site, whereas Arg375, Lys377, Asn405, Asp406, Asp425 and Ser444 may fix the diphosphate group and Gln373 may interact with a pyrimidine moiety, possibly via two hydrogen bonds to the ring N-3 and the 4-amino group [39].
Table 5. Active centre residues of Salmonella Typhimurium HMP kinase, B. subtilis TMP synthase and the corresponding domains of Z. mays THI3 bifunctional enzyme.
For bacterial enzymes, the residues found in the closest neighbourhood of the bound ligands in the crystal structures [32,33,39] are listed, except the residues of the putative ATP-binding site of Salmonella Typhimurium HMP kinase, modelled through its homology with other members of the ribokinase family [32]. The corresponding putative active centre residues of Z. mays THI3 domains were identified by sequence alignment with the ClustalW program and structural modelling with the use of the SWISS-MODEL server.
| Active centre | Ligand | Salmonella Typhimurium | B. subtilis | Z. mays |
|---|---|---|---|---|
| HMP kinase | HMP | Gly11 | Gly65 | |
| Ala18 | Ala72 | |||
| Asp23 | Asp77 | |||
| Val42 | Thr96 | |||
| Glu44 | Gln98 | |||
| Met80 | Met134 | |||
| Val107 | Val161 | |||
| Cys213 | Cys268 | |||
| ATP | Lys176 | Lys230 | ||
| Asp187 | Asp242 | |||
| Arg202 | Arg257 | |||
| Thr211 | Thr266 | |||
| Lys237 | Lys292 | |||
| TMP synthase | HMP-PP | Gln57 | Gln373 | |
| Arg59 | Arg375 | |||
| Lys61 | Lys377 | |||
| Asn92 | Asn405 | |||
| Ser130 | Ser444 | |||
| Lys159 | Lys473 | |||
| HET-P | Thr156 | Thr470 | ||
| Thr158 | Thr472 | |||
| Gly188 | Gly500 | |||
| Ile208 | Val523 | |||
| Ser209 | Ser524 | |||
| Mg2+ | Asp93 | Asp406 | ||
| Asp112 | Asp425 |
Figure 5. The models of overall structures and arrangements of putative active centre residues of THI3 HMP/HMP-P kinase (A) and TMP synthase (B) domains, overlaid on the corresponding features of three-dimensional structures of Salmonella Typhimurium HMP/HMP-P kinase [32] and B. subtilis TMP synthase [33].
The models of THI3 domains were obtained with the SWISS-MODEL server and all structures were visualized using the Swiss-PdbViewer and PyMOL programs. The ribbon diagrams of the overall architectures of the THI3 domains (in violet) and the corresponding bacterial enzymes (in orange) are shown in the background. Stick representations of the active-site residues in the bacterial proteins and corresponding putative active site residues of THI3 domains are marked with similar colours, lighter for THI3. Additionally, the THI3 residues are indicated by continuous arrows and labelled with normal font, and the bacterial protein residues with dashed arrows and italic font. Dark grey stick models of substrates, HMP in A, and HMP-PP and HET-P in B, are shown in the positions they occupy in the crystal structures of bacterial enzymes.
We performed a preliminary experimental verification of the above models of the active centres of Z. mays HMP kinase/TMP synthase by mutations at selected residues. We exchanged two residues, Gln98 and Met134, at the HMP kinase site and three residues, Gln373, Ser444 and Thr472 at the TMP synthase site. The enzymatic properties of Q98L, M134K and S444A mutants exactly followed the above structural predictions. The first two exclusively express full TMP synthase activity, and the latter only has HMP kinase activity. The last type mutation (S130A) had been extensively exploited during the structural studies of the B. subtilis TMP synthase to prove the essential role of the Ser130 residue in the catalytic mechanism [33,39]. The full TMP synthase activity of the T472D mutant is consistent with the predicted escape of Thr472 from the bound ligand. A lack of effect of the Q373L mutation is more difficult to explain, although the model of the TMP synthase active site shows a possible rotation of the amide group which may prevent the formation of hydrogen bonds with the aminopyrimidine moiety. Unfortunately there is no data available on the effects of similar mutations in the bacterial enzyme. Two double-mutants of the THI3 protein, Q98L/Q373D and M134K/T472D, were additionally studied, with a single residue substituted at each active centre. As expected, both exhibited full TMP synthase activity and decreased HMP kinase activities, although the HMP kinase of the M134K/T472D mutant exceeds the predictions from the single mutant data. A systematic study of the mutations of all residues which were preliminarily classified in the present study as occurring at active centres of THI3 protein is currently being carried out in our laboratory on the level of purified mutant proteins.
Several features of the recombinant THI3 bifunctional enzyme of Z. mays described in the present study implicate putative biological functions of the natural product of the thi3 gene. In vivo, the full-length THI3 polypeptide would be targeted to chloroplasts, within which the entire thiamine biosynthesis pathway is known to occur [14]. The matured two-domain enzyme with HMP(-P) and TMP synthase activities could catalyse two consecutive steps of thiamine biosynthesis that lead from HMP-P to TMP. An additional ability of THI3 to phosphorylate HMP in the presence of ATP does not appear to be important in vivo as no mechanisms by which HMP could enter chloroplasts have ever been discussed. This reaction is well known in various micro-organisms as a salvage pathway. Bacterial TMP synthases were often suspected to play a regulatory function, primarily because of their susceptibility to inhibition by ATP, ADP and inorganic diphosphate [37]. The unique fusion of HMP-P kinase and TMP synthase activities into one molecule of higher plant protein such as THI3 may result in a more selective response to ATP as the TMP synthesis is quenched by HMP-PP (which is formed in the presence of ATP on the same enzyme and is immediately consumed for TMP formation). It may be that ATP levels regulate the yield of entire thiamine synthesis in higher plants through the inhibitory action on the bifunctional enzyme HMP(-P) kinase/TMP synthase. The THI3 orthologues occur in several higher plant species, strongly suggesting a specific role of the combination of the two activities into one protein for the thiamine biosynthesis in higher plants.
Acknowledgments
This work was partly supported by grant 2P04C 017 27 from the Ministry of Science and Higher Education, Poland (to M. R.-K.).
References
- 1.Friedrich W. Handbuch der Vitamine. Munich: Urban and Schwarzenberg; 1987. Thiamin (Vitamin B1, aneurin) pp. 240–258. [Google Scholar]
- 2.Yamada K., Kawasaki T. Properties of the thiamine transport system in Escherichia coli. J. Bacteriol. 1980;141:254–261. doi: 10.1128/jb.141.1.254-261.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iwashima A., Nosaka K., Nishimura H., Enjo F. Thiamin transporters in yeast. Methods Enzymol. 1997;279:109–117. doi: 10.1016/s0076-6879(97)79015-3. [DOI] [PubMed] [Google Scholar]
- 4.Begley T. P., Downs D. M., Ealick S. E., McLafferty F. W., van Loon A. P., Taylor S., Campobasso N., Chiu H. J., Kinsland C., Reddick J. J., Xi J. Thiamin biosynthesis in prokaryotes. Arch. Microbiol. 1999;171:293–300. doi: 10.1007/s002030050713. [DOI] [PubMed] [Google Scholar]
- 5.Settembre E., Begley T. P., Ealick S. E. Structural biology of enzymes of the thiamin biosynthesis pathway. Curr. Opin. Struct. Biol. 2003;13:739–747. doi: 10.1016/j.sbi.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 6.Nosaka K. Recent progress in understanding thiamin biosynthesis and its genetic regulation in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2006;72:30–40. doi: 10.1007/s00253-006-0464-9. [DOI] [PubMed] [Google Scholar]
- 7.Baker L. J., Dorocke J. A., Harris R. A., Timm D. E. The crystal structure of yeast thiamin pyrophosphokinase. Structure. 2001;9:539–546. doi: 10.1016/s0969-2126(01)00615-3. [DOI] [PubMed] [Google Scholar]
- 8.Kawasaki Y. Copurification of hydroxyethylthiazole kinase and thiamine-phosphate pyrophosphorylase of Saccharomyces cerevisiae: characterization of hydroxyethylthiazole kinase as a bifunctional enzyme in the thiamine biosynthetic pathway. J. Bacteriol. 1993;175:5153–5158. doi: 10.1128/jb.175.16.5153-5158.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jurgenson C. T., Chatterjee A., Begley T. P., Ealick S. E. Structural insights into the function of the thiamine biosynthetic enzyme Thi4 from Saccharomyces cerevisiae. Biochemistry. 2006;45:11061–11070. doi: 10.1021/bi061025z. [DOI] [PubMed] [Google Scholar]
- 10.Haas A. L., Laun N. P., Begley T. P. Thi20, a remarkable enzyme from Saccharomyces cerevisiae with dual thiamin biosynthetic and degradation activities. Bioorg. Chem. 2005;33:338–344. doi: 10.1016/j.bioorg.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 11.Voskoboyev A. I., Ostrovsky Y. M. Thiamin pyrophosphokinase: structure, properties and role in thiamin metabolism. Ann. N. Y. Acad. Sci. 1982;378:161–176. doi: 10.1111/j.1749-6632.1982.tb31195.x. [DOI] [PubMed] [Google Scholar]
- 12.Dorrestein P. C., Huili Zhai H., Taylor S. V., McLafferty F. W., Begley T. P. The biosynthesis of the thiazole phosphate moiety of thiamin (Vitamin B1): the early steps catalyzed by thiazole synthase. J. Am. Chem. Soc. 2004;126:3091–3096. doi: 10.1021/ja039616p. [DOI] [PubMed] [Google Scholar]
- 13.Machado C. R., Praekelt U. M., de Oliveira R. C., Barbosa A. C. C., Byrne K. L., Meacock P. A., Menck C. F. M. Dual role for the yeast THI4 gene in thiamine biosynthesis and DNA damage tolerance. J. Mol. Biol. 1997;273:114–121. doi: 10.1006/jmbi.1997.1302. [DOI] [PubMed] [Google Scholar]
- 14.Belanger F. C., Leustek T., Chu B., Kriz A. L. Evidence for the thiamine biosynthetic pathway in higher-plant plastids and their developmental regulation. Plant Mol. Biol. 1995;29:809–821. doi: 10.1007/BF00041170. [DOI] [PubMed] [Google Scholar]
- 15.Machado C. R., Costa de Oliveira R. L., Boiteux S., Praekelt U. M., Meacock P. A., Menck C. F. M. Thi1, a thiamine biosynthetic gene in Arabidopsis thaliana, complements bacterial defects in DNA repair. Plant Mol. Biol. 1996;31:585–593. doi: 10.1007/BF00042231. [DOI] [PubMed] [Google Scholar]
- 16.Ribeiro A., Praekelt U., Akkermans A. D. L., Meacock P. A., van Kammen A., Bisseling T., Pawlowski K. Identification of agthi1, whose product is involved in biosynthesis of the thiamine precursor thiazole, in actinorhizal nodules of Alnus glutinosa. Plant J. 1996;10:361–368. doi: 10.1046/j.1365-313x.1996.10020361.x. [DOI] [PubMed] [Google Scholar]
- 17.Kim Y. S., Nosaka K., Downs D. M., Kwak J. M., Park D., Chung I. K., Nam H. G. A Brassica cDNA clone encoding a bifunctional hydroxymethylpyrimidine kinase/thiamin-phosphate pyrophosphorylase involved in thiamin biosynthesis. Plant Mol. Biol. 1998;37:955–966. doi: 10.1023/a:1006030617502. [DOI] [PubMed] [Google Scholar]
- 18.Godoi P. H., Galhardo R. S., Luche D. D., van Sluys M. A., Menck C. F., Oliva G. Structure of the thiazole biosynthetic enzyme THI1 from Arabidopsis thaliana. J. Biol. Chem. 2006;281:30957–30966. doi: 10.1074/jbc.M604469200. [DOI] [PubMed] [Google Scholar]
- 19.Frohman M. A. Rapid amplification of complementary DNA ends for generation of full-length complementary DNAs: thermal RACE. Methods Enzymol. 1993;218:340–356. doi: 10.1016/0076-6879(93)18026-9. [DOI] [PubMed] [Google Scholar]
- 20.Chong S., Williams K. S., Wotkowicz C., Xu M.-Q. Modulation of protein splicing of the Saccharomyces cerevisiae vacuolar membrane ATPase intein. J. Biol. Chem. 1998;273:10567–10577. doi: 10.1074/jbc.273.17.10567. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli U. K. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 22.DiBella E. P., Hennessey D. J. Isomeric S-methylthiamins. J. Am. Chem. Soc. 1961;83:2017–2021. [Google Scholar]
- 23.Morimoto H., Hayashi N., Naka T., Kato S. Bildungsmechanismus von 4-amino-5-aminomethyl-2-methyl-pyrimidin aus 3-äthoxy-2-(diathoxymethyl)propionitril und äcetamidin. Chem. Ber. 1973;106:893–901. [Google Scholar]
- 24.Brown G. M. Preparation of the mono- and pyrophosphate esters of 2-methyl-4-amino-5-hydroxymethylpyrimidine for thiamine biosynthesis. Methods Enzymol. 1970;18(1):162–164. [Google Scholar]
- 25.Lee B. L., Ong H. Y., Ong C. N. Determination of thiamine and its phosphate esters by gradient-elution high-performance liquid chromatography. J. Chromatogr. 1991;567:71–80. doi: 10.1016/0378-4347(91)80311-y. [DOI] [PubMed] [Google Scholar]
- 26.Bisswanger H. Weinheim: Wiley-VCH; 2002. Enzyme kinetics, Principles and methods. [Google Scholar]
- 27.Altchul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson J. D., Higgins D. G., Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clamp H., Cuff J., Searle S. M., Barton G. J. The Jalview alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 30.Emanuelsson O., Nielsen H., von Heijne G. ChloroP, a neutral network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marchler-Bauer A., Bryant S. H. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:327–331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng G., Bennett E. M., Begley T. P., Ealick S. E. Crystal structure of 4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate kinase from Salmonella Typhimurium at 2.3 Å resolution. Structure. 2002;10:225–235. doi: 10.1016/s0969-2126(02)00708-6. [DOI] [PubMed] [Google Scholar]
- 33.Chiu H. J., Reddick J. J., Begley T. P., Ealick S. E. Crystal structure of thiamin phosphate synthase from Bacillus subtilis at 1.25 Å resolution. Biochemistry. 1999;38:6460–6470. doi: 10.1021/bi982903z. [DOI] [PubMed] [Google Scholar]
- 34.Schwede T., Kopp J., Guex N., Peitsch M. C. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wallace T. P., Howe C. J. Plant organellar targeting sequences. In: Croy R. R. D., editor. Plant Molecular Biology Labfax. Oxford: Bios Scientific Publishers; 1993. pp. 287–292. [Google Scholar]
- 36.Reddick J. J., Kinsland C., Nicewonger R., Christian T., Downs D. M., Winkler M. E., Begley T. P. Overexpression, purification and characterization of two pyrimidine kinases involved in the biosynthesis of thiamin: 4-amino-5-hydroxymethyl-2-methylpyrimidine kinase and 4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate kinase. Tetrahedron. 1998;54:15983–15991. [Google Scholar]
- 37.Kawasaki T. Thiamine phosphate pyrophosphorylase. Methods Enzymol. 1979;62:69–73. doi: 10.1016/0076-6879(79)62194-8. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y., Taylor S. V., Chiu H. J., Begley T. P. Characterization of the Bacillus subtilis thisC operon involved in thiamine biosynthesis. J. Bacteriol. 1997;179:3030–3035. doi: 10.1128/jb.179.9.3030-3035.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peapus D. H., Chiu H. J., Campobasso N., Reddick J. J., Begley T. P., Ealick S. E. Structural characterization of the enzyme–substrate, enzyme–intermediate, and enzyme–product complexes of thiamin phosphate synthase. Biochemistry. 2001;40:10103–10114. doi: 10.1021/bi0104726. [DOI] [PubMed] [Google Scholar]