Abstract
In the present study we investigated promoter regions of the PHGPx [phospholipid hydroperoxide GPx (glutathione peroxidase)] gene and transcription factors involved in TNFα (tumour necrosis factor α)-induced up-regulation of PHGPx in non-differentiated HL60 cells. Non-differentiated HL60 cells displayed up-regulation of non-mitochondrial and mitochondrial PHGPx mRNA in response to TNFα stimulation. The promoter activity was up-regulated by TNFα stimulation in cells transfected with a luciferase reporter vector encoding the region from −282 to −123 of the human PHGPx gene compared with the non-stimulated control. The up-regulated promoter activity was effectively abrogated by a mutation in the C/EBP (CCAAT/enhancer-binding protein)-binding sequence in this region. ChIP (chromatin immunoprecipitation) assays demonstrated that C/EBPϵ bound to the −247 to −34 region in HL60 cells, but C/EBPα, β, γ and δ did not. The binding of C/EBPϵ to the promoter region was increased in HL60 cells stimulated with TNFα compared with that of the non-stimulated control. An increased binding of nuclear protein to the C/EBP-binding sequence was observed by EMSA (electrophoretic mobility-shift assay) in cells stimulated with TNFα, and it was inhibited by pre-treatment with an anti-C/EBPϵ antibody, but not with other antibodies. The C/EBPϵ mRNA was expressed in PMNs (polymorphonuclear cells), non-differentiated HL60 cells and neutrophil-like differentiated HL60 cells displaying TNFα-induced up-regulation of PHGPx mRNA, but not in macrophage-like differentiated HL60 cells, HEK-293 cells (human embryonic kidney-293 cells) and other cell lines exhibiting no up-regulation. The up-regulation of PHGPx mRNA, however, was detected in HEK-293 cells overexpressing C/EBPϵ as a result of TNFα stimulation. These results indicate that C/EBPϵ is a critical transcription factor in TNFα-induced up-regulation of PHGPx expression.
Keywords: CCAAT/enhancer-binding protein (C/EBP), HL60 cell, phospholipid hydroperoxide glutathione peroxidase (PHGPx), promoter activity, tumour necrosis factor α (TNFα)
Abbreviations: C/EBP, CCAAT/enhancer-binding protein; ChIP, chromatin immunoprecipitation; COX, cyclo-oxygenase; CREB, cAMP-response-element-binding protein; DMEM, Dulbecco's modified Eagle's medium; EMSA, electrophoretic mobility-shift assay; FBS, foetal bovine serum; GPx, glutathione peroxidase; HEK-293 cell, human embryonic kidney-293 cell; ICAM-1, intercellular adhesion molecule 1; IL, interleukin; JNK, c-Jun N-terminal kinase; LPS, lipopolysaccharide; MAPK, mitogen-activated protein kinase; MEK, MAPK/ERK (extracellular-signal-regulated kinase) kinase; NAC, N-acetylcysteine; NF-κB, nuclear factor κB; NF-Y, nuclear factor-Y; PDTC, pyrrolidinecarbodithioate; PHGPx, phospholipid hydroperoxide GPx; PMN, polymorphonuclear cell; RBL2H3, rat basophilic leukaemia 2H3; ROS, reactive oxygen species; RT, reverse transcriptase; TNFα, tumour necrosis factor α
INTRODUCTION
PMNs (polymorphonuclear cells), and neutrophils in particular, are known to play important roles in innate immune defence systems. They kill invading pathogens via the generation of ROS (reactive oxygen species) and release of lytic enzymes stored in granules [1,2]. PMNs also produce several inflammatory cytokines [e.g. TNFα (tumour necrosis factor α) and IL (interleukin)-1β], chemokines [e.g. IL-8 and GRO (growth-regulated oncogene], and lipid mediators to modulate inflammatory and immune responses [3–6]. In an inflammatory response, PMNs are thought to be exposed to these molecules produced by themselves and other inflammatory cells. The ROS-mediated damage to intracellular molecules is considered to be limited by cellular antioxidant enzymes such as superoxide dismutases and GPxs (glutathione peroxidases). There are four types of isoenzymes known for GPxs, and all of them contain a selenocysteine residue at the active site that reduces hydrogen peroxide and lipid hydroperoxide. PHGPx (phospholipid hydroperoxide GPx) is an isoenzyme of GPxs which is widely distributed in many tissues and cells. We have investigated several functions of PHGPx by overexpressing the enzyme in RBL2H3 (rat basophilic leukaemia 2H3) cells. PHGPx-overexpressing RBL2H3 cells are resistant to necrosis and apoptosis caused by various oxidative stresses [7–9], and have reduced production of lipid mediators such as leukotrienes, PGD2 (prostaglandin D2) and PAF (platelet-activating factor) [10–12]. Furthermore, overexpression of PHGPx in human dermal fibroblasts and rabbit abdominal aortic smooth-muscle cells resulted in impaired activation of NF-κB (nuclear factor κB) [13,14], a key molecule in the transcriptional regulation of inflammatory protein factors such as TNFα and IL-8 [15,16]. These findings suggest that the expression levels of PHGPx could modulate various functions in PMNs.
Our recent study [17] demonstrated that the expression level of PHGPx was up-regulated in human peripheral PMNs and neutrophil-like differentiated HL60 cells treated with TNFα. The TNFα-induced up-regulation of PHGPx in neutrophil-like differentiated HL60 cells was inhibited by incubation with the antioxidants NAC (N-acetylcysteine) and PDTC (pyrrolidinecarbodithioate) NF-κB and Src kinase inhibitors, indicating the involvement of NF-κB and/or Src kinase activation through ROS signalling in the up-regulation of PHGPx. The up-regulation of PHGPx, however, was not suppressed by treatment with MAPK (mitogen-activated protein kinase) cascade inhibitors, which are downstream targets of Src kinases, and no binding sequence for NF-κB exists in the 5′ UTR (untranslated region) of the promoter region in the human PHGPx gene. Therefore we were unable to clearly demonstrate the signalling pathways and transcription factors involved in TNFα-induced up-regulation of PHGPx expression in neutrophil-like differentiated HL60 cells [17].
The sequences for the PHGPx gene have been reported in the human, mouse, rat and pig genomes [18–21]. There are three types of PHGPx; mitochondrial, non-mitochondrial and nucleolar types, and the mRNA of these molecules is transcribed from one gene by alternative transcription using different promoter regions and transcriptional start codons [8,22]. The responsible promoter regions associated with the basal transcription of the PHGPx gene in mice and humans have been identified by promoter assays. In the mouse PHGPx gene, the 5′-flanking regions ranging from −60 to −9 bp, −233 to −158 bp and +342 to +375 bp are critical for the basal transcription of non-mitochondrial, mitochondrial and nucleolar PHGP mRNA respectively. Furthermore, the regions from −176 to −126 and from −115 to −56 bp (non-mitochondrial PHGPx), from −358 to −281 bp (mitochondrial PHGPx) and from +244 to +271 bp (nucleolar PHGPx) are involved in enhancement of promoter activity [22]. For the human PHGPx gene, the 5′-flanking region spanning from −212 to −121 bp, which contains a NF-Y (nuclear factor-Y)-binding region (CCAAT box), is important for the basal transcription of the PHGPx gene, although the type of PHGPx was not specified [23].
The lack of detailed experimental results in TNFα-induced up-regulation of PHGPx expression in HL60 cells prompted us to investigate the regions responsible for up-regulation in the 5′-flanking region of the human PHGPx gene, and also the transcription factors associated with TNFα-induced up-regulation of PHGPx expression.
MATERIALS AND METHODS
Reagents
Recombinant human TNFα was obtained from R&D Systems (Minneapolis, MN, U.S.A.). AMV reverse transcriptase XL and random 9-mers were from TaKaRa (Shiga, Japan). RNase inhibitor and dNTP mixture were from TOYOBO (Osaka, Japan). NAC, PDTC and PMA were from Sigma–Aldrich. Bay 11-7082, PD98059, SB20358, SP600125 and PP2 were from Calbiochem. SYBR Green and TaqMan PCR master mix were from Applied Biosystems. HotStar Taq master mix was from Qiagen and FuGENE™ was from Roche. Anti-C/EBP (CCAAT/enhancer-binding protein)α (14AA), anti-C/EBPβ (Δ198), anti-C/EBPγ (H-50), anti-C/EBPδ (M-17), anti-C/EBPϵ (C-22), anti-CREB (cAMP-response-element-binding protein) (X-12) and anti-NF-κB (C-20) antibodies were from Santa Cruz Biotechnology. Anti-(NF-Y) antibody was from Rockland Immunochemicals (Gilbertsville, PA, U.S.A.).
Cell culture
HL60 and HEK-293 cells (human embryonic kidney-293 cells) were obtained from the Japanese Collection of Research Bioresources Cell Bank (Osaka, Japan) and were grown in RPMI 1640 medium and DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% (v/v) FBS (foetal bovine serum) respectively. ECV304, an endothelial cell line, and A498 cells were obtained from the A.T.C.C. Differentiation of HL60 cells into neutrophil-like cells was induced by culturing HL60 cells in a CO2 incubator for 7 days in the presence of 1.25% DMSO. Differentiation into macrophage-like cells was performed by culturing HL60 cells for 3 days in the presence of 10 nM PMA.
RNA isolation and PCR
Total RNA was extracted by cell lysis using ISOGEN (Wako Pure Chemical Industries, Osaka, Japan) and purified according to the manufacturer's instructions. Briefly, cells were homogenized in 800 μl ISOGEN. After the addition of 200 μl chloroform and phase separation, RNA was precipitated with 800 μl isopropyl alcohol and washed with ethanol. The RNA was dissolved in RNase-free distilled water and total RNA (0.5 or 1 μg) was subjected to first-strand cDNA synthesis with 0.25 unit/μl AMV reverse transcriptase XL, 2.5 μM random 9-mers, 1 unit/μl RNase inhibitor and 1 mM dNTP mixture. Thermal cycling conditions of 30 °C for 10 min, 42 °C for 30 min, 99 °C for 5 min and 5 °C for 5 min were used for the reverse transcription. cDNA and each primer were mixed with SYBR Green master mix and quantitative PCR was performed using the ABI PRISM 7700 Sequence Detection System (Applied Biosystems). The following specific primers were used for amplification of total PHGPx and 18S rRNA (internal control): total PHGPx sense, 5′-AAGGACATCGACGGGCACATGG-3′ and antisense, 5′-TTGGATCTTCATCCACTTCCA-3′; 18S rRNA sense, 5′-GCCCGAAGCGTTTACTTTGAA-3′ and antisense, 5′-GGCATCGTTTATGGTCGGAAC-3′.
To detect non-mitochondrial, mitochondrial and nucleolar types of PHGPx, TaqMan PCR was performed with the following primers and probes: non-mitochondrial PHGPx sense, 5′-TGGCCTGGCCGGGACCA-3′ and antisense, 5′-ATGTCCTTGGCGGAAAACTCGT-3′ and TaqMan probe, 5′-GTCCCGGGACGACTGGCGCTG-3′; mitochondrial PHGPx sense, 5′-AGCCTCGGCCGCCTTTGCCG-3′ and antisense, 5′-ATGTCCTTGGCGGAAAACTCGT-3′ and TaqMan probe, 5′-CCTGGCCTGGCCGGGACCATGTGC-3′; nucleolar PHGPx sense, 5′-AGCTTGCGACCGGAGATCCA-3′ and antisense, 5′-AAAACTCGTGCATGGAGCG-3′ and TaqMan probe, 5′-AATGTCCCAAGTCCCAGGACCCG-3′. The relative quantity of each of the three PHGPx types was calculated from a standard curve generated from diluted standard cDNA samples obtained from non-differentiated HL60 cells. The quantity was normalized with that of 18S rRNA.
For conventional PCR, HotStar Taq master mix, cDNA and each primer were mixed and PCR was then performed on a Hybaid Express thermal cycler (Hybaid) with the following thermal cycling protocol: 1 cycle at 95 °C for 15 min and 25–35 cycles of 95 °C for 30 s and 65 °C for 90 s. The following specific primers were used: C/EBPϵ sense, 5′-GGATCCGCCATGTCCCACGGGACCTACTACGAGTGTG-3′ and antisense, 5′-GAATTCTCAGCTGCAACCCCCCACGCCCTTGATGAGG-3′; NF-Y sense, 5′-GAATTCGCCATGGAGCAGTATACAGCAAACAGCAATAG-3′ and antisense, 5′-GCGGCCGCTTAGGACACTCGGATGATCTGTGTCATTGC-3′; CREB sense, 5′-GAATTCGCCATGACCATGGAATCTGGAGCCGAGAACCAGC-3′ and antisense, 5′-GCGGCCGCTTAATCTGATTTGTGGCAGTAAAGGTCCTTAAG-3′. After PCR, 10 μl of the PCR products were subjected to electrophoresis and the DNA was visualized by ethidium bromide staining.
Construction of luciferase reporter vectors encoding the promoter region of the PHGPx gene
The pGL3-Basic reporter vector (Promega) containing the luciferase reporter gene was used to analyse the promoter activity of a subcloned promoter-deletion mutant. For the promoter assay of the mouse PHGPx gene, the reporter vectors (deletion mutants) containing various lengths of the promoter regions of the mouse PHGPx gene [22] were used.
The deletion mutants of the human PHGPx gene were constructed as follows. The promoter regions ranging from −687 to −33 bp of human PHGPx were amplified by PCR using human genomic DNA as the template, and then subcloned into the pTZ-18R vector. The following modified specific primers (with an additional KpnI restriction site at the 5′-end and additional BglII and KpnI sites at the 3′-end) were used: sense, 5′-ATTGGTACCACGCATGTGATCCCAGCAC-3′ and antisense, 5′-TAGGGTACCAGATCTCTCGTCCAGCCGACCAATGG-3′. The PCR fragments were purified from a 1% (w/v) agarose gel, subsequently cleaved with KpnI and cloned into the pGL-3 Basic reporter vector.
The deletion mutants with various lengths of the promoter regions were prepared by PCR using the constructed reporter vector containing the region of PHGPx from −687 to −33 bp, and cloned into the pGL-3 vector by the same method stated above. Deletion mutants inserted into the pGL-3 vector were confirmed by digestion with KpnI. The direction of these inserted deletion mutants was also checked by digestion with KpnI and BglII.
Vectors with a mutation in the C/EBP-binding sequence or CCAAT box (NF-Y-binding sequence) in the human gene were constructed with the QuikChange® site-directed mutagenesis kit (Stratagene), and the DNA sequences of mutated vectors were confirmed by sequence analysis. The sequences of the mutation site (underlined) of the vector are as follows: wild-type, 5′-CCCATTGGCTGACGTCGGC-3′; mutant 1, 5′-CCCATTGGCTAGCGTCGGC-3′; mutant 2, 5′-CCCATATGCTGACGTCGGC-3′; and mutant 3, 5′-CGAGTTGGCTGACGTCGGC-3′.
Transfection of reporter vectors and reporter gene assays
The luciferase reporter vector was transiently transfected by electroporation. Briefly, 250 μl of a HL60 cell suspension at a density of 8×107 cells/ml was placed in a cuvette, into which 10 μg of the constructed reporter vector and 0.5 μg of phRL-TK vector (Promega) (internal control to determine transfection efficiency) were added. The vectors were electroporated into the cells with the Gene Pulser (Bio-Rad) at conditions of 300 V and 950 μF.
The transfected HL60 cells were cultured for 16 h in the presence or absence of 50 ng/ml TNFα, and then collected and lysed with 200 μl passive lysis buffer (Promega). After centrifugation at 20000 g for 1 min at room temperature (approx. 20 °C), the supernatant was collected and assayed for firefly and renilla luciferase activity. The activity was measured using the dual-luciferase reporter assay system (Promega) according to the manufacturer's protocol with a luminescence counter (PerkinElmer). Firefly luciferase activity from the constructed vector was normalized to the renilla luciferase activity from the phRL-TK vector, and the data were expressed as a ratio of the pGL3-Basic vector activity without the PHGPx promoter region.
EMSA (electrophoretic mobility-shift assay)
Nuclear proteins were extracted with the nuclear extraction kit (Active Motif, Carlsbad, CA, U.S.A.) according to the manufacturer's protocol. The following probe sequence was used for the EMSA assays: 5′-CCCATTGGCTGACGTCGGCGCGAGCGCTCAACACCGACGCG-3′. A double-stranded oligonucleotide probe was generated by annealing equimolar concentrations of complementary oligonucleotides. The probe was radiolabelled with [γ-32P]ATP using the Megalabel DNA 5′-end labelling kit (TaKaRa). Binding reactions were carried out for 30 min at 4 °C in 20 μl of binding buffer [100 mM Hepes, pH 7.9, 250 mM KCl, 5 mM EDTA, 25 mM DTT (dithiothreitol) and 2 mg/ml BSA] containing 10000–20000 c.p.m. of the radiolabelled probe, 1 μg of poly (dI-dC) [poly(deoxyinosinic-deoxycytidylic) acid] and 5 μg of the extracted nuclear proteins. For the competition assay, the unlabelled probe (at 100-fold molar excess of the labelled probe) and an anti-C/EBPα, C/EBPβ, C/EBPγ, C/EBPδ or C/EBPϵ antibody (4 μg) were pre-incubated with the reaction mixture for 1 h at 4 °C before the binding reaction.
The DNA–protein complexes were separated by non-denaturing PAGE (5% gels) in 0.25×Tris/acetate/EDTA electrophoresis buffer [10 mM Tris/7 mM acetate/0.5 mM EDTA (pH 8.0)] at 200 V. The gel was transferred on to 3 mm filter paper and dried under vacuum. The dried gel was visualized by autoradiography with the BAS-2000II Bio Imaging Analyzer (Fujifilm).
ChIP (chromatin immunoprecipitation) assays
ChIP assays were performed using the ChIP™ express enzymatic magnetic ChIP kits (Active Motif) according to the manufacturer's protocol. Briefly, HL60 cells (4.5×107 cells) cultured with or without 50 ng/ml TNFα for 8 h, were treated with 1% (v/v) formaldehyde for 10 min at 37 °C and lysed in 1 ml of lysis buffer. The chromatin samples were sheared with the enzymatic shearing cocktail. Anti-C/EBPα, C/EBPβ, C/EBPγ, C/EBPδ or C/EBPϵ antibody (30 μg/ml) and Protein-A–magnetic beads were added to the chromatin samples and incubated for 4 h at 4 °C. The beads were collected using a magnetic stand, washed repeatedly and resuspended in the binding buffer. The beads were then washed, eluted using the elution buffer and the cross-linking was reversed by protease digestion using 0.5 μg/μl proteinase K. After collection of the supernatant, PCR was performed using 5 μl of the supernatant and the following thermal cycling protocol: 1 cycle at 94 °C for 3 min, 45 cycles at 94 °C for 20 s, 59 °C for 30 s and 72 °C for 30 s. The following specific primers were used: sense: 5′-TAGACACAAGCGAGCATGCGCA-3′ (−247 to −226) and antisense: 5′-TCGTCCAGCCGACCAATGGACG-3′ (−55 to −34). The PCR product was electrophoresed through a 3% (w/v) agarose gel and visualized by ethidium bromide staining.
Overexpression of transcription factors in HEK-293 cells
The coding regions of the transcription factors C/EBPγ, δ and ϵ, NF-Y, CREB and NF-κB were amplified by PCR [using a human cDNA library (Human Bone Marrow QUICK-Clone cDNA; Clontech) and HL60 cDNA prepared by RT (reverse transcriptase)-PCR as templates] and then subcloned into pCR Blunt II TOPO (Invitrogen). The sequences of primers used to amplify C/EBPϵ, NF-Y and CREB were indicated above. The following primers for C/EBPγ, C/EBPδ and NF-κB with additional restriction enzyme sites at the 5′- and 3′-end were used: C/EBPγ sense, 5′-TTTGAATTCGGGAGAGTGCCCAAATGACAAG-3′ and antisense, 5′-ACCGCGGCCGCCTACTGTCCTGCTTGTCGCC-3′; C/EBPδ sense, 5′-TTTGAATTCGACGCCGCCATGACGGCCGCGCTCTT-3′ and antisense, 5′-ACCGTCGACTTACCGGCAGTCTGCTGTC-3′; NF-κB sense, 5′-GCTAGCGCCATGGACGAACTGTTCCCCCTCATCTTCCCG-3′ and antisense, 5′-CTCGAGTTAGGAGCTGATCTGACTCAGCAGGGCTGAG-3′.
The pCR Blunt II TOPO vector containing the insert was digested with appropriate restriction enzymes. The fragments then were purified by electrophoresis in a 1% (w/v) agarose gel, digested and cloned into the pCI-neo expression vector (Promega).
HEK-293 cells (5×104 cells) were placed into each well of a 12-well plate before transfection. For transient transfection, 1 μg of the expression vector was mixed with 3 μl of FuGENE™ transfection reagent in 100 μl of DMEM without FBS and subsequently allowed to stand for 20 min at room temperature. The mixture was added into the well and incubated for 24 h to overexpress the transcription factors. The overexpressing HEK-293 cells were stimulated with 50 ng/ml TNFα for 24 h and then subjected to RT-PCR.
RESULTS
Comparison of the TNFα-induced up-regulation of PHGPx mRNA expression between neutrophil-like differentiated and non-differentiated HL60 cells
We have shown that neutrophil-like differentiated HL60 cells up-regulated PHGPx expression on stimulation with TNFα [17]. In the present study, we compared the TNFα-induced up-regulation in PHGPx mRNA expression between neutrophil-like and non-differentiated HL60 cells, because neutrophil-like differentiated HL60 cells could not be transfected with the vectors to detect PHGPx promoter activity using either transfection reagents or electroporation.
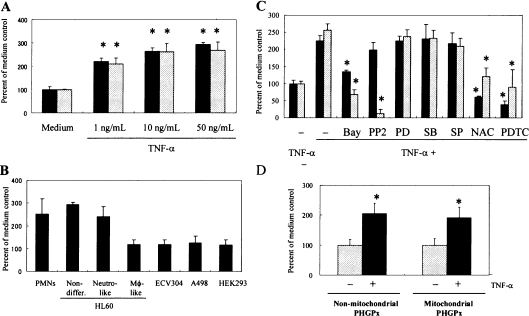
Non-differentiated HL60 cells showed a similar up-regulation in PHGPx mRNA expression on stimulation with TNFα in a dose-dependent manner (Figure 1A). This up-regulation of PHGPx mRNA expression was also observed in PMNs, but not in macrophage-like differentiated HL60 cells, ECV304, A498 or HEK-293 cells (Figure 1B).
Figure 1. Comparison of the TNFα-induced up-regulation of PHGPx mRNA expression between neutrophil-like differentiated and non-differentiated HL60 cells.
(A) Non-differentiated HL60 (black bars) and neutrophil-like differentiated HL60 cells (grey bars) were incubated for 24 h in RPMI 1640 medium containing 10% (v/v) FBS with TNFα at the concentrations indicated. Total cellular RNA was isolated from the cells, and the mRNA levels of PHGPx and 18S rRNA were detected by quantitative PCR. The relative quantity of PHGPx mRNA was normalized with that of 18S rRNA. Results are means±S.D. of the percentage of the medium control. Data were analysed statistically by Dunnett's test (n=4, *P<0.05 significant increase against the medium control). Reproducibility of these results was confirmed in three experiments. (B) PMNs, non-differentiated, neutrophil-like (neutro-like) differentiated and macrophage-like (MΦ-like) differentiated HL60 cells, ECV304, A498 and HEK-293 cells were cultured for 24 h in RPMI 1640 medium containing 10% (v/v) FBS with or without 50 ng/ml TNFα. The RNA isolation and quantitative PCR were performed using the same methods described in (A). Reproducibility of these results was confirmed in three experiments. (C) Non-differentiated HL60 (black bars) and neutrophil-like HL60 cells (grey bars) were cultured for 24 h in the presence or absence of 50 ng/ml TNFα with or without 3 μM Bay 11-7082 (Bay), 1 nM PP2, 30 μM PD98059 (PD), 10 μM SB20358 (SB), 300 nM SP600125 (SP), 50 mM NAC or 50 μM PDTC. RNA isolation, quantitative PCR and data analysis were performed as described in (A). Reproducibility of these results was confirmed in three experiments. (D) Non-differentiated HL60 cells were cultured for 24 h in RPMI 1640 medium containing 10% (v/v) FBS with (black bars) or without (grey bars) 50 ng/ml TNFα. The RNA isolation was performed as described in (A). The mRNA level of non-mitochondrial and mitochondrial PHGPx was detected by TaqMan PCR with the relevant specific primer and probe. The relative quantity of each PHGPx mRNA was normalized with that of 18S rRNA. Results are means±S.D. of the percentage of the medium control. Data were analysed statistically by Student's t test (n=4, *P<0.05 significant increase compared with the medium control). Reproducibility of these results was confirmed in three experiments.
Previous work has shown that the TNFα-induced up-regulation of PHGPx mRNA in neutrophil-like differentiated HL60 cells was inhibited by treatment with NF-κB (Bay 11-7082), Src kinase (PP2) and ROS (NAC and PDTC) inhibitors, but not with MEK [MAPK/ERK (extracellular-signal-regulated kinase) kinase] (PD98059), p38 MAPK (SB203580) and JNK (c-Jun N-terminal kinase) (SP600125) inhibitors [17]. We compared the inhibitory effect of these inhibitors between non-differentiated and neutrophil-like differentiated HL60 cells to confirm similarity in the signalling pathway for TNFα-induced up-regulation of PHGPx mRNA. The NF-κB and ROS inhibitors showed suppressing effects on the up-regulation of PHGPx mRNA in both HL60 cell types, but the Src kinase inhibitor had no effect on non-differentiated HL60 cells. The up-regulation of PHGPx mRNA was not inhibited by treatment with MEK, p38 MAPK or JNK inhibitors in either HL60 cell type (Figure 1C).
We then examined changes in the mRNA levels of non-mitochondrial, mitochondrial and nucleolar types of PHGPx in non-differentiated HL60 cells stimulated with TNFα using TaqMan PCR. The mRNA expression level of non-mitochondrial and mitochondrial PHGPx was up-regulated in cells stimulated with TNFα (Figure 1D). However, no mRNA of nucleolar PHGPx was detected using TaqMan PCR (results not shown).
These findings indicated that the up-regulation of PHGPx mRNA expression in non-differentiated HL60 cells was characterized as a similar response to that of neutrophil-like differentiated HL60 cells, except for the response to the Src kinase inhibitor. Furthermore, non-mitochondrial and mitochondrial PHGPx mRNA was up-regulated in non-differentiated HL60 cells on stimulation with TNFα.
Functional promoter analysis of the 5′-flanking region of the mouse and human PHGPx gene
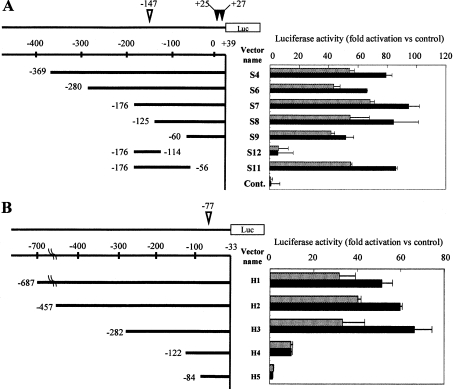
Promoter activity was examined in non-differentiated HL60 cells transfected with the vectors encoding the deletion mutant of either the mouse or human PHGPx promoter region. Luciferase activity of HL60 cells was measured after incubation with or without 50 ng/ml TNFα for 16 h (Figure 2). The translational first start codon (ATG) for the mitochondrial PHGPx gene is expressed as 0. The transcriptional start sites for mitochondrial and non-mitochondrial PHGPx are shown by inverted white and black triangles in Figures 2(A) and 2(B) respectively.
Figure 2. Functional analysis of the PHGPx promoter region in mice and humans.
HL60 cells were transfected by electroporation with the constructed reporter vectors encoding the deletion mutants of the promoter region of the mouse (A) or human (B) PHGPx gene. Firefly luciferase activity of HL60 cells was measured after cultivating with (black bars) or without (grey bars) 50 ng/ml TNFα for 16 h. The phRL-TK vector was used as the internal control to determine transfection efficiency. Firefly luciferase activity from the luciferase reporter vector was normalized to the renilla luciferase activity from the phRL-TK vector, and this was expressed as a ratio of the activity of the pGL3-Basic vector without the PHGPx promoter region. Results are means±S.D. The transcriptional start sites for mitochondrial and non-mitochondrial PHGPx are shown as inverted open and closed triangles respectively. Reproducibility of these results was confirmed in three experiments.
In the reporter assay with the reporter vectors for mouse PHGPx (Figure 2A), promoter activity was detected in the reporter vector (S4), which encoded the promoter region from −369 to +39 bp. Sequential deletions of the 5′ region up to −60 bp (S6 to S9 vectors) had no effect on the promoter activity of the vectors. Promoter activity of the vectors with the promoter region from −125 to +39 bp or longer (S4 to S8) were up-regulated approx. 1.5-fold higher in HL60 cells stimulated with TNFα than in non-stimulated cells. However, the TNFα-induced up-regulation of promoter activity was attenuated in the S9 vector (−60 to +39 bp). The S11 vector (−176 to −56 bp), in which the 3′ region of the S7 vector from −55 to +39 was deleted, retained the promoter activity and the response to TNFα. In contrast, the S12 vector encoding the region from −176 to −14 bp, in which the 3′ region of the S7 vector was further deleted, displayed a decrease in promoter activity and did not respond to TNFα stimulation. These results indicated that the promoter region from −113 to −56 bp was critical for TNFα-induced up-regulation of mouse PHGPx promoter activity.
Reporter vectors encoding the human PHGPx promoter region spanning from −687 to −33 bp were constructed, and the promoter assay was carried out in HL60 cells transfected with the reporter vectors (Figure 2B). Promoter activity was detected in the H1 vector containing the promoter region from −687 to −33 bp, and a sequential deletion of the 5′ promoter region ranging from −457 to −282 bp (H2 and H3 vectors) had no effect on the promoter activity. The promoter activity of these vectors (H1 to H3 vectors) was up-regulated from 1.5- to 2-fold in cells stimulated with TNFα compared with the activity of non-stimulated cells. The promoter activity and the TNFα-induced up-regulation, however, were attenuated in the H4 vector, which contained the region from −122 to −33 bp. These results indicated that the promoter region from −282 to −123 bp was critical for the TNFα-induced up-regulation of PHGPx promoter activity in humans.
Mutational analysis of C/EBP- and NF-Y-binding sequences in the promoter region of the human PHGPx gene
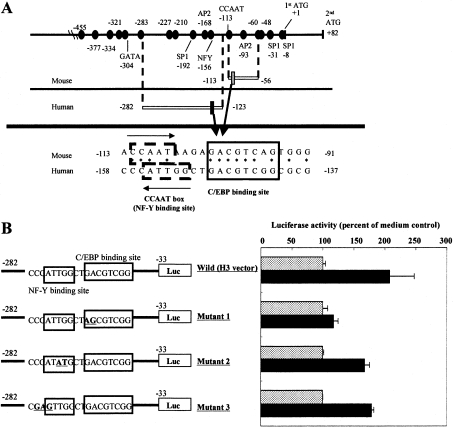
Since the region responsible for the TNFα-induced up-regulation of the promoter activity in humans was distinct from that of mice, the sequence of the region necessary for TNFα-induced up-regulation in humans was compared with that of mice (Figure 3A). Highly homologous sequences were found between the PHGPx regions responsible for TNF-α-induced up-regulation in mice and humans. In a search for a binding site for transcription factors by computer analysis using the TRANSFAC database, the homologous sequence was matched to a C/EBP-binding sequence. In addition, a CCAAT box (a NF-Y-binding site) was also identified as being present immediately upstream of the C/EBP-binding sequence (Figure 3A).
Figure 3. Mutational analysis of C/EBP- and NF-Y-binding sequences in the promoter region of the human PHGPx gene.
(A) Sequences that were highly conserved between human and mouse promoter regions are represented as closed circles. When the sequence of the responsible promoter region from −158 to −137 in humans (black square) was compared with that from −113 to −91 in mice (grey square), highly homologous sequences and binding domains for C/EBP and NF-Y (CCAAT box) were found in the regions. Asteriks indicate identical bases between mouse and human PHGPx. (B) HL60 cells were transfected with the human H3 reporter vector with a mutation in the region responsible for the TNFα-induced up-regulation of PHGPx promoter activity (bold and underlined). Firefly luciferase activity of HL60 cells was measured after incubation for 16 h with (black bars) or without (grey bars) 50 ng/ml TNFα. The phRL-TK vector was used as the internal control to determine transfection efficiency. Firefly luciferase activity from the reporter vector was normalized to the renilla luciferase activity from the phRL-TK vector. Results are means±S.D. of the percentage of the non-stimulated control. Reproducibility of these results was confirmed in three experiments.
To determine the contribution of the C/EBP- and NF-Y-binding sequences to the TNFα-induced up-regulation of promoter activity, we constructed reporter vectors with a mutation in the C/EBP- or NF-Y-binding sequences (Figure 3B). The TNFα-induced up-regulation of promoter activity was attenuated by the mutation in the C/EBP-binding sequence, but not by the mutation in the NF-Y-binding sequence. These findings suggest that the C/EBP-binding sequence is critical for TNFα-induced up-regulation of promoter activity.
Effect of signalling pathway inhibitors on the TNFα-induced up-regulation of promoter activity
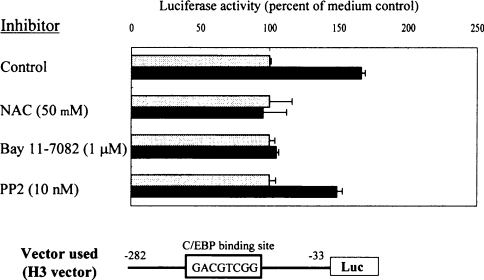
The TNFα-induced up-regulation of PHGPx mRNA in non-differentiated HL60 cells was inhibited by treatment with antioxidants (NAC and PDTC) and NF-κB (Bay 11-7082) inhibitors, but not with the Src kinase (PP2) inhibitor (Figure 1C). To examine whether these inhibitors are effective on the up-regulation of PHGPx promoter activity by treatment with TNFα, HL60 cells transfected with the H3 vector were cultured with or without TNFα in the presence of NAC, Bay 11-7082 or PP2, and promoter activity was measured (Figure 4).
Figure 4. Effects of signalling pathway inhibitors on TNFα-induced up-regulation of PHGPx promoter activity.
HL60 cells were transfected with the human H3 reporter vector. Firefly luciferase activity was measured in HL60 cells cultured for 16 h with (black bars) or without (grey bars) 50 ng/ml TNFα in the presence or absence of NAC (50 mM), Bay 11-7082 (1 μM) or PP2 (10 nM). The phRL-TK vector was used as the internal control to determine transfection efficiency. Firefly luciferase activity from the reporter vector was normalized to the renilla luciferase activity from the phRL-TK vector. Results are means±S.D. of the percentage of the non-stimulated control. Reproducibility of these results was confirmed in three experiments.
The TNFα-induced up-regulation of the promoter activity was inhibited by treatment with 50 mM NAC and 1 μM Bay 11-7082, but not with 10 nM PP2.
Binding of C/EBPϵ to the human PHGPx promoter region
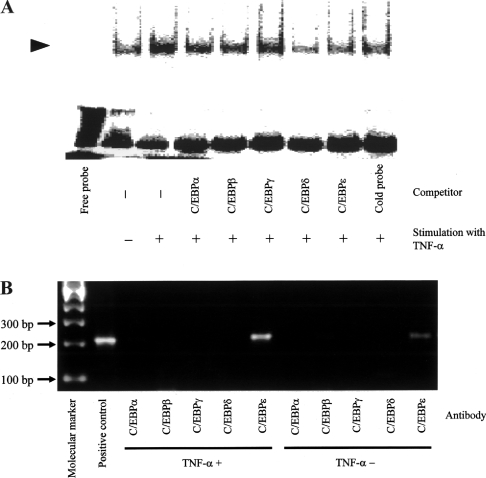
In the promoter assay, TNFα-induced up-regulation of PHGPx promoter activity in non-differentiated HL60 cells was diminished by the mutation in the C/EBP-binding sequence. It has been previously reported that C/EBPϵ, a member of the C/EBP family, is expressed in PMNs and neutrophil-like differentiated HL60 cells, but not in macrophages [24–26]. Conversely, another C/EBP family member, C/EBPβ, is expressed in macrophage-like differentiated HL60 cells, but not in non-differentiated and neutrophil-like differentiated HL60 cells [27]. Therefore we examined whether C/EBPϵ could bind to the C/EBP-binding sequence. Non-differentiated HL60 cells were cultured with or without 50 ng/ml TNFα for 8 h and nuclear proteins were extracted. The binding of proteins to the probe containing the C/EBP-binding sequence was detected by EMSA. To confirm the specificity of the protein binding to the probe, the anti-C/EBPα, C/EBPβ, C/EBPγ, C/EBPδ or C/EBPϵ antibody or the non-radiolabelled probe was mixed with the nuclear protein extracts before the binding reaction with the radiolabelled probe as a competition experiment (Figure 5A).
Figure 5. Binding of C/EBPϵ to the promoter region of the human PHGPx gene.
(A) HL60 cells were incubated with or without 50 ng/ml TNFα for 8 h, and nuclear protein was then extracted. Proteins binding to the DNA probe containing the C/EBP-binding sequence were detected by EMSA. To identify a protein binding to the probe, anti-C/EBPα, C/EBPβ, C/EBPγ, C/EBPδ or C/EBPϵ antibody or the non-radiolabelled probe was mixed and incubated for 1 h at 4 °C with the nuclear protein extract before performing the binding reaction with the radiolabelled probe as a competition experiment. The arrowhead indicates DNA–protein complex. Reproducibility of these results was confirmed in two experiments. (B) HL60 cells incubated for 8 h with or without 50 ng/ml TNFα, fixed with 1% (v/v) formaldehyde, lysed and subjected to immunoprecipitation using the anti-C/EBPα, C/EBPβ, C/EBPγ, C/EBPδ or C/EBPϵ antibody. DNA was released from immunoprecipitates by proteinase K treatment, and the DNA was amplified by PCR using primers corresponding to the promoter region from −247 to −34 of the PHGPx gene. Reproducibility of these results was confirmed in two experiments.
Protein binding to the probe was increased in cells stimulated with TNFα, when compared with non-stimulated cells. The increased binding of the protein was diminished by pre-treatment with the anti-C/EBPδ and C/EBPϵ antibodies and the non-radiolabelled probe as competitors, whereas pre-treatment with the anti-C/EBPα, C/EBPβ and C/EBPγ antibodies was ineffective in disrupting the binding of the protein to the probe (Figure 5A).
We did not find any supershifted bands in the competition assay with the anti-C/EBPδ and C/EBPϵ antibodies. Although the causes of decreased binding of the nuclear proteins to the probe without any supershifted bands are unclear, it might be due to steric hindrance in binding of the nuclear proteins to the probe as a result of the binding of the antibodies to the proteins under the present experimental conditions. Similar reduced binding of nuclear proteins to a probe without any supershifted bands has been reported in EMSA by treatment with anti-C/EBPα and C/EBPβ antibodies [28,29]. These results indicated the formation of complexes between the probe and C/EBPδ or C/EBPϵ.
We next examined whether any of the C/EBP isoforms, especially C/EBPδ and C/EBPϵ, bound to the promoter region (−247 to −34) in chromatin. ChIP assays were performed using chromatin from HL60 cells cultured with or without 50 ng/ml TNFα for 8 h (Figure 5B). When PCR using primer sets for the human PHGPx promoter region (−247 to −34) containing the C/EBP-binding site (−148 to −141) was performed in the presence of HL60 genomic DNA as a positive control, a PCR product of the appropriate size (214 bp) was generated. Likewise, when chromatin was incubated with an antibody against C/EBPϵ, a PCR product corresponding to this segment was also detected, indicating that C/EBPϵ could bind to the C/EBP-binding site. When PCR was performed on immunoprecipitates generated using antibodies against C/EBPα, C/EBPβ, C/EBPδ and C/EBPγ, no cDNA product was detected, indicating the absence of binding of these C/EBP isoforms to the promoter region from −247 to −34 (Figure 5B). A similar result was also observed in nonstimulated HL60 cells. In cells stimulated with TNFα, the cDNA product was increased when compared with that of the non-stimulated cells, suggesting increased binding of C/EBPϵ to the C/EBP sites of the PHGPx promoter region (Figure 5B). These results demonstrated that C/EBPϵ only bound to the human PHGPx promoter region.
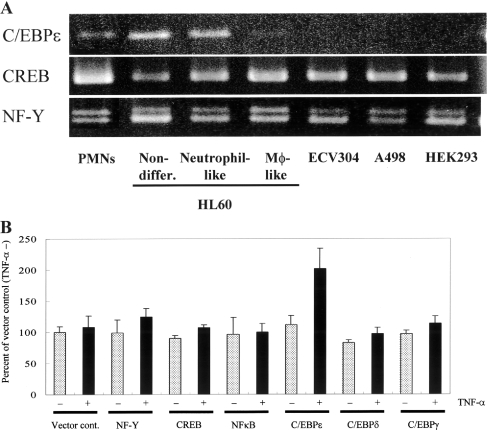
The expression level of C/EBPϵ mRNA in various cell lines, and the TNFα-induced up-regulation of PHGPx mRNA expression in HEK-293 cells overexpressing C/EBPϵ
Since TNFα-induced up-regulation of PHGPx mRNA expression was noted only in some cell types, especially neutrophils (Figure 1B), we examined the relationship between the expression of C/EBPϵ, CREB and NF-Y mRNA, and the TNFα-induced up-regulation of PHGPx mRNA in several cells by RT-PCR (Figure 6A). C/EBPϵ mRNA was detected in responder cells, peripheral PMNs, neutrophil-like differentiated HL60 and non-differentiated HL60 cells displaying TNFα-induced up-regulation of PHGPx mRNA, but it was not detected in non-responder cells, including macrophage-like differentiated HL60, ECV304, A498 and HEK-293 cells. A clear relationship exists between TNFα-induced up-regulation of PHGPx mRNA and the expression of C/EBPϵ. Expression levels of other transcription factors, CREB and NF-Y, were also measured in the responder and non-responder cells, but no relationship was observed between the up-regulation of PHGP mRNA and the expression level of CREB or NF-Y.
Figure 6. Expression level of C/EBPϵ mRNA in various cell lines, and TNFα-induced up-regulation of PHGPx mRNA expression in HEK-293 cells overexpressing C/EBPϵ.
(A) Total cellular RNA was isolated from the cells, and the mRNA levels of C/EBPϵ, CREB and NF-Y were detected by RT-PCR. After PCR, 10 μl of the PCR products were subjected to electrophoresis and the DNA was visualized by ethidium bromide staining. Reproducibility of these results was confirmed in three experiments. (B) HEK-293 cells transiently overexpressing a transcription factor (NF-Y, CREB, NF-κB, C/EBPϵ, C/EBPδ or C/EBPγ) were incubated for 24 h with (black bars) or without (grey bars) 50 ng/ml TNFα. Total cellular RNA was isolated from the cells, and mRNA levels of PHGPx and 18S rRNA as an internal control, were detected by quantitative PCR. Results are means±S.D. of the percentage of the vector control incubated without TNFα. Reproducibility of these results was confirmed in two experiments.
HEK-293 cells showed neither TNFα-induced up-regulation of PHGPx mRNA (Figure 1B) nor C/EBPϵ mRNA expression (Figure 6A). To investigate the involvement of C/EBPϵ in TNFα-induced up-regulation of PHGPx mRNA expression, HEK-293 cells were transfected with the C/EBPδ, C/EBPγ, C/EBPϵ, NF-Y, CREB or NF-κB expression vectors, and then cultured for 24 h to overexpress the transcription factors. The cells were then further cultured with or without 50 ng/ml TNFα for 24 h, and the expression levels of PHGPx mRNA in the cells with or without TNFα stimulation were measured by quantitative PCR (Figure 6B).
HEK-293 cells transfected with the control vector did not show any up-regulation of PHGPx mRNA on stimulation with TNFα. Up-regulation of PHGPx mRNA, however, was detected in HEK-293 cells overexpressing C/EBPϵ and stimulated with TNFα. In contrast, no TNFα-induced up-regulation of PHGPx mRNA was detected in HEK-293 cells overexpressing C/EBPδ, C/EBPγ, NF-Y, CREB or NF-κB.
DISCUSSION
The mRNA of three types of PHGPx: mitochondrial, non-mitochondrial, and nucleolar types, is transcribed from one gene by alternative transcription using different promoter regions, which are located upstream of exon Ia for the mitochondrial and non-mitochondrial types, and upstream of exon Ib for the nucleolar type [8,22]. In the present study, HL60 cells stimulated with TNFα displayed up-regulation of mitochondrial and non-mitochondrial PHGPx mRNA expression, but no nucleolar PHGPx mRNA was detected. Earlier work had indicated that strong promoter activity was detected in the reporter vector (S4) encoding the promoter region from −369 to +39 bp which contained the transcriptional start site for mitochondrial (−147) and non-mitochondrial (+25 and +27) PHGPx. This region also includes highly homologous sequences between mice and human, and several binding sites for transcription factors, whereas the S10 vector with the region from −8 to +39 bp showed no promoter activity [22]. From these findings, we performed functional promoter analysis using the S4 vector and six other vectors which were sequential deletions of the promoter region of the S4 vector (S6 to S9, S11 and S12). For the functional promoter assay of the human PHGPx gene, the H1 vector with the region from −687 to −33 and the other four vectors (H2 to H5) which were sequential deletions of the promoter region of the H1 vector, were used because the S11 vector (−176 to −56), lacking the downstream sequence from −56 to +39, and the S4 vector showed up-regulation of promoter activity on stimulation with TNFα in the promoter assay of the mouse PHGPx gene (Figure 2A). The promoter region in the human vectors has the transcriptional start site for mitochondrial PHGPx (−77), but not that of non-mitochondrial PHGPx (+1 to +33). These vectors, however, are thought to be appropriate for analysis of promoter activity of both types of PHGPx because the S11 vector, which does not have the transcriptional start site for non-mitochondrial PHGPx, can measure the promoter activity of non-mitochondrial PHGPx [22].
The C/EBPs are a family of transcription factors [30]. All C/EBP family members contain a characteristic basic domain in the conserved C-terminus, which is important for DNA binding and interaction with certain other proteins, and a dimerization domain that contains a leucine zipper and is responsible for the formation of stable, covalently-linked homodimers and heterodimers of different C/EBP-like proteins. C/EBP family members have been reported to have different functions and display differing expression profiles. It is noteworthy that C/EBPϵ is expressed primarily in myeloid and lymphoid cells, and preferentially expressed during granulocyte differentiation, but not macrophage differentiation. Furthermore, the non-differentiated and neutrophil-like differentiated HL60 cells express C/EBPϵ, but expression in the monocytic cell line U937 is negative, or at levels less than that of HL60 cells [24–26], suggesting neutrophil-specific expression of C/EBPϵ. PMNs from C/EBPϵ-deficient mice show a reduction in phagocytotic killing, abnormalities in migration, abnormal CD11b integrin and L-selectin expression and altered IL-1Ra (IL-1 receptor antagonist) and TNFα expression in response to inflammation [31]. Gombart et al. [32] have reported that the down- and up-regulation of many genes involved in cell adhesion and chemotaxis, cytoskeletal organization, signal transduction and immune and inflammatory responses is detectable by microarray analysis in PMNs from C/EBPϵ-deficient mice. This indicates that C/EBPϵ plays a critical role in modulating the functions of PMNs. In the present study, we found a close association between the expression of C/EBPϵ and TNFα-induced up-regulation of PHGPx mRNA (Figures 1B and 6A). Reddy et al. [33] reported that increased binding of C/EBPϵ to the promoter region of 5-lipoxygenase-activating protein was induced in cells treated with TNFα. As with C/EBPϵ, C/EBPβ has been reported to show increased binding to a promoter region of ICAM-1 (intercellular adhesion molecule 1) and COX (cyclo-oxygenase)-2, which are closely associated with inflammatory responses, on stimulation with TNFα and LPS (lipopolysaccharide), and play a critical role in the gene expression of those molecules [34,35]. However, although C/EBPβ mRNA is expressed in macrophage-like HL60 cells, it is not expressed in non-differentiated and neutrophil-like differentiated HL60 cells [27]. Expression of C/EBPδ is detected in the intestine, lung and adipose tissue, with high levels of expression detected in all tissues following LPS stimulation. C/EBPγ is ubiquitously expressed, with the highest levels found in non-differentiated and progenitor cells. C/EBPα is detected in several tissues such as the liver, mammary gland, skin and the haematopoietic system, and the expression of C/EBPα is also relatively high in early myeloid progenitors and decreases during granulocytic differentiation [24]. However, we confirm that C/EBPα, C/EBPβ and C/EBPδ mRNA was not expressed in non-differentiated HL60 cells, although the mRNA of C/EBPγ and C/EBPϵ was detected (results not shown). Furthermore, the TNFα-induced up-regulation of PHGPx mRNA was detected in HEK-293 cells overexpressing C/EBPϵ, but not in cells overexpressing C/EBPδ or C/EBPγ (Figure 6B). These findings strongly indicated that C/EBPϵ is an essential molecule for TNFα-induced up-regulation of PHGPx mRNA.
C/EBPϵ has not been reported to be activated through ROS signalling pathways, whereas the involvement of ROS signalling in the activation of C/EBPβ has been demonstrated. NF-κB and C/EBPβ are key transcription factors in the up-regulation of ICAM-1 expression induced by TNFα stimulation, and the binding of C/EBPβ to a promoter region of the ICAM-1 gene is reduced by treatment with antioxidants, PDTC and dimethylthiourea [36]. C/EBPβ plays a key role in the elevation of IL-6 production in lung epithelial cells stimulated by asbestos, and C/EBPβ activation was inhibited by an antioxidant, tetramethylurea. Furthermore, treatment with H2O2 activated C/EBPβ in lung epithelial cells [37]. In the present study, the TNFα-induced up-regulation of PHGPx promoter activity by activation of C/EBPϵ was inhibited by treatment with NAC (Figure 4), indicating that ROS signalling pathways were involved in up-regulation via the activation of C/EBPϵ, similar to C/EBPβ. In addition, Bay 11-7082 also inhibited the TNFα-induced up-regulation of promoter activity, suggesting that the NF-κB pathway was implicated in the up-regulation of the PHGPx promoter (Figure 4). However, the reporter vector encoding the human PHGPx promoter region (from −282 to −33; H3 vector) has no NF-κB-binding sequence. Therefore the involvement of NF-κB in TNFα-induced up-regulation of PHGPx mRNA is unclear. NF-κB has been reported to induce transcriptional activation of several proteins, not only by direct binding to its binding sequences in promoter regions, but also by interacting with other transcription factors with the basic-leucine zipper domain through the RDH domain in NF-κB [38]. For example, C/EBP- and NF-κB-binding sequences are closely located in the promoter region of the COX-2 and IL-8 genes, and an interaction between both transcription factors is important in the induction of COX-2 and IL-8 gene transcription [39,40]. An NF-κB and C/EBPδ complex is reported to be involved in the transactivation of serum amyloid-A protein induced by LPS stimulation [28]. Furthermore, transactivation via the complex is observed when NF-κB binding is abrogated by a mutation in the binding sequence [40]. Therefore it is postulated that a complex between NF-κB and C/EBPϵ is involved in TNFα-induced up-regulation of PHGPx promoter activity in the H3 vector without the presence of a NF-κB-binding sequence.
Three previous studies have only addressed the up-regulation of PHGPx expression in spermatogenesis [41,42] and embryogenesis [18], and its down-regulation in stimulation of cytokines IL-4 and IL-13 [43]. The mechanisms and transcription factors affecting these interesting changes of PHGPx, however, are still not well known. Tramer et al. [44] recently reported that overexpression of a transcription factor, CREMt (cAMP-response element modulator-tau) in NIH-3T3 cells induced the up-regulation of nucleolar PHGPx promoter activity. In the present study, we originally identified C/EBPϵ as a critical transcription factor in the up-regulation of promoter activity and expression level of mRNA for non-mitochondrial and mitochondrial PHGPx. Our previous studies have shown that the intracellular PHGPx level is closely associated with the production of eicosanoids in RBL2H3 cells. The levels of leukotrienes and prostaglandins are decreased in PHGPx-overexpressing RBL2H3 cells compared with those in a control cell line. This decreased production of eicosanoids is attributed to a reduction of activity of lipoxygenases and COXs by PHGPx [10–12]. Furthermore, PMNs collected from the peritoneal cavity of rats given an intraperitoneal injection of sodium casein show an increase in the expression level of PHGPx, followed by a reduced production of leukotriene B4 and 5-HETE (5-hydroxyeicosatetraenoic acid) after culturing these PMNs in vitro for a further 24 h [45]. The results obtained suggests that the PHGPx expression level in activated PMNs would modulate the progression of inflammation via the suppression of eicosanoid production from PMNs at an inflammatory site. Therefore it is possible that C/EBPϵ, which is a critical molecule in TNFα-induced up-regulation of PHGPx, may be a new target molecule for anti-inflammation therapy in the future.
Acknowledgments
We thank Mr Tsuyoshi Yamamoto and Ms Naomi Tsunashima for their expert technical assistance. This work was supported in part by a Kitasato University Research Grant for Young Researchers, by Special Coordination Funds for the Promotion of Science and Technology, by grants-in-aid (No. 17590067) from the Ministry of Education, Science and Culture of Japan, and by PRESTO from the Japan Science and Technology Agency.
References
- 1.Root R. K., Cohen M. S. The microbicidal mechanisms of human neutrophils and eosinophils. Rev. Infect. Dis. 1981;3:565–598. doi: 10.1093/clinids/3.3.565. [DOI] [PubMed] [Google Scholar]
- 2.Babior B. M. Oxygen-dependent microbial killing by phagocytosis. N. Engl. J. Med. 1978;298:659–668. doi: 10.1056/NEJM197803232981205. [DOI] [PubMed] [Google Scholar]
- 3.Scapini P., Lapinet-Vera J. A., Gasperini S., Calzetti F., Bazzoni F., Cassatella M. A. The neutrophil as a cellular source of chemokines. Immunol. Rev. 2000;177:195–203. doi: 10.1034/j.1600-065x.2000.17706.x. [DOI] [PubMed] [Google Scholar]
- 4.Ford-Hutchinson A. W., Bray M. A., Doig M. V., Shipley M. E., Smith M. J. H. Leukotriene B, a potent chemokinetic and aggregating substance released from polymorphonuclear leukocytes. Nature. 1980;286:264–265. doi: 10.1038/286264a0. [DOI] [PubMed] [Google Scholar]
- 5.Goldstein I. M., Malmsten C. L., Kindahl H., Kaplan H. B., Radmark O., Samuelsson B., Weissmann G. Thromboxane generation by human peripheral blood polymorphonuclear leukocytes. J. Exp. Med. 1978;148:787–792. doi: 10.1084/jem.148.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunican A. L., Leuenroth S. J., Grutkoski P., Ayala A., Simms H. H. TNF-α-induced suppression of PMN apoptosis is mediated through interleukin-8 production. Shock. 2000;14:284–288. doi: 10.1097/00024382-200014030-00007. [DOI] [PubMed] [Google Scholar]
- 7.Arai M., Imai H., Koumura T., Toshida M., Emoto K., Umeda M., Chiba N., Nakagawa Y. Mitochondrial phospholipid hydroperoxide glutathione peroxidase plays a major role in preventing oxidative injury to cells. J. Biol. Chem. 1999;274:4924–4933. doi: 10.1074/jbc.274.8.4924. [DOI] [PubMed] [Google Scholar]
- 8.Imai H., Nakagawa Y. Biological significance of phospholipid hydroperoxide glutathione peroxidase (PHGPx, GPx4) in mammalian cells. Free Radical. Biol. Med. 2003;34:145–169. doi: 10.1016/s0891-5849(02)01197-8. [DOI] [PubMed] [Google Scholar]
- 9.Nomura K., Imai H., Koumura T., Arai M., Nakagawa Y. Mitochondrial phospholipid hydroperoxide glutathione peroxidase suppresses apoptosis mediated by a mitochondrial death pathway. J. Biol. Chem. 1999;274:29294–29302. doi: 10.1074/jbc.274.41.29294. [DOI] [PubMed] [Google Scholar]
- 10.Imai H., Narashima K., Arai M., Sakamoto H., Chiba N., Nakagawa Y. Suppression of leukotriene formation in RBL-2H3 cells that overexpressed phospholipid hydroperoxide glutathione peroxidase. J. Biol. Chem. 1998;273:1990–1997. doi: 10.1074/jbc.273.4.1990. [DOI] [PubMed] [Google Scholar]
- 11.Sakamoto H., Imai H., Nakagawa Y. Involvement of phospholipid hydroperoxide glutathione peroxidase in the modulation of prostaglandin D2 synthesis. J. Biol. Chem. 2000;275:40028–40035. doi: 10.1074/jbc.M003191200. [DOI] [PubMed] [Google Scholar]
- 12.Sakamoto H., Tosaki T., Nakagawa Y. Overexpression of phospholipid hydroperoxide glutathione peroxidase modulates acetyl-CoA, 1-O-alkyl-2-lysosn-glycero-3-phosphocholine acetyltransferase activity. J. Biol. Chem. 2002;277:50431–50438. doi: 10.1074/jbc.M204190200. [DOI] [PubMed] [Google Scholar]
- 13.Wenk J., Schuller J., Hinrichs C., Syrovets T., Azoitei N., Podda M., Wlaschek M., Brenneisen P., Schneider L. A., Sabiwalsky A. Overexpression of phospholipid-hydroperoxide glutathione peroxidase in human dermal fibroblasts abrogates UVA irradiation-induced expression of interstitial collagenase/matrix metalloproteinase-1 by suppression of phosphatidylcholine hydroperoxide-mediated NFκB activation and interleukin-6 release. J. Biol. Chem. 2004;279:45634–45642. doi: 10.1074/jbc.M408893200. [DOI] [PubMed] [Google Scholar]
- 14.Brigelius-Flohe R., Maurer S., Lotzer K., Bol G., Kallionpaa H., Lehtolainen P., Viita H., Yla-Herttuala S. Overexpression of PHGPx inhibits hydroperoxide-induced oxidation, NFκB activation and apoptosis and affects oxLDL-mediated proliferation of rabbit aortic smooth muscle cells. Atherosclerosis. 2000;152:307–316. doi: 10.1016/s0021-9150(99)00486-4. [DOI] [PubMed] [Google Scholar]
- 15.Hsu M. H., Wang M., Browning D. D., Mukaida N., Ye R. D. NF-κB activation is required for C5a-induced interleukin-8 gene expression in mononuclear cells. Blood. 1999;93:3241–3249. [PubMed] [Google Scholar]
- 16.May M. J., Ghosh S. Signal transduction through NF-κB. Immunol. Today. 1998;19:80–88. doi: 10.1016/s0167-5699(97)01197-3. [DOI] [PubMed] [Google Scholar]
- 17.Hattori H., Imai H., Furuhama K., Sato O., Nakagawa Y. Induction of phospholipid hydroperoxide glutathione peroxidase in human polymorphonuclear neutrophils and HL60 cells stimulated with TNFα. Biochem. Biophys. Res. Commun. 2005;337:464–473. doi: 10.1016/j.bbrc.2005.09.076. [DOI] [PubMed] [Google Scholar]
- 18.Imai H., Hirao F., Sakamoto T., Sekine K., Mizukura Y., Saito M., Kitamoto T., Hayasaka M., Hanaoka K., Nakagawa Y. Early embryonic lethality caused by targeted disruption of the mouse PHGPx gene. Biochem. Biophys. Res. Commun. 2003;305:278–286. doi: 10.1016/s0006-291x(03)00734-4. [DOI] [PubMed] [Google Scholar]
- 19.Kelner M. J., Montoya M. A. Structural organization of the human selenium-dependent phospholipid hydroperoxide glutathione peroxidase gene (GPX4): chromosomal localization to 19p13.3. Biochem. Biophys. Res. Commun. 1998;249:53–55. doi: 10.1006/bbrc.1998.9086. [DOI] [PubMed] [Google Scholar]
- 20.Imai H., Sumi D., Hanamoto A., Arai M., Sugiyama A. Molecular cloning and functional expression of a cDNA for rat phospholipid hydroperoxide glutathione peroxidase: 3′-untranslated region of the gene is necessary for functional expression. J. Biochem. 1995;118:1061–1067. doi: 10.1093/jb/118.5.1061. [DOI] [PubMed] [Google Scholar]
- 21.Brigelius-Flohe R., Aumannm K. D., Blockerm H., Gross G., Kiess M., Kloppel K. D., Maiorino M., Roveri A., Schuckelt R., Usani F. Phospholipid-hydroperoxide glutathione peroxidase. Genomic DNA, cDNA, and deduced amino acid sequence. J. Biol. Chem. 1994;269:7342–7348. [PubMed] [Google Scholar]
- 22.Imai H., Saito M., Kirai N., Hasegawa J., Konishi K., Hattori H., Nishimura M., Naito S., Nakagawa Y. Identification of the positive regulatory and distinct core regions of promoters, and transcriptional regulation in three types of mouse phospholipid hydroperoxide glutathione peroxidase. J. Biochem. 2006;140:573–590. doi: 10.1093/jb/mvj186. [DOI] [PubMed] [Google Scholar]
- 23.Huang H. S., Chen C. J., Chang W. C. The CCAAT-box binding factor NF-Y is required for the expression of phospholipid hydroperoxide glutathione peroxidase in human epidermoid carcinoma A431 cells. FEBS Lett. 1999;455:111–116. doi: 10.1016/s0014-5793(99)00866-2. [DOI] [PubMed] [Google Scholar]
- 24.Lekstrom-Himes J., Xanthopoulos K. G. Biological role of the CCAAT/enhancer-binding protein family of transcription factors. J. Biol. Chem. 1998;273:28545–28548. doi: 10.1074/jbc.273.44.28545. [DOI] [PubMed] [Google Scholar]
- 25.Morosetti R., Park D. J., Chumakov A. M., Grillier I., Shiohara M., Gombart A. F., Nakamaki T., Weinberg K., Koeffler H. P. A novel, myeloid transcription factor, C/EBPϵ, is upregulated during granulocytic, but not monocytic, differentiation. Blood. 1997;90:2591–2600. [PubMed] [Google Scholar]
- 26.Yamanaka R., Kim G. D., Radomska H. S., Lekstrom-Himes J., Smith L. T., Antonson P., Tenen D. G., Xanthopoulos K. G. CCAAT/enhancer binding protein ϵ is preferentially up-regulated during granulocytic differentiation and its functional versatility is determined by alternative use of promoters and differential splicing. Proc. Natl. Acad. Sci. U.S.A. 1997;94:6462–6467. doi: 10.1073/pnas.94.12.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Natsuka S., Akira S., Nishio Y., Hashimoto S., Sugita T., Isshiki H., Kishimoto T. Macrophage differentiation-specific expression of NF-IL6, a transcription factor for interleukin-6. Blood. 1992;79:460–466. [PubMed] [Google Scholar]
- 28.Ray A., Hannink M., Ray B. K. Concerted participation of NF-κB and C/EBP heteromer in lipopolysaccharide induction of serum amyloid A gene expression in liver. J. Biol. Chem. 1995;270:7365–7374. doi: 10.1074/jbc.270.13.7365. [DOI] [PubMed] [Google Scholar]
- 29.Takeshita F., Suzuki K., Sasaki S., Ishii N., Klinman D. M., Ishii K. J. Transcriptional regulation of the human TLR9 gene. J. Immunol. 2004;173:2552–2561. doi: 10.4049/jimmunol.173.4.2552. [DOI] [PubMed] [Google Scholar]
- 30.Ramji D. P., Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem. J. 2002;365:561–575. doi: 10.1042/BJ20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lekstrom-Himes J., Xanthopoulos K. G. CCAAT/enhancer binding protein ϵ is critical for effective neutrophil-mediated response to inflammatory challenge. Blood. 1999;93:3096–3105. [PubMed] [Google Scholar]
- 32.Gombart A. F., Krug U., O'Kelly J., An E., Vegesna V., Koeffler H. P. Aberrant expression of neutrophil and macrophage-related genes in a murine model for human neutrophil-specific granule deficiency. J. Leukocyte Biol. 2005;78:1153–1165. doi: 10.1189/jlb.0504286. [DOI] [PubMed] [Google Scholar]
- 33.Reddy K. V., Serio K. J., Hodulik C. R., Bigby T. D. 5-lipoxygenase-activating protein gene expression. Key role of CCAAT/enhancer-binding proteins (C/EBP) in constitutive and tumor necrosis factor (TNF) α-induced expression in THP-1 cells. J. Biol. Chem. 2003;278:13810–13818. doi: 10.1074/jbc.M211102200. [DOI] [PubMed] [Google Scholar]
- 34.Roebuck K. A., Rahman A., Lakshminarayanan V., Janakidevi K., Malik A. B. H2O2 and tumor necrosis factor-α activate intercellular adhesion molecule 1 (ICAM-1) gene transcription through distinct cis-regulatory elements within the ICAM-1 promoter. J. Biol. Chem. 1995;270:18966–18974. doi: 10.1074/jbc.270.32.18966. [DOI] [PubMed] [Google Scholar]
- 35.Gorgoni B., Caivano M., Arizmendi C., Poli V. The transcription factor C/EBPβ is essential for inducible expression of the cox-2 gene in macrophages but not in fibroblasts. J. Biol. Chem. 2001;276:40769–40777. doi: 10.1074/jbc.M106865200. [DOI] [PubMed] [Google Scholar]
- 36.Krunkosky T. M., Martin L. D., Fischer B. M., Voynow J. A., Adler K. B. Effects of TNF-α on expression of ICAM-1 in human airway epithelial cells in vitro: oxidant-mediated pathways and transcription factors. Free. Radical Biol. Med. 2003;35:1158–1167. doi: 10.1016/s0891-5849(03)00498-2. [DOI] [PubMed] [Google Scholar]
- 37.Simeonova P. P., Toriumi W., Kommineni C., Erkan M., Munson A. E., Rom W. N., Luster M. I. Molecular regulation of IL-6 activation by asbestos in lung epithelial cells: role of reactive oxygen species. J. Immunol. 1997;159:3921–3928. [PubMed] [Google Scholar]
- 38.Perkins N. D. Achieving transcriptional specificity with NF-κB. Int. J. Biochem. Cell Biol. 1997;29:1433–1448. doi: 10.1016/s1357-2725(97)00088-5. [DOI] [PubMed] [Google Scholar]
- 39.Stein B., Baldwin A. S., Jr Distinct mechanisms for regulation of the interleukin-8 gene involve synergism and cooperativity between C/EBP and NF-κB. Mol. Cell. Biol. 1993;13:7191–7198. doi: 10.1128/mcb.13.11.7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen J., Zhao M., Rao R., Inoue H., Hao C. M. C/EBPβ and its binding element are required for NFκB-induced COX2 expression following hypertonic stress. J. Biol. Chem. 2005;280:16354–16359. doi: 10.1074/jbc.M411134200. [DOI] [PubMed] [Google Scholar]
- 41.Imai H., Suzuki K., Ishizaka K., Ichinose S., Oshima H., Okayasu I., Emoto K., Umeda M., Nakagawa Y. Failure of the expression of phospholipid hydroperoxide glutathione peroxidase in the spermatozoa of human infertile males. Biol. Reprod. 2001;64:674–683. doi: 10.1095/biolreprod64.2.674. [DOI] [PubMed] [Google Scholar]
- 42.Maiorino M., Wissing J. B., Brigelius-Flohe R., Calabrese F., Roveri A., Steinert P., Ursini F., Flohe L. Testosterone mediates expression of the selenoprotein PHGPx by induction of spermatogenesis and not by direct transcriptional gene activation. FASEB J. 1998;12:1359–1370. doi: 10.1096/fasebj.12.13.1359. [DOI] [PubMed] [Google Scholar]
- 43.Schnurr K., Borchert A., Kuhn H. Inverse regulation of lipid-peroxidizing and hydroperoxyl lipid-reducing enzymes by interleukins 4 and 13. FASEB J. 1999;13:143–154. doi: 10.1096/fasebj.13.1.143. [DOI] [PubMed] [Google Scholar]
- 44.Tramer F., Vetere A., Martinelli M., Paroni F., Marsich E., Boitani C., Sandri G., Panfili E. cAMP-response element modulator-tau activates a distinct promoter element for the expression of the phospholipid hydroperoxide/sperm nucleus glutathione peroxidase gene. Biochem. J. 2004;383:179–185. doi: 10.1042/BJ20040974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hattori H., Imai H., Hanamoto A., Furuhama K., Nakagawa Y. Up-regulation of phospholipid hydroperoxide glutathione peroxidase in rat casein-induced polymorphonuclear neutrophils. Biochem. J. 2005;389:279–287. doi: 10.1042/BJ20050006. [DOI] [PMC free article] [PubMed] [Google Scholar]