Abstract
GH5BG, the cDNA for a stress-induced GH5 (glycosyl hydrolase family 5) β-glucosidase, was cloned from rice (Oryza sativa L.) seedlings. The GH5BG cDNA encodes a 510-amino-acid precursor protein that comprises 19 amino acids of prepeptide and 491 amino acids of mature protein. The protein was predicted to be extracellular. The mature protein is a member of a plant-specific subgroup of the GH5 exoglucanase subfamily that contains two major domains, a β-1,3-exoglucanase-like domain and a fascin-like domain that is not commonly found in plant enzymes. The GH5BG mRNA is highly expressed in the shoot during germination and in leaf sheaths of mature plants. The GH5BG was up-regulated in response to salt stress, submergence stress, methyl jasmonate and abscisic acid in rice seedlings. A GUS (glucuronidase) reporter tagged at the C-terminus of GH5BG was found to be secreted to the apoplast when expressed in onion (Allium cepa) cells. A thioredoxin fusion protein produced from the GH5BG cDNA in Escherichia coli hydrolysed various pNP (p-nitrophenyl) glycosides, including β-D-glucoside, α-L-arabinoside, β-D-fucoside, β-D-galactoside, β-D-xyloside and β-D-cellobioside, as well as β-(1,4)-linked glucose oligosaccharides and β-(1,3)-linked disaccharide (laminaribiose). The catalytic efficiency (kcat/Km) for hydrolysis of β-(1,4)-linked oligosaccharides by the enzyme remained constant as the DP (degree of polymerization) increased from 3 to 5. This substrate specificity is significantly different from fungal GH5 exoglucanases, such as the exo-β-(1,3)-glucanase of the yeast Candida albicans, which may correlate with a marked reduction in a loop that makes up the active-site wall in the Candida enzyme.
Keywords: environmental stress, fascin-like domain, β-glucosidase, glycosyl hydrolase family 5, recombinant protein expression, rice (Oryza sativa)
Abbreviations: ABA, abscisic acid; CDS, coding sequence; Exg, exo-β-(1,3)-glucanase; DP, degree of polymerization; GH, glycosyl hydrolase; GH5, glycosyl hydrolase family 5; GUS, glucuronidase; IMAC, immobilized-metal affinity chromatography; IPTG, isopropyl β-D-thiogalactoside; LB, Luria–Bertani; pNP, p-nitrophenyl; pNPG, pNP β-D-glucopyranoside; PGO, peroxidase/glucose oxidase; X-Glu, 5-bromo-4-chloroindol-3-yl β-glucuronide
INTRODUCTION
GHs (glycosyl hydrolases), enzymes that catalyse the hydrolysis of glycosidic bonds between sugars and other moieties, can be classified into more than 100 families [1] [for up-to-date information, see the Carbohydrate-Active Enzymes database (CAZY) at http://www.cazy.org/CAZY/index.html]. On the basis of their three-dimensional structures, GHs can be grouped into clans of related structures [1]. Clan A is the largest group and contains 17 families, the structures of which contain a core (β/α)8 barrel with two catalytic amino acid residues, an acid/base and a nucleophile, on the ends of strands 4 and 7 of the barrel, respectively [2–4].
GH5 (GH family 5) is one clan A family, originally identified as cellulase family A [5], that contains enzymes with a wide range of catalytic activities, including cellulases, chitosanases, endoglucanases, exoglucanases, exoxylanases, endoxylanases, β-mannanases, and endoglycoceramidase. The GH5 enzymes that have been investigated are primarily from micro-organisms [6–12], though a β-mannanase from a plant was recently described [13]. Since the variation in the protein sequences is high, the endoglucanse families were split into six subfamilies, A1–A6 [14,15], and two more subfamilies of mannanases, A7 and A8, were subsequently added [16] with sequence identity between subfamilies generally being 20% or less. The Exgs [exo-β-(1,3)-glucanases] have been noted to form a distinct subfamily, though no number was designated [9]. Despite the high variation in the protein sequences and enzyme activities of the family members, they all possess eight conserved residues (including two glutamate residues acting as catalytic acid/base and nucleophile) around the active site, which distinguish GH5 from other GH families [9,17].
There are now 22 known GH5 three-dimensional crystal structures: endoglucanases from Acidothermus cellulolyticus, Bacillus agaradhaerens, Bacillus sp., Clostridium cellulyticum, Clostridium thermocellum, Erwinia chrysanthemi, Pseudoalteromonas haloplanktis, Thermobifida fusca and Thermoascus aurantiacus; exo-β-(1,3)-glucanases from C. albicans and Saccharomyces cerevisiae; mannanases from Bacillus sp., Cellvibrio mixtus, T. fusca, Thermotoga maritima, Hypocrea jecorina (anamorph Trichoderma reesei), Solanum lycopersicum (=Lycopersicon esculentum, tomato) and Mytilus edulis (common mussel); xylanase from H. jecorina and E. chrysanthemi; xyloglucanase from Paenibacillus pabuli; and endoglycoceramidase from Rhodococcus sp. ([4]; http://www.CAZY.org/CAZY/index.html). This abundance of structural data is necessary, since the similar overall structure of GH5 has resulted in several distinct activities, as indicated. Though these enzymes may have similar (β/α)8 barrel structures, differences in the loops at the ends of the β-strands of this barrel result in active site clefts ranging from long grooves to slot-like pockets [9,18].
Exoglucanases are generally secreted enzymes with both hydrolase and transferase activities towards β-glucans [19]. Exgs may act in the metabolism of cell-wall glucan by cleaving a single glucose moiety from the non-reducing end of β-1,3-glucans [20]. Most GH5 exoglucanases that have been studied are fungal Exgs, including those from C. albicans [9,21], S. cerevisiae [22], Agaricus bisporus [23], Lentinula edodes (medicinal shiitake mushroom) [24], and Pichia pastoris [25]. Cutfield et al. [9] reported the structure of C. albicans Exg to be a distorted (β/α)8 barrel structure with a deep active-site pocket. The geometry of the pocket fits for cleavage of β-1,3-, but not β-1,4-, glycosidic linkages. Using active-site labelling and mutagenesis experiments, Glu192 and Glu292 in the mature C. albicans Exg protein were identified as the proton donor and nucleophile respectively [19,21].
There has been no previous report of characterization of a GH5 exoglucanase from plants. However, several genes encoding proteins similar to fungal Exgs are found in the genomic sequences from rice (Oryza sativa L.). In the present study we cloned the cDNA of a putative GH5 glucan-1,3-β-glucosidase containing a fascin (an actin-bundling protein)-like domain near its N-terminus from germinating rice based on genomic data and assessed its function by recombinant protein expression and its expression by Northern blot. The catalytic activity indicated that the enzyme is a β-glucosidase. This is the first report of a GH5 β-glucosidase from a plant that contains a fascin-like domain.
EXPERIMENTAL
Plant materials and growth conditions
Rice (spp. indica cv. KDML105 and spp. japonica cv. Yukihikari) seeds were germinated in the dark from day 0 to day 3 and in 12 h light/12 h dark from day 4 to day 7 at 28 °C on germinating paper moistened with sterile distilled water. For expression analysis, whole Yukihikari seedlings were harvested and some were dissected into separate parts (shoot, root and endosperm) and kept at −70 °C. Some 14-day-old rice seedlings were transferred to soil and grown for an additional 4 weeks. Rice plants were harvested and separated into six parts: flower, stem, root, node, leaf blade and leaf sheath. Some 7-day-old rice seedlings were exposed to abiotic stresses and plant hormones for an additional 2 days under the following conditions: salt stress (0.3 M NaCl), osmotic stress (0.3 M sorbitol), drought (no water), flooding (full submergence of seedlings 1 cm below surface of distilled water), cold stress (5 and 12 °C), heat stress (37 °C), 0.1 mM methyl jasmonate, 0.1 mM ABA (abscisic acid) and 1 mg/ml Ethephon [the plant growth regulator (2-chloroethyl)phosphonic acid]. All plant samples were kept at −70 °C for RNA isolation.
Cloning of GH5BG cDNA
Total RNA was isolated from 100 mg of 5–6-day-old rice cv. KDML105 seedlings with Trizol Reagent, and 5 μg of total RNA was used as the template to synthesize the first-strand cDNA with SuperScript II reverse transcriptase according to the manufacturer's (Invitrogen, Carlsbad, CA, U.S.A.) protocol. The GenBank® rice genome contig accession number AC107314 (deduced protein sequence GenBank AC AAM08614) and AK065000 cDNA sequences [26] were used to design the primers to amplify a full-length CDS (coding sequence) cDNA and a cDNA encoding the mature protein of rice glycosyl hydrolase family 5 β-glucosidase (designated GH5BG). The 5′ sense primer AK065000f (5′-GCTGAAAAATCTTCGTCTTC- ATC-3′) and the antisense primer AAM08614EcoRIr (5′-CCATCCAACTGGAATTCTCACTG-3′) were used to amplify a 774 bp-5′ PCR fragment. The 5′ sense primer AM08614EcoRIf (5′-CGCAGTGAGAATTCCAGTTG-3′) and the antisense primer AK065000r (5′-CTTCACAAGAGAAAGTTACACTC-3′) were used to amplify a 1016 bp-3′ PCR fragment. The amplification for 5′ and 3′ PCR fragments was done with Pfu DNA polymerase (Promega, Madison, WI, U.S.A.) with the first-strand cDNA as the template. Finally, the AK065000f and AK065000r primers were used to amplify a full-CDS cDNA with the 5′ and 3′ PCR cDNA fragments as template in overlapping PCR. A full-length product was cloned into the EcoRV site of pBlueScript II SK+ (Stratagene, La Jolla, CA, U.S.A.) and sequenced.
Protein sequence alignments were done with the ClustalX implementation of ClustalW [27,28] and manually adjusted with the Gendoc program [29]. In order to properly align the C. thermocellum cellulase sequences with those of the Exgs, a structural alignment was made between the C. albicans Exg (1CZ1) and C. thermocellum cellulase (CEC1) structures with Pymol [30], examining them to make sure the β-strands and α-helices of the central β/α-barrel were properly aligned. Protein analyses were done at the Expasy proteomics server (http://www.expasy.org/), and the signal sequence and cellular location were predicted with SignalP [31] and PSORT [32] respectively.
Recombinant protein expression in E. coli
The cDNA encoding the predicted mature protein of the GH5BG was PCR-amplified with the cloned full-length cDNA as the template, the AAM08614matNcoIf (5′-CACCATGG TCTCCGATGGGAGGACG-3′) and AAM08614XhoIstopr (5′-CCCTCGAGCTAGCTTTTGAGAGAGATGATCC-3′) primers and Pfu DNA polymerase to introduce an NcoI site at the 5′ end and an XhoI site at the 3′ end. The amplification was done as described above, but with a 45 °C annealing temperature. The cDNA product was digested with NcoI and XhoI, cloned into pENTR4 Gateway entry vector that had been digested with the same restriction enzymes, and subcloned into the pET32a+/DEST Gateway expression vector [33] by LR Clonase recombination by the recommended protocol (Invitrogen) and thoroughly sequenced. The recombinant pET32a+/DEST-GB5BG plasmid was transformed into E. coli strain OrigamiB (DE3) [34], and positive clones were selected on LB (Luria–Bertani) agar containing 15 μg/ml kanamycin, 12.5 μg/ml tetracycline and 50 μg/ml ampicillin.
To produce the protein, selected clones were grown in the selection media at 37 °C until the attenuance (D600) reached 0.5–0.6. They were then induced with 0.5 mM IPTG (isopropyl β-D-thiogalactoside) at 20 °C for 12 h. Induced cultures were harvested by centrifugation at 3000 g at 4 °C for 10 min. The cell pellets were resuspended in freshly prepared extraction buffer (50 mM sodium phosphate, pH 8.0, 200 μg/ml lysozyme, 1% Triton X-100, 1 mM PMSF and 40 μg/ml DNase I) and incubated at room temperature (25 °C) for 30 min. The soluble protein was recovered by centrifugation at 12000 g at 4 °C for 10 min. The expressed thioredoxin–GH5BG fusion protein was purified by IMAC (immobilized-metal affinity chromatography) with BD TALON™ cobalt resin according to the manufacturer's instructions (Clontech, Palo Alto, CA, U.S.A.). The fractions with pNPG (p-nitrophenyl β-D-glucopyranoside) hydrolysis activity were pooled and concentrated with 10-kDa-cut-off centrifugal-ultrafiltration membranes (YM-10; Amicon, Beverly, MA, U.S.A.). All of the protein samples were analysed by SDS/PAGE using standard methods [35].
Enzyme assays and kinetic analysis
Kinetic parameters were calculated from triplicate assays using five to seven substrate concentrations at 37 °C in 50 mM sodium acetate, pH 5.0. The purified thioredoxin–GH5BG recombinant protein was tested against pNP (p-nitrophenyl) derivatives of monosaccharides and cellobioside to determine sugar specificity. In a 100 μl reaction assay volume, 1.47–2.94 pmol of enzyme was incubated with substrate in 50 mM sodium acetate, pH 5.0, at 37 °C, except for the assay with pNP β-D-cellobioside, in which 29.4 pmol of enzyme was used. At the end of the reaction time, 70 μl of 0.4 M sodium carbonate was added to stop the reaction, and the absorbance of the liberated pNP was measured at 405 nm. The enzyme was tested with oligosaccharides including cello-oligosaccharides with DP (degree of polymerization) values of 2–6, laminarioligosaccharides with DPs of 2–5 and gentiobiose. In a 50 μl reaction volume, 0.74 pmol of enzyme was incubated with substrate in 50 mM sodium acetate, pH 5.0, for 5 min at 37 °C, except for the assay with cellobiose, in which 14.7 pmol of enzyme was used. The reactions were stopped by boiling, and the glucose released was quantified by the PGO (peroxidase/glucose oxidase) assay method [36,37]. The products of GH5BG hydrolysis of cello- and laminari-oligosaccharides were detected by TLC. In a 50 μl reaction mixture, 7.4 pmol enzyme was incubated with 5 mM substrate in 50 mM sodium acetate, pH 5.0, for 30 min at 37 °C. A 5 μl sample of the reaction mixture was spotted on silica-gel 60 F254 plates (Merck, Darmstadt, Germany) and chromatographed vertically with solvent consisting of ethyl acetate, acetic acid and water (2:1:1, by vol.). The products were detected by spraying with developer solution (ethanolic 10% H2SO4) and baked at 120 °C for 5 min to visualize the sugar.
The enzyme was also tested for hydrolysis of polysaccharides. In the assay, 1–5 μg of enzyme was incubated separately with 0.5% (w/v) laminarin and barley (Hordeum vulgare) β-glucans in 50 mM sodium acetate, pH 5.0, at 37 °C for 30–60 min. The reaction was stopped by the addition of p-hydroxybenzoic acid hydrazide reagent, and the increase in reducing sugars was measured colorimetrically as described by Lever [38]. Protein assays were performed using the Bio-Rad (Richmond, CA, U.S.A.) protein assay kit using BSA as a standard. The kinetic parameters Km and Vmax (at pH 5.0 and 37 °C) were calculated by linear regression of Lineweaver–Burk plots with the Enzfitter (Elsevier Biosoft, Cambridge, U.K.) computer program. Note that the activity values for disaccharides were determined by dividing the amount of glucose released by 2, since two glucose molecules are released per molecule of disaccharide hydrolysed. The amount of product for the oligosaccharide substrates is given in terms of total glucose released, since release of more than one glucose per substrate molecule due to sequential cleavage should be negligible for V0.
The pH optimum was determined by measuring the release of pNP from pNPG in different 50 mM buffers ranging in pH from 3.5 to 10 in 0.5 pH unit increments for 10 min {formate, pH 3.5–4.5; sodium acetate, pH 4.0–5.5; sodium phosphate, pH 5.5–8; Tris, pH 7.5–9.0; Caps [3-(cyclohexylamino)propane-1-sulfonic acid], pH 9.0–10}.
Northern-blot analysis
GH5BG gene-specific probe was amplified using rice genomic DNA as the template with the AAM08614_Cterf (5′-GAATGTGCAGGGAGCATC-3′) and AAM08614_3UTRr (5′-CTTTAATTCAGCTTCAC-3′) primers derived from the C-terminal part of the CDS and 3′-untranslated region of the gene respectively. The rice 18 S rRNA probe (342 bp) was PCR-amplified using first-strand cDNA synthesized from RNA extracted from 6-day-old rice seedlings as template with the 18 S rice forward (5′-AAGTTTGAGGCAATAACAG-3′) and reverse (5′-CCTCTAAATGATAAGGTTC-3′) primers derived from the GenBank accession number AF069218 sequence. The amplification for both probes was done with Taq DNA polymerase (Roche Diagnostics, Indianapolis, IN, U.S.A.).
Total RNA was isolated from different parts of rice (cv. Yukihikari) plants at various developmental stages and environmental conditions by the method of Bachem et al. [39]. A 20 μg portion of total RNA from each sample was denatured and electrophoresed on a formaldehyde/1.2%-agarose gel and transferred on to Hybond N+ nylon membrane (Amersham-Pharmacia, Uppsala, Sweden) by standard procedures [34]. RNA blots were hybridized separately with the [α-32P]dCTP-labelled gene-specific probe for GH5BG (396 bp), and 18 S rRNA (342 bp) for 16 h at 65 °C. The blots were then washed once in 0.1% SDS/2×SSC for 20 min at 65 °C and twice in 0.1% SDS/0.1×SSC for 20 min at 65 °C (1×SSC is 0.15 M NaCl/0.015 M sodium citrate). The membranes were then exposed to X-ray film for signal detection for 4–6 days at −80 °C.
Construction of GUS-fusion plasmid and particle bombardment of onion cells
The cDNA encoding full-length protein of GH5BG was PCR-amplified with the cloned full-length cDNA as the template, AAM08614 full attB forward primer (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGCGCCATTTTGAGCTCCTC-3′) and AAM08614 full attB reverse primer (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTAGCTTTTGAGAGAGATGATCC-3′) and Pfu DNA polymerase to introduce an attB1 and attB2 site at the 5′ end and 3′ end respectively. The PCR product was clone into the pDONR-Zeo vector with BP clonase according to the Gateway Technology (Invitrogen) protocol. The recombination reaction was transformed into DH5α E. coli, and positive clones were selected with 50 μg/ml zeocin LB agar plates in the dark. The GH5BG cDNA was subcloned from the recombinant pDONR-Zeo-GH5BG into pMDC139 (provided by Dr Mark Curtis, Institute of Plant Biology and Zürich-Basel Plant Science Centre, University of Zürich, Zürich, Switzerland [40]) by LR clonase recombination (Invitrogen).
The resulting pMDC139-GH5BG constructs were delivered into epidermal layers of onion bulbs by particle bombardment with a Gene Gun Machine PDA-1000/He (Bio-Rad). After bombardment, the transformed cells were incubated at 25 °C for 48 h in complete darkness. After that, the epidermal layers were stained overnight with X-Glu (5-bromo-4-chloroindol-3-yl β-glucuronide) at 37 °C and the blue colour product derived from the hydrolysis of X-Glu by β-glucuronidase was observed using optical microscopy.
RESULT AND DISCUSSION
GH5BG cDNA cloning and sequence analysis
With the completion of high-quality drafts of the rice genome, analysis of GH5 members in rice has been reported in the CAZY homepage (http://www.cazy.org/CAZY/). A total of 20 GH5 genes putatively encoding seven cellulases, nine endo-β-mannanases, three glucan 1,3-β-glucosidases and one 1,3-β-glucanase have been identified in the rice databases (see CAZY). The putative glucan 1,3-β-glucosidases encoded by these genes include Genbank accession numbers (AC) AAM08614, AAM08620, and AAV43969. A BLAST comparison of AAM08614 with the others showed that AAM08620 contains three repeats of homologous sequences, each of which has 71% identity with AAM08614. The AAV43969 sequence is 69% identical with the AAM08614 sequence. The amino acid sequence of the putative 1,3-β-glucanase BAD10703 is also 49% identical with AAM08620, which has only 28–33% identity with the glucan-1,3-β-glucosidases of fungi, so BAD10703 is more closely related to AAM08620, despite its different annotation. Therefore there appear to be four putative rice glucan 1,3-β-glucanase genes, of which AAM08614 was chosen for investigation.
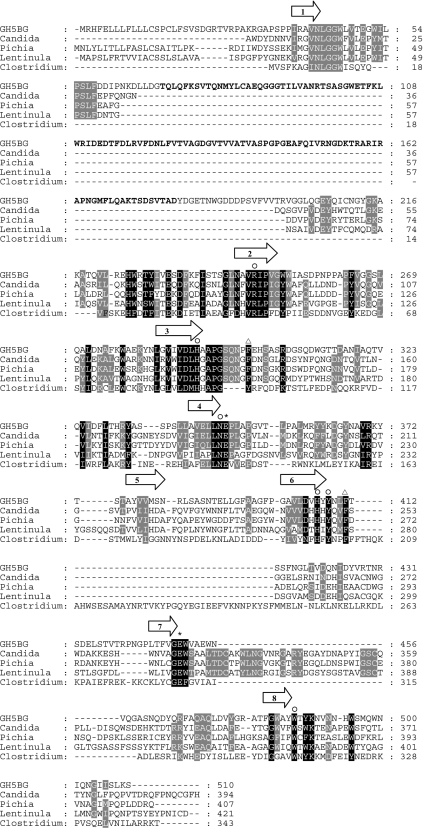
A GH5 glucan-1,3-β-glucosidase cDNA, designated GH5BG, was cloned from rice seedlings by reverse-transcription PCR with KDML105 rice seedling RNA and primers derived from the GenBank accession number AC107314 (rice genomic contig from which AAM08614 is derived) and AK065000 (full-length cDNA [26]) sequences. A specific PCR product of 1680 bp was produced, and its sequence overlapped those of AC107314 and AK065000. The full-length cDNA sequence contains a 1530-nucleotide open reading frame encoding a 510-amino-acid precursor protein. The protein sequence was predicted to contain a 19-amino-acid-long prepeptide and a 491-amino-acid-long mature protein and to be secreted out of the cell. Its predicted pI is 5.28. The mature protein includes two domains, a fascin-like domain (amino acids 70–180) and a glucan-1,3-β-glucosidase domain (amino acids 37–60 and 208–496) (Figure 1). The fascin-like domain found at the N-terminus of this enzyme is not commonly found in plant enzymes, but it aligned well with the N-terminus of fascin found in a Hawaiian sea urchin (Tripneustes gratilla), the fruitfly Drosophilia, the South African clawed frog Xenopus, mouse and human [41–45]. A BLAST search analysis in GenBank® revealed that the fascin-like domain was found in five plant enzymes, at the N-terminus of three GH5 proteins from Medicago truncatula (annual barrel medic) (AC ABP02833, ABP03047 and ABP03050), a Vitus vinifera (the common grape vine) hypothetical protein fragment (CAN78586) and the previously described putative rice GH5 glucan-1,3-β-glucosidases AC AAM08614 (GH5BG), AAM08620, AAV43969 and BAD10703. These proteins all displayed highest similarity to each other (36–72% identity), suggesting they are all part of the same subgroup of GH5. In human, fascin contributes to the bundle of F-actin [46]. However, this action requires a cytoplasmic location, whereas GH5BG is predicted to be extracellular, so it is unlikely to interact with intracellular actin molecules. In fact, all the plant sequences with the fascin-like domain, except AAM08620, contain signal sequences for secretion, so the fascin-like domain may have been adapted for binding other molecules or to bind actin in the case of cell lysis.
Figure 1. Alignment of the protein sequence of rice GH5BG with exo-β-1,3-glucanases and endo-β-1,4-glucanase.
GH5BG is rice GH5BG, Candida is exo-β-(1,3)-glucanase from Candida albicans (AC CAA39908), Lentinula is exo-β-(1,3)-glucanase from Lentinula edodes (AC AB192344), Pichia is exo-β-(1,3)-glucanase from Pichia pastoris (AC AY954499), and Clostridium is endo-β-(1,4)-glucanase from Clostridium thermocellum (AC AAA23220). The alignment was generated with the ClustalX implementation of ClustalW [27,28] and analysed and manually adjusted with Gendoc [29]. Alignment of the C. thermocellum sequence relied on the structural alignment of the 1CEC structural model with the C. albicans Exg 1CZ1 structure. The positions of the β-strands of the central (β/α)8 barrel are indicated by arrows above the alignment. The asterisks identify the two catalytic glutamate residues, the invariant GH family 5 residues are marked by the symbol ○ above the column, and the black and grey shadings highlight other identities between sequences. The two phenylalanine residues found at the +1 subsite of C. albicans Exg are marked by triangles above the column. The region of rice GH5BG homologous with fascin is indicated by bold text.
Functional expression of recombinant GH5BG and substrate specificity
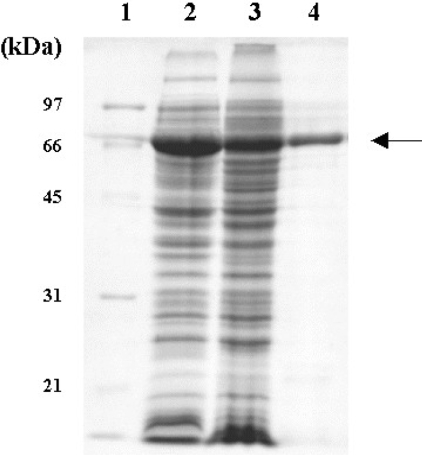
The GH5BG cDNA was expressed in redox-deficient E. coli Origami (DE3) as a catalytically active thioredoxin fusion protein. Induced cultures of Origami (DE3) expressing GH5BG thioredoxin fusion proteins had intense bands at 68 kDa in total cell lysates on SDS/PAGE (Figure 2). The fusion protein was purified and found to hydrolyse pNPG with an optimal pH of 5.0. The activity of the purified GH5BG towards artificial glycosides and oligosaccharides is summarized in Table 1, and Figure 3 shows the cleavage of disaccharides and oligosaccharides by GH5BG as indicated by TLC of the products. GH5BG hydrolysed pNP glycosides of the monosaccharides, β-D-glucoside, β-D-fucoside, β-D-galactoside, β-D-xyloside and α-L-arabinoside, but was unable to hydrolyse pNP β-D-mannoside. Among pNP glycosides, pNP β-D-fucoside was hydrolysed twice as efficiently as pNPG (on the basis of kcat/Km values), whereas pNP β-D-galactoside and pNP α-L-arabinoside were hydrolysed with 27%, and pNP β-D-xyloside with 15%, of the efficiency of pNPG hydrolysis. These results indicate there is low stringency at the −1 subsite of GH5BG, where the non-reducing glycosyl moiety is bound, which is somewhat similar to the situation in many GH1 and GH3 β-glucosidases such as rice BGlu1 [37,47]. The enzyme could hydrolyse glycosides of β-D-glucose, β-D-fucose, β-D-galactose, β-D-xylose and α-L-arabinoside, which are epimers with equatorial OH-2, but not β-D-mannoside, which has an axial projection at OH-2. Therefore, the epimerization of OH-2 is critical for binding of the sugar residue to the −1 subsite. However, the equatorial or axial projection at OH-4 and the conversion of CH2OH-6 of D-glucose into H- in D-xylose or CH3- in D-fucose are not critical to the capacity of the substrates to bind to the active site. The specificity for epimerization at OH-2 may reflect steric hindrance of the axial projection or the geometry of Asn346 and the catalytic nucleophile, which hydrogen-bond to this position in β-glucoside substrates, but not in β-mannosides, where they are nonetheless important for stabilization of the transition state [9,16,48].
Figure 2. SDS/PAGE of GH5BG–thioredoxin fusion protein expressed in E. coli strain Origami B (DE3) after incubation in the presence of 0.5 mM IPTG at 20 °C for 12 h.
Lanes: 1, standard marker (Bio-Rad); 2, total protein of E. coli cells containing pET32a(+)/DEST-GH5BG; 3, soluble fraction of E. coli cells containing pET32a(+)/DEST-GH5BG; 4, purified thioredoxin–GH5BG. The arrow points to the thioredoxin–GH5BG.
Table 1. Kinetic parameters of rice GH5BG in the hydrolysis of pNP glycosides, disaccharides and oligosaccharides.
Insufficient activity was detected to enable parameters to be determined for the pNP glycoside pNP β-D-mannoside or the oligosaccharides laminaritriose, laminaritetraose, laminaripentaose, laminarin and barley (1→3),(1→4)-β-glucans.
| Substrate | kcat (s−1) | Km (mM) | kcat/Km (s−1·mM−1) |
|---|---|---|---|
| pNP glycosides | |||
| pNP β-D-glucoside | 36.1±0.7 | 0.47±0.03 | 77±4 |
| pNP β- D-fucoside | 24.5±0.5 | 0.17±0.07 | 144±3 |
| pNP β-D-galactoside | 27±3 | 1.30±0.10 | 20.7±0.5 |
| pNP β-D-xyloside | 3.2±0.3 | 0.27±0.05 | 11.9±1.5 |
| pNP α-L-arabinoside | 2.88±0.08 | 0.14±0.02 | 21±3 |
| pNP β-D-cellobioside | 2.07±0.09 | 6.23±0.17 | 0.34±0.01 |
| Disaccharides | |||
| Cellobiose | 4.3±0.8 | 16.4±1.9 | 0.27±0.02 |
| Laminaribiose | 36±5 | 7.0±1.1 | 5.05±0.07 |
| Oligosaccharides | |||
| Cellotriose | 41±5 | 4.53±0.01 | 9.1±1.2 |
| Cellotetraose | 38±2 | 4.09±0.17 | 9.3±0.9 |
| Cellopentaose | 35.5±0.4 | 3.4±0.4 | 10.4±0.4 |
| Cellohexaose | 9.7±0.8 | 2.2±0.5 | 4.5±0.5 |
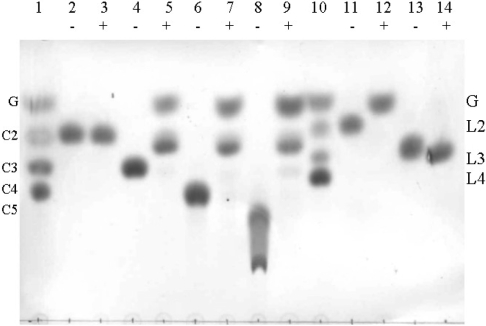
Figure 3. Hydrolysis of disaccharides and oligosaccharide substrates by thioredoxin–GH5BG detected by TLC.
The thioredoxin–GH5BG was incubated with 5 mM substrates for 30 min and the products were detected by the carbohydrate staining method described in the Experimental section. Samples were incubated with (+) or without (−) enzyme in 50 mM sodium acetate, pH 5.0, for 30 min at 37 °C prior to being spotted on silica-gel 60 F254 TLC plates, developed and charred with 10% H2SO4 in ethyl alcohol. Lanes: 1, glucose (G) and cello-oligosaccharides of DP 2–4 (C2–C4) marker; 2 and 3, cellobiose; 4 and 5, cellotriose; 6 and 7, cellotetraose; 8 and 9, cellopentaose; 10, standard laminari-oligosaccharides of DP 2–4 (L2–L4); 11 and 12, laminaribiose; 13 and 14, laminaritriose.
For disaccharides, GH5BG hydrolysed laminaribiose (β-1,3) with a relatively high efficiency of 5.05 mM−1·s−1, but it was 20-fold less efficient with cellobiose (β-1,4). Hydrolysis of gentiobiose (β-1,6-linked) was not detectable. It also hydrolysed cello-oligosaccharides with DP values of 3–6 at relatively high rates, with a catalytic efficiency (kcat/Km) of 9–10 mM−1·s−1 for DP 3–5 and a decrease to half of this value for cellohexaose, owing to a decrease in the kcat for this substrate. On the other hand, GH5BG could not hydrolyse laminari-oligosaccharides with DP values of 3–5, laminarin or barley 1,3-1,4-β-glucans. Having rates of hydrolysis of oligomeric substrates which remain approximately constant or decrease with increasing DP is a characteristic often observed with β-glucosidases, unlike polysaccharide exohydrolases, in which the hydrolytic rate increases with oligosaccharide length [49].
The marked preference for β-1,4-linked oligosaccharides and β-1,3-linked disaccharide of rice GH5BG is different from fungal GH5 exo-β-1,3-glucanases, which prefer to hydrolyse 1,3-glucans (laminarin) [9,24,25]. This difference in substrate specificity must be the result of differences in the structures and/or positions of amino acid residues in the active site between rice GH5BG and the fungal enzymes. The geometry of the pocket of C. albicans Exg allows hydrolysis of longer β-1,3-linked oligosaccharides and is not well suited to the cleavage of 1,4-glycosidic linkages [9]. By contrast, GH5BG exhibited a marked preference for β-1,4-linked oligosaccharides and cannot hydrolyse extended β-1,3-linked oligosaccharides. Thus the geometry of the pocket may be quite different from that of C. albicans Exg.
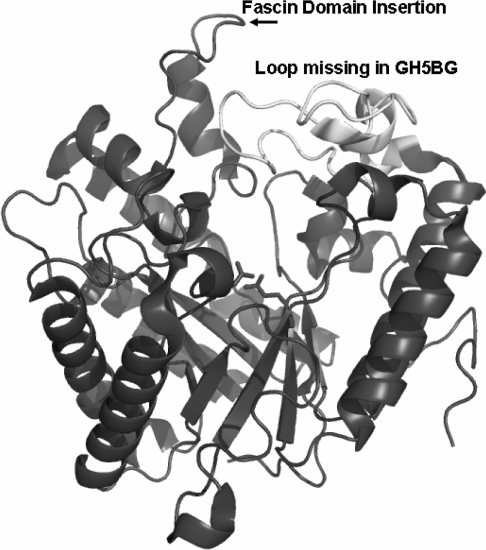
The sequence alignment between GH5BG with C. albicans Exg and a GH5 endo-1,4-β-glucanase (cellulase) from C. thermocellum [18] in Figure 1 shows that, although rice GH5BG is somewhat more similar to C. albicans Exg, it shares some features more similar to those of the cellulase. In addition to being more similar at some residues near the active site, such as Tyr407, Asn409 and Tyr488, it lacks the extended loop at the end of strand 7 of the β-barrel, which forms a wall along one side of the active site in the fungal exoglucanases, as shown in Figure 4. This loop is partially replaced by an extended loop at the end of β-strand 6 in the C. thermocellum cellulase, but its absence contributes to the more groove-like active-site cleft in the cellulase. Rice GH5BG lacks both these loops, which might result in a more open active site, allowing it to accept β-1,4-linked glucan oligosaccharides as well as laminaribiose. However, as shown in Figures 1 and 4, the extended loop into which the fascin-like domain is inserted at the end of strand 1 of the β-barrel is adjacent to these missing loops, so the loop and fascin-like domain may replace these loops in the wall of the active site. Although homology modelling can place the fascin-domain-containing loop at this location, the structural effect it has on the active site cannot be determined accurately without structural data. Nevertheless, the unique structure of the rice GH5BG and the related rice exoglucanase-like genes, along with GH5BG's distinct substrate preferences, suggest that GH5BG and its closely related plant homologues could be considered a separate subfamily of GH5 distinct from the exo-β-(1,3)-glucanases, to which they are most closely related.
Figure 4. Active site of Candida albicans Exg structural model with differences in the loops around the active site found in rice GH5BG highlighted.
The 1CZ1 structure [9] is shown as a ribbon diagram coloured dark grey, with the loop after β-strand 7 of the (β/α)8 barrel shown in white and labelled to draw attention to its absence in rice GH5BG. The insertion of the fascin-like domain after the first helix of the extended loop after strand 1 of the β-barrel is indicated by the label. The catalytic acid/base (left) and catalytic nucleophile (right) are displayed in stick representation to indicate the location of the active site. The image was generated with Pymol [30].
Comparison of the deduced amino acid sequence of rice GH5BG with those of fungal GH5 exoglucanases revealed that Glu-347, which lies in the conserved NEP motif, is likely to be the catalytic acid/base and Glu-450, which lies in the conserved GEW motif, is likely the catalytic nucleophile [9,24,25]. Similar to other GH5 members, rice GH5BG contains eight invariant residues, these being Arg247, His291, Asn346, Glu347, His406, Tyr408, Glu450 and Trp486. These residues contribute hydrogen-bond interactions to the non-reducing terminal sugar residue at the −1 subsite found in C. albicans Exg. Since these residues are conserved, the geometry of the −1 subsite is not likely to account for the differences in substrate specificity between GH5BG and the fungal enzymes. However, differences are seen at the +1 subsite, in which amino acid residues Trp229, Leu304 and Asn305 of C. albicans Exg are not conserved in GH5BG. C. albicans Exg residues Leu304 and Asn305 are located in the extended loop after strand 7 of the β-barrel and surround Glu292 (the catalytic nucleophile) together with Ala296, Asp299 and Gly306 [19]. These residues are conserved among fungal GH5 exo-β-(1,3)-glucanases and many GH5 members with this extended loop, but not GH5BG. However, GH5BG does contain Phe300 and Phe411, corresponding to Phe144 and Phe258 in C. albicans Exg, which were found to be located at the +1 subsite near the entrance to the active-site pocket [9]. Indeed, these residues are found in the same position when a homology model of GH5BG was built with the C. albicans Exg structure (1CZX) as template. Cutfield et al. [9] suggested that the role of the aromatic side chains of these two phenylalanine residues is to direct substrates into the pocket and that it acts as a clamp for acceptor molecules participating in transfer reactions. The geometry of the +1 substite might have a high stringency requirement for the stereochemistry of the linkage and the orientation of a second sugar. Hrmova et al. [50] suggested that the narrow phenyl groups would restrain the position of the second sugar in productive binding for hydrolysis more than the larger tryptophan indole rings found in a similar position with respect to the active site in barley GH family 3 exoglucanase, which can accept β-1,3- and β-1,4- linkages. If the orientation of the non-reducing glucose residue in the −1 site is the same in each case, the orientation of the sugar in the +1 site should be different, depending on whether it is linked through its O-3, O-4 or O-6. So, the β-1,4- and β-1,6- linked disaccharides, and longer β-1,3-linked oligosaccharides, may not maintain proper binding geometry.
However, this does not explain how the rice GH5BG can hydrolyse longer β-1,4-oligosaccharides and 1,3-linked disaccharides. The fact that the cellotriose is hydrolysed more efficiently than cellobiose implies that GH5BG has three subsites for binding β-1,4-linked glucosyl residues in the active site. Although GH5BG was designated a putative glucan exo-β-(1,3)-glucosidase on the basis of its sequence homology, its catalytic activity is somewhat like that of GH1 β-glucosidases, which show similar oligosaccharide preferences. Rice BGlu1 GH1 exoglucanase/β-glucosidase [47] and rice Os4bglu12 GH1 β-glucosidase [33] are enzymes that prefer to hydrolyse β-1,4-linked oligosaccharides and a broad range of pNP glycosides, but with differences in kcat/Km compared with GH5BG. Similarly to rice BGlu1, GH5BG cleaved the β-glucosidic bond between the two glucose residues in pNP β-D-cellobioside, thereby releasing glucose and pNPG, which was then rapidly hydrolysed (results not shown), but the kcat/Km of GH5BG for hydrolysis of pNP β-D-cellobioside is about ten times lower than that of rice BGlu1 [37,47]. Although GH5BG could hydrolyse both cellotriose and pNP β-D-cellobioside, it hydrolysed pNP β-D-cellobioside about 26 times less efficiently than cellotriose. This suggests that the pNP β-D-cellobioside, unlike cellotriose, cannot bind well to the third subsite in the active-site cleft of GH5BG.
Expression of GH5BG in rice tissues and in response to environmental conditions
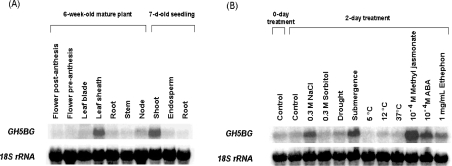
The transcript level of GH5BG was high in the shoot of 7-day-old seedlings and very low in the root and endosperm. In 6-week-old mature plants, GH5BG transcripts were detected at significant levels in leaf sheaths, while a low signal was seen in other mature plant parts (Figure 5).
Figure 5. Northern-blot analysis of GH5BG transcript levels in (A) 7-day-old rice seedlings and 6-week-old mature plant tissues and (B) 7-day-old rice seedlings grown a further 2 days with various abiotic stresses and plant hormones.
GH5BG, RNA blots were probed with α-32P-labelled GH5BG gene-specific probe; 18S rRNA indicates the same blot probed with an α-32P-labelled 18 S rRNA cDNA probe. A 20 μg portion of total RNA from the appropriate tissues was loaded in each lane.
To determine the effects of environmental conditions on rice GH5BG gene expression during seedling growth, the transcript levels of GH5BG were compared between 9-day-old seedlings that had been exposed to various conditions for 2 days and 9-day-old rice seedlings grown at 28 °C (control) (Figure 5). The GH5BG transcript level was up-regulated in response to salt stress, submergence stress, 0.1 mM methyl jasmonate and 10 mM ABA in rice seedlings and increased slightly in response to ethephon. The mRNA transcripts were detected at levels similar to those of the control when the seedlings were treated with sorbitol, drought, cold and heat stresses. The up-regulation of GH5BG in response to various environmental conditions may correlate with a need to recycle cell-wall oligosaccharides in these processes or to the function of other, as yet unidentified, substrates.
Cellular localization of the GH5BG protein
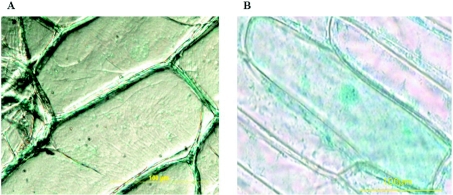
In onion cells transformed with pMDC139-GH5BG-GUS, β-glucuronidase activity was observed outside the cell, whereas the blue-coloured product was diffusely distributed throughout the control cells transformed with control pMDC139 plasmid (Figure 6). So the GH5BG tagged at its C-terminus with GUS reporter (GH5BG-GUS) was secreted to the apoplast (the free diffusional space outside the plasma membrane), which is consistent with its predicted subcellular localization. Together with the preference of GH5BG to hydrolyse cello-oligosaccharides and laminaribiose, its extracellular location implies it may play a role in cell-wall recycling.
Figure 6. Extracellular localization of the GH5BG protein.
The epidermal layers of onion bulbs were transformed with (A) pMDC139-GH5BG and (B) control pMDC139 plasmid without insert by particle bombardment. After 48 h incubation at 25 °C in complete darkness, the transformed onion cells were stained overnight with X-Glu at 37 °C, and the location of blue-coloured product was observed by optical microscopy.
In summary, the cDNA of a putative GH5 glucan-1,3-β-glucosidase containing a fascin-like domain at the N-terminus was cloned from rice seedlings. A recombinant thioredoxin–GH5BG fusion protein produced in E. coli showed high hydrolytic activity toward various kinds of pNP glycosides and exhibited a marked preference for β-(1,4)-linked oligosaccharides and laminaribiose [β-(1,3)-linked disaccharide]. The substrate specificity of GH5BG is different from that of fungal GH5 exo-β-(1,3)-glucanases, which is likely to be due to differences in the structures of the loops and types of amino acids around the active site, indicating that GH5BG, along with three closely related rice enzymes, could be considered a new subfamily of GH5. GH5BG is expressed in rice leaves and seedling shoots, whereas its expression is induced by stress-related hormones, submergence and salt in whole seedlings. The protein appears to be secreted outside the cell, where it may be involved in release of glucose from cell-wall-derived oligosaccharides under these conditions.
Acknowledgments
This work was supported by a grant (MRG4880066) from the Commission on Higher Education and the Thailand Research Fund. Additional support was provided by grant BT-B-06-RG-19-4608 from the National Science and Technology Development Agency of Thailand, the National Center for Genetic Engineering and Biotechnology, Thailand Research Fund grant RTA4780006, Suranaree University of Technology and the National Research Council of Thailand. T. A. was supported by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan for the Green Technology Project.
References
- 1.Henrissat B., Davies G. T. Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 1997;7:637–644. doi: 10.1016/s0959-440x(97)80072-3. [DOI] [PubMed] [Google Scholar]
- 2.Jenkins J., Leggio L. L., Harris G., Pickersgill R. β-Glucosidase, β-galactosidase, family A cellulases, family F xylanases and two barley glycanases form a superfamily of enzymes with 8-fold β/α architecture and with two conserved glutamates near the carboxy-terminal ends of β-strands four and seven. FEBS Lett. 1995;362:281–285. doi: 10.1016/0014-5793(95)00252-5. [DOI] [PubMed] [Google Scholar]
- 3.Henrissat B., Callebaut I., Fabrega S., Lehn P., Mornon J.-P., Davies G. Conserved catalytic machinery and the prediction of a common fold for several families of glycosyl hydrolases. Proc. Natl. Acad. Sci. U.S.A. 1995;92:7090–7094. doi: 10.1073/pnas.92.15.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coutinho P. M., Henrissat B. Carbohydrate-active enzymes: an integrated database approach. In: Gilbert H. J., Davies G., Henrissat B., Svensson B., editors. Recent Advances in Carbohydrate Bioengineering. Cambridge: Royal Society of Chemistry; 1999. pp. 3–12. [Google Scholar]
- 5.Gilkes N. R., Henrissat B., Kilburn D. G., Miller R. C., Jr, Warren R. A. J. Domains in microbial β-1,4-glycanases: sequence conservation, function, and enzyme families. Microbiol. Rev. 1991;55:303–315. doi: 10.1128/mr.55.2.303-315.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsai C. F., Qiu X., Liu J. H. A comparative analysis of two cDNA clones of the cellulase gene family from anaerobic fungus Piromyces rhizinflata. Anaerobe. 2003;9:131–140. doi: 10.1016/S1075-9964(03)00087-8. [DOI] [PubMed] [Google Scholar]
- 7.Tanabe T., Morinaga K., Fukamizo T., Mitsutomi M. Novel chitosanase from Streptomyces griseus HUT 6037 with transglycosylation activity. Biosci. Biotechnol. Biochem. 2003;67:354–364. doi: 10.1271/bbb.67.354. [DOI] [PubMed] [Google Scholar]
- 8.Reinhold-Hurek B., Maes T., Gemmer S., van Montagu M., Hurek T. An endoglucanase is involved in infection of rice roots by the non-cellulose-metabolizing endophyte Azoarcus sp. strain BH72. Mol. Plant Microbe Interact. 2006;19:181–188. doi: 10.1094/MPMI-19-0181. [DOI] [PubMed] [Google Scholar]
- 9.Cutfield S. M., Davies G. T., Murshudov G., Anderson B. F., Moody P. C., Sullivan P. A., Cutfield J. F. The structure of the exo-β-1,3-glucanase from Candida albicans in native and bound forms: relationship between a pocket and groove in family 5 glycosyl hydrolases. J. Mol. Biol. 1999;294:771–783. doi: 10.1006/jmbi.1999.3287. [DOI] [PubMed] [Google Scholar]
- 10.Mitreva-Dautova M., Roze E., Overmars H., Graaff L., Schots A., Helder J., Goverse A., Bakker J., Smant G. A symbiont-independent endo-1,4-β-xylanase from the plant-parasitic nematode Meloidogyne incognita. Mol. Plant Microbe Interact. 2006;19:521–529. doi: 10.1094/MPMI-19-0521. [DOI] [PubMed] [Google Scholar]
- 11.Perret S., Bélaich A., Fierobe H.-P., Bélaich J.-P., Tardif C. Towards designer cellulosomes in Clostridia: mannanase enrichment of the cellulosomes produced by Clostridium cellulolyticum. Bacteriology. 2004;186:6544–6552. doi: 10.1128/JB.186.19.6544-6552.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caines M. E., Vaughan M. D., Tarling C. A., Hancock S. M., Warren R. A., Withers S. G., Strynadka N. C. Structural and mechanistic analyses of endo-glycoceramidase II, a membrane-associated family 5 glycosidase in the apo and GM3 ganglioside-bound forms. J. Biol. Chem. 2007;282:14300–14308. doi: 10.1074/jbc.M611455200. [DOI] [PubMed] [Google Scholar]
- 13.Hrmova M., Burton R. A., Biely P., Lahnstein J., Fincher G. B. Hydrolysis of (1,4)-β-D-mannans in barley (Hordeum vulgare L.) is mediated by the concerted action of (1,4)-β-D-mannan endohydrolase and β-D-mannosidase. Biochem. J. 2006;399:77–90. doi: 10.1042/BJ20060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Béguin P. Molecular biology of cellulose degradation. Annu. Rev. Microbiol. 1990;44:219–248. doi: 10.1146/annurev.mi.44.100190.001251. [DOI] [PubMed] [Google Scholar]
- 15.Lo Leggio L., Parry N. J., van Beeumen J., Claeyssens M., Bhat M. K., Pickersgill R. W. Crystallization and preliminary X-ray analysis of the major endoglucanase from Thermoascus aurantiacus. Acta Crystallogr. D. 1997;53:599–604. doi: 10.1107/S0907444997005404. [DOI] [PubMed] [Google Scholar]
- 16.Hilge M., Gloor S. M., Rypniewski W., Sauer O., Heightman T. D., Zimmermann W., Winterhalter K., Piontek K. High-resolution native and complex structures of thermostable β-mannanase from Thermomonospora fusca substrate specificity in glycosyl hydrolase family 5. Structure. 1998;6:1433–1444. doi: 10.1016/s0969-2126(98)00142-7. [DOI] [PubMed] [Google Scholar]
- 17.Sakon J., Adney W. S., Himmel M. E., Thomas S. R., Karplus P. A. Crystal structure of thermostable family 5 endocellulase E1 from Acidothermus cellulolyticus in complex with cellotetraose. Biochemistry. 1996;35:10648–10660. doi: 10.1021/bi9604439. [DOI] [PubMed] [Google Scholar]
- 18.Dominguez R., Souchon H., Spinelli S., Dauter Z., Wilson K. S., Chauvaux S., Beguin P., Alzari P. M. A common fold and similar active site in two distinct families of β-glycanases. Nature Struct. Biol. 1995;2:569–576. doi: 10.1038/nsb0795-569. [DOI] [PubMed] [Google Scholar]
- 19.Mackenzie L. F., Brooke G. S., Cutfield J. F., Sullivan P. A., Withers S. G. Identification of Glu-330, as the catalytic nucleophile of Candida albicans exo-β-glucanase. J. Biol. Chem. 1997;272:3161–3167. doi: 10.1074/jbc.272.6.3161. [DOI] [PubMed] [Google Scholar]
- 20.Stubbs H. J., Brasch D. J., Emerson G. W., Sullivan P. A. Hydrolase and transferase activities of the β-1,3-exoglucanase of Candida albicans. Eur. J. Biochem. 1999;263:889–895. doi: 10.1046/j.1432-1327.1999.00581.x. [DOI] [PubMed] [Google Scholar]
- 21.Chambers R. S., Walden A. R., Brooke G. S., Cutfield J. F., Sullivan P. A. Identification of a putative active site residue in the exo-β-(1,3)-glucanase of Candida albicans. FEBS Lett. 1993;327:366–369. doi: 10.1016/0014-5793(93)81022-r. [DOI] [PubMed] [Google Scholar]
- 22.Vazquez de Aldana C. R., Correa J., San Segundo P., Bueno A., Nebreda A. R., Mendez E., del Rey F. Nucleotide sequence of the exo-1,3-β-glucanase-encoding gene, EXG1, of the yeast Saccharomyces cerevisiae. Gene. 1991;97:173–182. doi: 10.1016/0378-1119(91)90049-h. [DOI] [PubMed] [Google Scholar]
- 23.van de Rhee M. D., Mendes O., Werten M. W., Huizing H. J., Mooibroek H. High efficient homologous integration via tandem exo-β-1,3-glucanase genes in the common mushroom, Agaricus bisporus. Curr. Genet. 1996;30:166–173. doi: 10.1007/s002940050116. [DOI] [PubMed] [Google Scholar]
- 24.Sakamoto Y., Irie T., Sato T. Isolation and characterization of a fruit body-specific exo-β-1,3-glucanase-encoding gene, exg1, from Lentinula edodes. Curr. Genet. 2005;47:244–252. doi: 10.1007/s00294-005-0563-7. [DOI] [PubMed] [Google Scholar]
- 25.Xu Z., Shih M., Poulton J. E. An extracellular exo-β-(1,3)-glucanase from Pichia pastoris: purification, characterization, molecular cloning and functional expression. Protein Expression Purif. 2006;47:118–127. doi: 10.1016/j.pep.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 26.Kikuchi S., Satoh K., Nagata T., Kawagashira N., Doi K., Kishimoto N., Yazaki J., Ishikawa M., Yamada H., Ooka H., et al. Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science. 2003;301:376–379. doi: 10.1126/science.1081288. [DOI] [PubMed] [Google Scholar]
- 27.Jeanmougin F., Thompson J. D., Gouy M., Higgins D. G., Gibson T. J. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- 28.Thompson J. D., Higgins D. G., Gibson T. J. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicholas K. B., Nicholas H. B. Genedoc: a tool for editing and annotating multiple sequence alignments ( http://www.psc.edu/biomed/gendoc) 1997
- 30.DeLano W. L. The PyMOL Molecular Graphics SystemDeLano Scientific, Palo Alto, CA, U.S.A, ( http://www.pymol.org) 2002
- 31.Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 32.Nakai K., Horton P. PSORT: a program for detecting the sorting signals of proteins and predicting their subcellular localization. Trends Biochem. Sci. 1999;24:34–35. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- 33.Opassiri R., Pomthong B., Onkoksoong T., Akiyama T., Esen A., Ketudat Cairns J. R. Analysis of rice glycosyl hydrolase family 1 and expression of Os4bglu12 β-glucosidase. BMC Plant Biol. 2006;6:33. doi: 10.1186/1471-2229-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook J., Fritsch E. F., Maniatis T. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 35.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 36.Babcock G. W., Esen A. Substrate specificity of maize β-glucosidase. Plant Sci. 1994;101:31–39. [Google Scholar]
- 37.Opassiri R., Ketudat Cairns J. R., Akiyama T., Wara-Aswapati O., Svasti J., Esen A. Characterization of a rice β-glucosidase highly expressed in flower and germinating shoot. Plant Sci. 2003;165:627–638. [Google Scholar]
- 38.Lever M. A new reaction for colorimetric determination of carbohydrates. Anal. Chem. 1972;47:273–279. doi: 10.1016/0003-2697(72)90301-6. [DOI] [PubMed] [Google Scholar]
- 39.Bachem C. W., van der Hoeven R. S., de Bruijn S. M., Vreugdenhil D., Zabeau M., Visser R. G. Visualization of differential gene expression using a novel method of RNA fingerprinting based on AFLP: analysis of gene expression during potato tuber development. Plant J. 1996;9:745–753. doi: 10.1046/j.1365-313x.1996.9050745.x. [DOI] [PubMed] [Google Scholar]
- 40.Curtis M. D., Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kane R. E. Actin polymerization and interaction with other proteins in temperature-induced gelation of sea urchin egg extracts. J. Cell Biol. 1976;171:704–714. doi: 10.1083/jcb.71.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paterson J., O'Hare K. Structure and transcription of the signed locus of Drosophila melanogaster. Genetics. 1991;129:1073–1084. doi: 10.1093/genetics/129.4.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holthuis J. C., Schoonderwoert V. T., Martens G. J. A vertebrate homolog of the actin-bundling protein fascin. Biochim. Biophys. Acta. 1994;1219:184–188. doi: 10.1016/0167-4781(94)90267-4. [DOI] [PubMed] [Google Scholar]
- 44.Edwards R. A., Herrera-Sosa H., Otto J., Bryan J. Cloning and expression of a murine fascin homolog from mouse brain. J. Biol. Chem. 1995;270:10764–10770. doi: 10.1074/jbc.270.18.10764. [DOI] [PubMed] [Google Scholar]
- 45.Ono S., Yamakita Y., Yamashiro S., Matsudaira P. T., Gnarra J. R., Obinata T., Matsumura F. Identification of an actin binding region and a protein kinase C phosphorylation site on human fascin. J. Biol. Chem. 1997;272:2527–2533. doi: 10.1074/jbc.272.4.2527. [DOI] [PubMed] [Google Scholar]
- 46.Adams J. C. Roles of fascin in cell adhesion and mobility. Curr. Opin. Cell Biol. 2004;16:590–596. doi: 10.1016/j.ceb.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 47.Opassiri R., Hua Y., Wara-Aswapati O., Akiyama T., Esen A., Ketudat Cairns J. R. β-Glucosidase, exo-β-glucanase and pyridoxine transglucosylase activities of rice BGlu1. Biochem. J. 2004;379:125–131. doi: 10.1042/BJ20031485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vincent F., Gloster T. M., Macdonald J., Morland C., Stick R. V., Dias F. M. V., Prates J. A. M., Fontes C. M. G. A., Gilbert H. J., Davies G. J. Common inhibition of both β-glucosidases and β-mannosidases by isofagomine lactam reflects different conformational itineraries for pyranoside hydrolysis. ChemBioChem. 2004;5:1596–1599. doi: 10.1002/cbic.200400169. [DOI] [PubMed] [Google Scholar]
- 49.Reese E. T., Maguire A. H., Parrish F. W. Glucosidases and exo-glucanases. Can. J. Biochem. 1968;46:25–34. doi: 10.1139/o68-005. [DOI] [PubMed] [Google Scholar]
- 50.Hrmova M., De Gori R., Smith B. J., Fairweather J. K., Driguez H., Varghese J. N., Fincher G. B. Structural basis for broad substrate specificity in higher plant β-D-glucan glucohydrolases. Plant Cell. 2002;14:1033–1052. doi: 10.1105/tpc.010442. [DOI] [PMC free article] [PubMed] [Google Scholar]