Abstract
PAI-1 and PAI-2 (plasminogen-activator inibitor types 1 and 2) are inhibitors of cell surface uPA (urokinase plasminogen activator). However, tumour expression of PAI-1 and PAI-2 correlates with poor compared with good patient prognosis in breast cancer respectively. This biological divergence may be related to additional functional roles of PAI-1. For example, the inhibition of uPA by PAI-1 reveals a cryptic high-affinity site within the PAI-1 moiety for the VLDLr (very-low-density-lipoprotein receptor), which sustains cell signalling events initiated by binding of uPA to its receptor. These interactions and subsequent signalling events promote proliferation of breast cancer cells. Biochemical and structural analyses show that, unlike PAI-1, the PAI-2 moiety of uPA–PAI-2 does not contain a high-affinity-binding site for VLDLr, although uPA–PAI-2 is still efficiently endocytosed via this receptor in breast cancer cells. Furthermore, global protein tyrosine phosphorylation events were not sustained by uPA–PAI-2 and cell proliferation was not affected. We thus propose a structurally based mechanism for these differences between PAI-1 and PAI-2 and suggest that PAI-2 is able to inhibit and clear uPA activity without initiating mitogenic signalling events through VLDLr.
Keywords: plasminogen-activator inhibitor type 1 (PAI-1), plasminogen-activator inhibitor type 2 (PAI-2), serpin (serine protease inhibitor), urokinase plasminogen activator (uPA), very-low-density lipoprotein receptor (VLDLr)
Abbreviations: ERK, extracellular-signal-regulated kinase; LDLR, low-density lipoprotein receptor; LRP, low-density lipoprotein receptor-related protein; PAI, plasminogen-activator inhibitor; RAP, receptor-associated protein; RCL, reactive centre loop; serpin, serine protease inhibitor; SPR, surface plasmon resonance; uPA, urokinase plasminogen activator; uPAR, urokinase plasminogen activator receptor; VLDLr, very-low-density lipoprotein receptor
INTRODUCTION
uPA (urokinase plasminogen activator) plays an important role in many physiological processes including metastasis, wound healing and angiogenesis, through the pericellular activation of plasminogen and degradation of the extracellular matrix [1]. The deregulation of uPA expression associated with metastatic cancer increases plasmin activity, catalysing extracellular matrix degradation and promoting migration [2]. Importantly, uPA can also promote metastasis through protease-independent mechanisms [3]. For example, binding of uPA to its cell-surface receptor, uPAR, often initiates mitogenic signalling responses [4]. As uPAR is not a transmembrane receptor, these and other potential cellular responses are facilitated by interactions with integrins [5] and associated co-receptors, including the EGFR (epidermal growth factor receptor) [6] and endocytosis receptors of the LDLR (low-density lipoprotein receptor) family [7].
The key physiological inhibitors of uPA, PAI-1 and PAI-2 (plasminogen-activator inhibitor types 1 and 2), belong to the serpin (serine protease inhibitor) superfamily (also known as SerpinE1 and SerpinB2 respectively). Despite only 24% amino acid sequence identity and their classification into different serpin classes, PAI-1 and PAI-2 fold into a well-conserved tertiary structure among serpins, which consists of three β-sheets, 8–9 α-helices and an RCL (reactive centre loop) containing the protease recognition site [8]. A structural feature unique to PAI-2 is an extended sequence of 33 amino acids between α-helices C and D known as the C–D loop (Ala65–Glu96) [9]. In a similar manner to other inhibitory serpins, PAI-1 and PAI-2 interact with their target protease through the RCL which ultimately leads to the formation of SDS-stable, equimolar covalent complexes in which the serpin adopts a more thermodynamically stable, relaxed conformation [8].
Although both PAI-1 and PAI-2 are efficient inhibitors of soluble or receptor-bound uPA [10,11], their apparent role(s) in breast cancer invasion and metastasis appear somewhat paradoxical. Clinical studies show that uPA–PAI-1 co-expression has level one evidence as a prognostic marker of progression in early breast cancer [12,13] and may have prognostic significance in ovarian, endometrial, bladder and other cancers [14]. In vitro studies have shown that uPA and PAI-1 are necessary for lung carcinoma cell invasion through Matrigel [15] and that PAI-1 deficiency inhibited invasion of transplanted malignant keratinocytes [16]. In contrast, high PAI-2 expression in breast carcinomas that also express uPA is correlated with prolonged relapse-free survival, whereas low levels of PAI-2 are associated with metastasis from various carcinomas [9,14,17]. Furthermore, a number of observations have shown that PAI-2 reduces tumour growth, invasion and metastasis using in vitro and in vivo models via inactivation of cell-surface uPA [9].
Both PAI-1 and PAI-2 can be internalized into cells through interactions with endocytosis receptors of the LDLR family, such as LRP (low-density lipoprotein receptor-related protein) and VLDLr (very-low-density lipoprotein receptor) [18–21]. Upon PAI-1/2 inhibition of uPAR-bound uPA, a covalent complex is formed with increased affinity for these receptors, resulting in an enhanced rate of uPA–serpin complex endocytosis [19,22]. In the case of PAI-1, specific arginine and lysine residues within (Arg76 and Lys80) and adjacent (Lys88, Arg118 and Lys122) to the helix D of PAI-1 have been shown to form part of a cryptic high-affinity binding site for LRP and VLDLr, exposed by complex formation with uPA [23,24]. The interaction of uPA–PAI-1 with LDLRs can indirectly affect signalling activity by regulating levels of uPA/uPAR on the cell surface [25] and also directly transmit signals through receptor cytoplasmic domains [24,26–28]. PAI-1 is also capable of stimulating cell migration independently of uPA, tPA (tissue plasminogen activator) and vitronectin. For example, a direct interaction between PAI-1 and LRP activates the Jak/STAT pathway, resulting in actin filament polarization, translocation of activated STAT1 into the nucleus and increased cell migration [29].
We have previously shown that specific and efficient internalization of PAI-2 is dependent on forming a complex with cell surface uPAR-bound uPA [10]. However, we also found that PAI-2, unlike PAI-1, cannot bind directly to LRP and that uPAR/uPA–PAI-2 endocytosis is mediated predominantly by binding sites within the uPA moiety of the complex [19]. In the present study, we not only show that this also applies to VLDLr but highlight novel differences in VLDLr binding mechanisms between uPA–PAI-1 and uPA–PAI-2, which may have important functional implications in cancer.
MATERIALS AND METHODS
Proteins and antibodies
Recombinant human PAI-2 (47 kDa form) was provided by PAI-2 Pty Ltd. Human HMW (high-molecular mass) uPA, anti-uPAR polyclonal antibody (#399r) and Spectrozyme PL substrate were from American Diagnostica. Anti-VLDLr (H-95) polyclonal antibody was from Santa Cruz Biotechnology. Anti-rabbit IgG-FITC was from Sigma–Aldrich. Glu-plasminogen was purified from human plasma, as previously described [30]. Anti-phosphotyrosine monoclonal antibody clone PY20 (#P11120) was from BD Biosciences. The Alexa Fluor® 488 labelling kit and Alexa Fluor® 488 polyclonal antibody were from Invitrogen.
Purified human RAP (receptor-associated protein), anti-LRP polyclonal antibody and anti-VLDLr blocking monoclonal antibodies (5F3 and 1H5, [31]) were a gift of Professor Dudley Strickland (Center for Vascular and Inflammatory Diseases, University of Maryland School of Medicine, MD, U.S.A.). Recombinant human VLDLr ligand-binding region was a gift of Professor Dieter Blaas (University of Vienna, Vienna, Austria). Recombinant PAI-1 14-1b stable variant and PAI-1R76E mutant on 14-1b backbone were provided by Professor Daniel Lawrence (University of Michigan, Michigan, U.S.A.) [24].
Cell culture
The MCF-7 epithelial breast cancer cell line was used for all experiments. Cells were grown, passaged and prepared for experiments as previously described [10].
Analysis of cell-surface antigen expression and internalization by flow cytometry
MCF-7 cells, grown to 80% confluency over a 48 h period, were detached using PBS/EDTA (5 mM), washed with ice-cold binding buffer (Phenol-Red-free Hanks buffered salt solution, pH 7.4, containing 1 mM CaCl2, 1 mM MgCl2 and 0.1% BSA) and centrifuged at 300 g at 4 °C. The cells were resuspended at 1×106 cells/ml in ice-cold binding buffer containing primary polyclonal antibodies or irrelevant isotype control antibody (5 μg/ml) and incubated for 45 min on ice. After three washes with ice-cold binding buffer, the cells were incubated with goat anti-rabbit IgG-FITC (1:50 dilution) for 45 min on ice. In all cases cell surface fluorescence was analysed by dual-colour flow cytometry as previously described [30].
Internalization assays using Alexa Fluor® 488 labelled PAI-2 or uPA and Alexa Fluor® 488 polyclonal quenching antibody were performed as previously described [19].
Preparation of uPA–serpin complexes
uPA–serpin complexes were prepared as previously described [28]. Briefly, uPA and serpin (PAI-1 or PAI-2) were incubated at a 1:1 molar ratio for 30 min at 37 °C. Complex formation was monitored by SDS/PAGE and Western blotting for uncomplexed uPA (results not shown). These analyses confirmed the complete inactivation of uPA.
SPR (surface plasmon resonance) analysis
VLDLr was immobilized to a CM5 BIAcore chip according to the manufacturer's instructions. Briefly, the chip was activated using a 1:1 mixture of 0.2 M N-ethyl-N′-(3-dimethylaminopropyl)carbodi-imide and 0.05 M N-hydroxysuccimide. VLDLr was coated to the chip at 40 μg/ml in 10 mM sodium acetate (pH 3) to a level of ∼10000 response units. Unoccupied binding sites were blocked using 1 M ethanolamine (pH 8.5). Ligands were diluted into running buffer [10 mM Hepes (pH 7.4), 140 mM NaCl, 1 mM CaCl2 and 0.05% Tween-20] before applying to the BIAcore chip at 20 μl/min. Regeneration of the chip was achieved using 100 mM H3PO4. For kinetic analysis, a blank cell was used as the reference cell and data were analysed using BIAevaluation software (Version 4).
Plasmin activity assay
MCF-7 cells were seeded in 96-well plates at 1×104 cells/well and cultured for 48 h without change of medium. Cells were washed and incubated in binding buffer containing purified human uPA (5 nM) for 30 min on ice. Cells were subsequently washed and incubated for 0, 1, 10 or 30 min at 37 °C in binding buffer containing PAI-1 or PAI-2 (5 nM). Cells were then washed and incubated with 0.5 μM human Glu-plasminogen for 10 min at room temperature (25 °C). Plasmin activity was then measured over 2 h at 37 °C using Spectrozyme PL substrate (0.4 mM final concentration). Colour development was recorded at 405 nm.
Confocal microscopy
MCF-7 cells were grown to ∼80% confluency in 8-well chamber slides and serum starved for 4 h. Cells were then incubated in the presence or absence of 100 nM RAP in binding buffer, at 37 °C for 15 min. The cells were then incubated with uPA, uPA–PAI-1, uPA–PAI-1R76E or uPA–PAI-2 (10 nM) in binding buffer, at 37 °C for 30 min. Following two washes with ice-cold PBS, the cells were fixed with 3.75% (w/v) paraformaldehyde, permeabilized with 0.2% Triton X-100, blocked with 1% BSA/PBS and probed with anti-phosphotyrosine antibodies (5 μg/ml), for 45 min at 4 °C in 1% BSA/PBS. Following a further two washes, the cells were incubated with anti-mouse IgG-FITC (1:200) and TO-PRO 3 (1:400) in 1% BSA/PBS for 45 min at 4 °C. After washing, the cells were analysed by confocal microscopy using a Leica TCS SP system.
Cell proliferation assay
MCF-7 cell proliferation assay was performed using the CellTiter 96® AQueous One Solution Cell Proliferation Assay (Promega) essentially as described previously [28].
Protein structure analysis
Protein structure analysis and Figure preparation was performed using the PyMol software package (DeLano Scientific). Structural co-ordinates were obtained from X-ray crystal structures of the relaxed conformations of both PAI-1 (PDB code 9PAI) [32] and PAI-2 C–D loop deletion mutant (PDB code 1JRR) [33].
RESULTS
uPA–PAI-2 endocytosis is mediated by uPAR and VLDLr
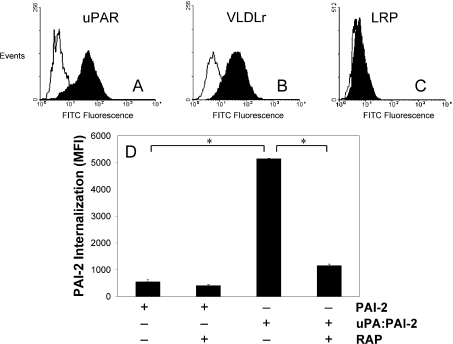
To confirm the suitability of MCF-7 cells for examining PAI-2 endocytosis and potential associated signalling events, the cell-surface expression of uPAR and VLDLr was analysed by flow cytometry. Both VLDLr and uPAR were detected on the surface of MCF-7 cells (Figures 1A and 1B). We have previously shown that LRP mediates PAI-2 endocytosis in PC-3 cells in an uPA/uPAR dependent manner [19]. However, LRP was not detected on MCF-7 cells (Figure 1C), confirming previous studies [25]. As megalin (LRP2) was also not detectable on these cells (results not shown), we concluded that VLDLr was the only LDLR family member of relevance on MCF-7 cells. Internalization assays were undertaken to confirm the involvement of receptor-bound uPA and VLDLr in the endocytosis of PAI-2. Relatively little PAI-2 internalization was observed in the absence of exogenous uPA, which was not sensitive to inhibition by RAP, a specific competitive inhibitor of LDLR binding (Figure 1D). Upon addition of uPA–PAI-2, significant RAP-sensitive internalization was observed (Figure 1D), indicating the presence of significant levels of unoccupied uPAR on these cells and once again highlighting the uPAR-binding dependency of subsequent uPA–PAI-2 endocytosis. This process was confirmed to be VLDLr-dependent, as pre-incubation with 10 μg/ml anti-VLDLr blocking antibodies 5F3 or 1H5 [31] caused a similar reduction in uPA–PAI-2 endocytosis as RAP (results not shown). This data, in the context of our previous data showing uPA/uPAR dependence [10], confirm that both uPAR and VLDLr are necessary for uPA–PAI-2 endocytosis in MCF-7 cells.
Figure 1. uPAR and VLDLr mediate the endocytosis of uPA–PAI-2 by MCF-7 cells.
MCF-7 cells were probed with 10 μg/ml of primary (A) uPAR, (B) VLDLr or (C) LRP polyclonal antibodies. These were detected using anti-rabbit IgG-FITC (1:50 dilution) and the cells analysed by flow cytometry, using propidium iodide to exclude non-viable cells. (D) MCF-7 cells were incubated in the presence or absence of RAP (200 nM) for 15 min at 37 °C, prior to analysis of PAI-2–Alexa Fluor®488 or uPA–PAI-2–Alexa Fluor® 488 internalization using the fluorescence quenching internalization assay (means±S.E.M., n=3; *P<0.05).
PAI-2 does not contain a high-affinity binding site for VLDLr
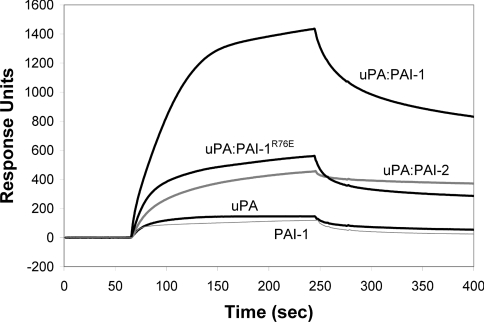
The PAI-1 component of the uPA–PAI-1 complex contains a cryptic high-affinity binding site for LRP and VLDLr, centered around the Arg76 residue within helix D [23,24]. We have previously shown that PAI-2 cannot directly bind LRP, indicating that it does not contain a similar binding site for LRP [19]. We utilized SPR analysis to characterize the binding of uPA–PAI-2 to VLDLr, and compared this with uPA–PAI-1 binding. Significantly stronger binding of uPA–PAI-1 to VLDLr compared with uPA–PAI-2 and uPA–PAI-1R76E was observed (Figure 2). Relatively lower binding of PAI-1 (Figure 2), and no binding of PAI-2 or PAI-1R76E (results not shown) to VLDLr was detected. uPA proteolytic activity was not necessary for binding of uPA to VLDLr as no difference in binding was observed following PMSF inactivation of uPA (results not shown).
Figure 2. SPR (BIAcore) analysis of PAI-1 and PAI-2 binding to VLDLr.
Sensorgrams showing the interaction between 100 nM PAI-1 (thin grey line), uPA, uPA–PAI-1, uPA–PAI-1R76E or uPA–PAI-2 (thick grey line) and immobilized VLDLr. Binding of 100 nM PAI-1R76E or PAI-2 to VLDLr was not detected (results not shown). The data shown are representative for at least three independent experiments.
Quantitative analysis showed that PAI-1 binding to VLDLr best fitted a 1:1 binding model, with a KD of ∼52 nM (Table 1). uPA and uPA–PAI-1 displayed complicated binding kinetics that best fitted a model where both uPA and uPA–PAI-1 contain two separate binding sites of higher and lower affinity capable of binding to VLDLr independently, but in a competitive manner (Table 1). The two sites within uPA bound to VLDLr with KD values of ∼209 and ∼31 nM. The two sites within uPA–PAI-1 bound to VLDLr with KD values of ∼85 nM and ∼1.5 nM (Table 1). Binding of uPA–PAI-1R76E or uPA–PAI-2 to VLDLr best fitted a 1:1 binding model with a KD of ∼10 and ∼5 nM respectively (Table 1).
Table 1. The kinetics parameters of PAI-1, PAI-1R76E, PAI-2, uPA and uPA–serpin complexes binding to VLDLr, measured by SPR.
aBinding data was fitted using the BIAevaluation 4.0 software. The binding model chosen represents that with the lowest φ2 value. Values are means±S.E.M., n=3.
| Analyte | Binding model | ka (M−1·s−1) | kd (S−1) | KDa (nM) | ϕ2 |
|---|---|---|---|---|---|
| PAI-1 | 1:1 | 9.90×104 (±6.83×104) | 4.17×10−3 (±1.19×10−3) | 51.8 (±21.9) | 8.12 |
| PAI-1R76E | No binding | − | − | − | − |
| PAI-2 | No binding | − | − | − | − |
| uPA | Heterologous analyte | 8.00×104 (±1.08×104) | 1.32×10−2 (±0.01×10−2) | 209 (±25.1) | 13.9 |
| 5.48×104 (±0.74×105) | 1.50×10−3 (±0.13× 0−3) | 31.2 (±5.69) | |||
| uPA–PAI-1 | Heterologous analyte | 1.50×105 (±0.20×105) | 1.30×10−2 (±0.10×10−2) | 84.8 (±1.69) | 19.6 |
| 1.00×105 (±0.40×104) | 1.30×10−4 (±0.18×10−3) | 1.51 (±0.27) | |||
| uPA–PAI-1R76E | 1:1 | 7.79×104 (±0.84×104) | 7.08×10−4 (±2.01×10−4) | 9.95 (±3.53) | 8.2 |
| uPA–PAI-2 | 1:1 | 1.73×105 (±0.62×105) | 5.61×10−4 (±0.98×10−4) | 4.68 (±0.90) | 1.9 |
Structural analysis of serpin–VLDLr binding
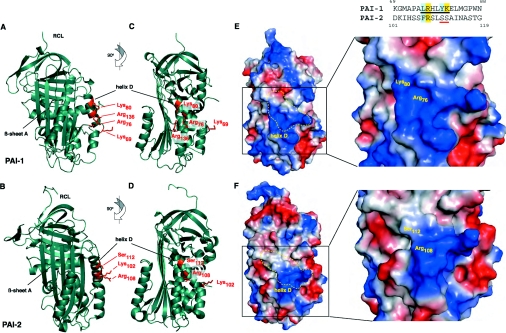
The Arg76 residue within helix D of PAI-1 is crucial for binding of PAI-1 and uPA–PAI-1 to LRP or VLDLr [24] (Figure 2). Although a homologous residue (Arg108) is conserved within helix D of PAI-2 [34], PAI-2 does not bind VLDLr (Table 1) or LRP [19] and uPA–PAI-2 binds with much lower affinity than uPA–PAI-1 (Figure 2). Many ligands interact with members of the LDLR family via complementary regions of positive electrostatic potential in the ligand and negative electrostatic potential in the receptor [35,36]. Therefore the differential binding of uPA–PAI-1 and uPA–PAI-2 to LDLR members may be heavily influenced by the charge of their respective helix D regions and adjacent residues.
At physiological pH, PAI-2 carries a relatively negative charge (predicted pI=5.4) compared with PAI-1 (predicted pI=7), as reflected in the comparative surface electrostatic potentials of the two molecules in the relaxed conformation (Figures 3E and 3F). Helix D of PAI-1 has a mostly basic (positive) charge, whereas helix D of PAI-2 is more neutral and surrounded by multiple acidic (negative) regions. Furthermore, Arg76 of PAI-1 is located in the middle of a basic cavity bounded by Lys80 and Arg136, whereas the corresponding residue in PAI-2 sits on the edge of a smaller basic region, with Lys80 being replaced by Ser112 (Figures 3E and 3F). Jensen et al. [37] have proposed a minimal binding motif in LRP ligands, comprising two basic residues separated by 2–5 residues and N-terminally flanked by hydrophobic residues. A sequence containing both Arg76 and Lys80 in helix D of PAI-1 fits this motif; however, this sequence is not conserved in PAI-2 (Figure 3G). Hence, both the electrostatic environment and surface topography of helix D, particularly surrounding the Arg76/108 residue, may explain the observed differences in binding of PAI-1 and PAI-2 to VLDLr and LRP.
Figure 3. Comparison of PAI-1 and PAI-2 VLDLr-binding interfaces.
(A–D) Ribbon representation showing secondary structure and position of key binding residues within or adjacent to α-helix D of PAI-1 and PAI-2. (E and F) Surface representation showing regions of positive electrostatic potential in blue, regions of negative potential in red and neutral regions in white. Surface position of helix D and key binding residues within or adjacent to this region are outlined in yellow. (G) Alignment of helix D amino acid sequence from PAI-1 and PAI-2. The putative minimal binding motif [37] in PAI-1 is underlined with necessary basic and hydrophobic residues highlighted in yellow and blue respectively. Analysis was performed using PyMol and X-ray crystal structures of the relaxed conformations of both PAI-1 (PDB code 9PAI, [32]) and PAI-2 C–D loop deletion mutant (PDB code 1JRR, [33]).
Serpin internalization is related to VLDLr affinity
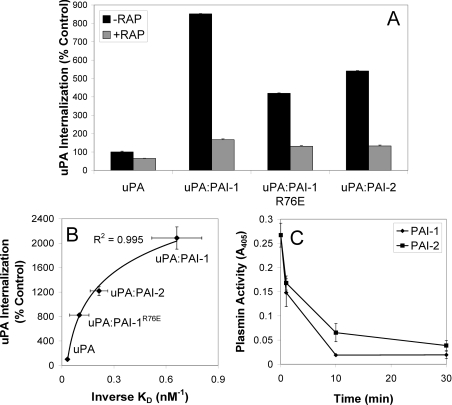
The effect of serpin inhibition and associated complex formation upon uPA internalization by MCF-7 cells was analysed using Alexa Fluor® 488-labelled uPA. As previously reported for PC-3 cells [19], we observed relatively little internalization of exogenous uPA after 1 h (Figure 4A). However, significantly increased uPA–PAI-1 internalization (∼8.5-fold) was observed in the same timeframe (Figure 4A). By comparison, a ∼4–5-fold increase in uPA–PAI-1R76E or uPA–PAI-2 internalization was observed compared with uPA alone (Figure 4A). RAP-mediated inhibition of uPA or uPA–serpin internalization confirmed the involvement of VLDLr in this process (Figure 4A). Hence, there was a very strong correlation between the affinity of uPA–serpin complexes for VLDLr and uPA–serpin internalization by MCF-7 cells (Figure 4B), indicating that VLDLr affinity may be the rate-limiting determinant of uPA/uPAR clearance from the cell surface. Even so, the rate of cell-surface uPA inhibition by PAI-1 and PAI-2 was very similar over a 30 min incubation period (Figure 4C).
Figure 4. VLDLr-mediated internalization of uPA–serpin complexes by MCF-7 cells.
(A) MCF-7 cells were incubated in the presence or absence of RAP (200 nM) for 15 min at 37 °C, prior to analysis of uPA–Alexa Fluor® 488 internalization using a fluorescence quenching internalization assay (means±S.E.M., n=3). (B) Relationship between the affinity of uPA and uPA–serpin complexes for VLDLr (Table 1, n=3) and internalization (n=2). (C) The inhibition of cell surface uPA activity by PAI-1 and PAI-2. MCF-7 cells pre-incubated with uPA (5 nM) at 4 °C for 30 min were washed and then incubated with PAI-1 or PAI-2 (5 nM) at 37 °C for the time periods indicated. uPA activity was measured indirectly by the addition of plasminogen (0.5 μM) and Spectrozyme-PL (0.4 mM) for 2 h. Absorbance was read at 405 nm. Activity in the absence of exogenous uPA was subtracted from all measurements (means±S.E.M., n=3).
PAI-2 does not induce mitogenic signalling in MCF-7 cells
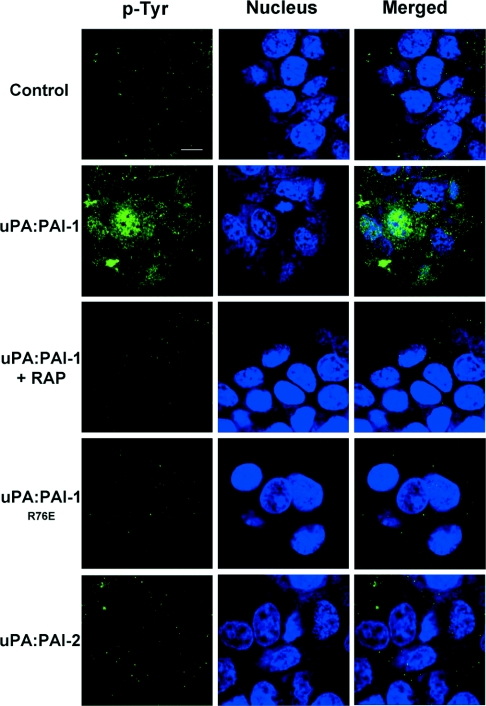
The high-affinity VLDLr-binding site in PAI-1 has previously been implicated in the initiation of signalling events by uPA–PAI-1 in breast cancer cells [28]. The absence of a corresponding high-affinity site in PAI-2 suggests potential differences in signalling capacities between these serpins. As a global indicator of intracellular signalling events [29], we utilized confocal microscopy to visualize protein tyrosine phosphorylation in MCF-7 cells after stimulation by uPA, uPA–PAI-1, uPA–PAI-1R76E and uPA–PAI-2 for 30 min. Stimulation by uPA–PAI-1 induced significant cytoplasmic and nuclear protein tyrosine phosphorylation, which was blocked by the addition of RAP (Figure 5). In stark contrast, protein tyrosine phosphorylation was not observed after a 30 min incubation with uPA, uPA–PAI-1R76E or uPA–PAI-2 (Figure 5).
Figure 5. uPA–PAI-2 does not induce nuclear/cytoplasmic protein tyrosine phosphorylation.
MCF-7 cells were serum starved for 4 h and incubated in the presence or absence of RAP (200 nM) for 15 min at 37 °C, then incubated with 10 nM uPA, uPA–PAI-1, uPA–PAI-1R76E or uPA–PAI-2 for 30 min at 37 °C. The cells were washed with ice-cold PBS, fixed with 3.75% (w/v) paraformaldehyde and permeabilized with 0.2% Triton X-100. After incubation with 10 μg/ml anti-phosphotyrosine monoclonal antibody (PY20), the cells were washed and incubated with goat anti-mouse IgG-FITC (1:200 dilution) and TO-PRO 3 (1:400). After washing, the cells were analysed by confocal microscopy. The scale bar represents 10 μm.
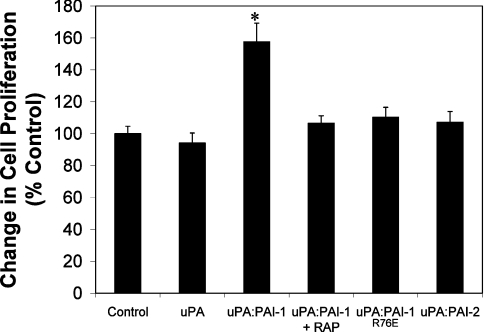
As uPA–PAI-1 is known to induce the proliferation of MCF-7 cells [28], we also compared the effect of uPA–PAI-1 with uPA–PAI-2 on MCF-7 cell proliferation. A ∼60% increase in proliferation of MCF-7 cells (relative to control) was observed after 36 h stimulation with uPA–PAI-1 (Figure 6). This effect was inhibited by RAP, again confirming a VLDLr-mediated effect. Consistent with the results above showing a lack of general protein tyrosine phosphorylation, no effect on cell proliferation was apparent following incubation with uPA, uPA–PAI-1R76E or uPA–PAI-2 (Figure 6).
Figure 6. uPA–PAI-2 does not stimulate cell proliferation of MCF-7 cells.
MCF-7 cells were cultured in RPMI/5% (v/v) fetal calf serum for 48 h, then treated with 10 nM uPA, uPA–PAI-1, uPA–PAI-1 plus RAP (200 nM), uPA–PAI-1R76E, uPA–PAI-2 or vehicle in RPMI containing 300 μg/ml glutamine, 5 μg/ml transferrin and 38 nM selenium. After culturing for 36 h, cell growth was determined using the MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] assay. Values of cell proliferation are relative to the proliferation of control cells (i.e. vehicle only). (means±S.E.M., n=6, *P<0.05).
DISCUSSION
In the present study we demonstrate that while VLDLr binds and mediates the endocytosis of uPA–PAI-1 and uPA–PAI-2 on breast cancer cells, clear structural differences exist which impact upon the binding mechanisms between these complexes and VLDLr. As the previously established LRP and VLDLr binding determinants in PAI-1 are absent in PAI-2, these may account for the lack of downstream signalling events associated with PAI-2 inhibition of uPA. PAI-2 thus appears to mainly result in the inhibition and clearance of cell surface uPA, which may potentially explain why increased PAI-2 expression levels relate to positive disease outcomes in breast cancer.
PAI-1 can promote invasion and metastasis independent of its inhibition of uPA-mediated proteolysis. For example, PAI-1 regulates cell migration through vitronectin binding, blocking cell attachment via uPAR and integrins [38]. Alternatively, uPA–PAI-1 stimulates pro-proliferative signalling events via a high-affinity interaction with VLDLr on MCF-7 cells [28], which also mediates endocytosis of this complex in various cell lines [7]. Given the lack of LRP expression on MCF-7 cells and the inhibition of uPA–PAI-2 endocytosis by blocking VLDLr (Figure 1), we undertook SPR analysis to characterize binding of uPA–PAI-1 and uPA–PAI-2 to VLDLr. These analyses confirmed a high-affinity interaction between uPA–PAI-1 and VLDLr. In contrast, we demonstrated that uPA–PAI-2 binds to VLDLr in a similar manner to uPA–PAI-1R76E, which lacks the high-affinity binding site in PAI-1 for VLDLr and LRP [24]. The mutation of Arg76 within helix D of PAI-1 results in a 10-fold reduction in the ability of uPA–PAI-1 and trypsin–PAI-1 to compete for binding to LRP [24]. Interestingly, in our studies this mutation also resulted in a complete abrogation of the binding of uncomplexed PAI-1 to VLDLr.
It should be noted that previous studies of uPA–PAI-1 binding to VLDLr, using solid-state binding assays, reported a 1:1 interaction with KD values of 14 and 15 nM [18,39]. Our SPR analyses of uPA–PAI-1 binding to VLDLr indicate a more complex interaction. However, if we force-fitted our data to a simple 1:1 interaction model, a similar KD of 24±3 nM was obtained, but with a greatly reduced significance of fit (ϕ2=38). Other studies have reported higher affinity 1:1 binding interactions between uPA–PAI-1 and VLDLr, with KD values of 0.8, 1.5 and 1.6 nM [40–42]. These values are almost identical with that which we obtained for the high-affinity VLDLr-binding site in PAI-1 (Table 1). Although the heterologous analyte model of uPA and uPA–PAI-1 binding to VLDLr does not provide a perfect fit, it is currently the best available model (lowest ϕ2 values) and suggests that a 1:1 model is not appropriate or accurate. Indeed, the presence of an independent, moderate-affinity binding site within the uPA moiety of uPA–PAI-1 (Table 1) has previously been suggested [24]. Furthermore, another study utilizing SPR to investigate the binding of uPA–PAI-1 to VLDLr focused on the initial fast-association and later slow-dissociation phases, while ignoring the slow-association and fast-dissociation phases, indicative of a second lower affinity site, present in the data [42]. It could also be expected that a heterologous analyte model would be observed for the binding of uPA–PAI-1R76E as only one residue has been mutated, although it is probable that the disruption of adjacent residues within this area has significantly altered the biochemistry of the interaction (as discussed below).
The strong correlation between the affinity of uPA–serpin complexes for VLDLr, together with its specific involvement in uPA–PAI-2 internalization by MCF-7 cells, indicates that VLDLr affinity may be the primary determinant of uPA–serpin/uPAR internalization in these cells. Comparison of structural characteristics of PAI-1 and PAI-2 in their relaxed conformations (i.e. mimicking the conformation in uPA–serpin complexes) [32,33], provides a clear explanation for the differential binding of PAI-1 and PAI-2 to VLDLr and LRP (Figure 3). Amino acid residues Arg76, Lys80 and Lys88 within and adjacent to helix D of PAI-1 contribute to binding of PAI-1 to LRP [43]. Along with Arg118 and/or Lys122 (within β-strand 1A), these residues have also been shown to contribute to the binding of the uPA complexed form of PAI-1 to LRP and VLDLr [23,43], with Arg76 forming part of a cryptic high-affinity binding site for LRP exposed by complex formation with uPA [24]. These residues conform with the proposed common binding motif for LRP ligands of two basic residues separated by 2–5 residues and N-terminally flanked by hydrophobic residues [37]. This motif is not conserved in PAI-2. Although the residue corresponding to Arg76 in PAI-1 is conserved within helix D of PAI-2 (Arg108), the residue corresponding to Lys80 is replaced by a serine residue (Ser112) in PAI-2 and the adjacent hydrophobic residue is not conserved (Figure 3). Furthermore, there are clear differences in the surface topography and overall electrostatic charge between PAI-1 and PAI-2. Previous studies have shown that mutation of basic residues within and adjacent to helix D of PAI-1, specifically the Arg76, Lys80 and Arg118 residues, can reduce affinity of uPA–PAI-1 for LDLR members [23,42]. This mechanism is further supported by a recent detailed description of the interaction between RAP domain 3 and LDLR type-A modules highlighting the importance of electrostatic and hydrophobic interactions between these binding determinants [35]. As our modelling is based on the structure of a C–D loop deletion mutant of PAI-2, it is difficult to predict the influence of the C–D loop on LRP/VLDLr binding. However, preliminary results suggest that deletion of the C–D loop does not alter the affinity of uPA–PAI-2 for VLDLr (results not shown).
uPA–PAI-2 and uPA–PAI-1R76E did not induce the significant global tyrosine phosphorylation observed following incubation of MCF-7 cells with uPA–PAI-1 (Figure 5). Furthermore, the proliferation of MCF-7 cells stimulated by uPA–PAI-1 was not observed following treatment with either uPA–PAI-2 or uPA–PAI-1R76E (Figure 6). The mitogenic signalling initiated by uPA–PAI-1 in MCF-7 cells has previously been attributed to the sustained activation of ERK 1/2 (extracellular-signal-regulated kinase 1/2) [28]. It was demonstrated that uPA binding to its receptor induces a transient pulse of ERK phosphorylation, although following PAI-1 inhibition, uPA–PAI-1 binds to VLDLr, resulting in the sustained activation of ERK [28]. The putative mechanisms of VLDLr-mediated cell signalling induced by uPA–PAI-1 are discussed in detail by Strickland et al. [7]. However, given the striking differences in binding mechanisms described above, these results suggest that the lack of a high-affinity VLDLr-binding site in PAI-2 prevents the initiation of these mitogenic signalling events. Indeed, we do not observe any additional ERK activation by uPA–PAI-2 over and above the transient activation initiated by uPA alone binding to uPAR on MCF-7 cells (results not shown).
Clearly, PAI-1 and PAI-2 have differential effects on uPA/uPAR endocytosis and mitogenic signalling. Although both PAI-1 and PAI-2 inhibit cell surface uPA and consequently decrease pericellular plasminogen activation capacity, PAI-1 has significant additional functional roles stimulating cell proliferation. We now propose a novel structural mechanism for this functional difference based on the absence of a high-affinity LDLR-binding site in PAI-2. Thus the poor prognosis for breast cancer patients with high uPA/PAI-1 protein levels may be associated with the ability of PAI-1 to initiate mitogenic signalling events through LDLRs such as VLDLr. In contrast, the favourable overall survival of patients with high PAI-2 protein expression may be due to uPA inhibition and clearance via LDLRs without the cell signalling events and increased metastatic potential associated with high PAI-1.
Acknowledgments
D. R. C. was a recipient of an Australian Postgraduate Award and a Cancer Institute NSW Scholar. D. N. S. is a Cancer Institute NSW Fellow. G. E. S. was a recipient of an Australian Postgraduate Award. M. R. is a Cancer Institute NSW Fellow. We wish to thank Dr Tilman Brummer, Dr Paul Timpson and Professor Roger Daly for helpful discussions.
References
- 1.Dano K., Romer J., Nielsen B. S., Bjorn S., Pyke C., Rygaard J., Lund L. R. Cancer invasion and tissue remodeling–cooperation of protease systems and cell types. APMIS. 1999;107:120–127. doi: 10.1111/j.1699-0463.1999.tb01534.x. [DOI] [PubMed] [Google Scholar]
- 2.Blasi F. Proteolysis, cell adhesion, chemotaxis, and invasiveness are regulated by the uPA-uPAR-PAI-1 system. Thromb. Haemostasis. 1999;82:298–304. [PubMed] [Google Scholar]
- 3.Han B., Nakamura M., Mori I., Nakamura Y., Kakudo K. Urokinase-type plasminogen activator system and breast cancer. Oncol. Rep. 2005;14:105–112. [PubMed] [Google Scholar]
- 4.Kjoller L. The urokinase plasminogen activator receptor in the regulation of the actin cytoskeleton and cell motility. Biol. Chem. 2002;383:5–19. doi: 10.1515/BC.2002.002. [DOI] [PubMed] [Google Scholar]
- 5.Reuning U., Magdolen V., Hapke S., Schmitt M. Molecular and functional interdependence of the urokinase-type plasminogen activator system with integrins. Biol. Chem. 2003;384:1119–1131. doi: 10.1515/BC.2003.125. [DOI] [PubMed] [Google Scholar]
- 6.Liu D., Aguirre Ghiso J., Estrada Y., Ossowski L. EGFR is a transducer of the urokinase receptor initiated signal that is required for in vivo growth of a human carcinoma. Cancer Cell. 2002;1:445–457. doi: 10.1016/s1535-6108(02)00072-7. [DOI] [PubMed] [Google Scholar]
- 7.Strickland D. K., Gonias S. L., Argraves W. S. Diverse roles for the LDL receptor family. Trends Endocrinol. Metab. 2002;13:66–74. doi: 10.1016/s1043-2760(01)00526-4. [DOI] [PubMed] [Google Scholar]
- 8.Silverman G. A., Bird P. I., Carrell R. W., Church F. C., Coughlin P. B., Gettins P. G., Irving J. A., Lomas D. A., Luke C. J., Moyer R. W., et al. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J. Biol. Chem. 2001;276:33293–33296. doi: 10.1074/jbc.R100016200. [DOI] [PubMed] [Google Scholar]
- 9.Kruithof E. K., Baker M. S., Bunn C. L. Biological and clinical aspects of plasminogen activator inhibitor type 2. Blood. 1995;86:4007–4024. [PubMed] [Google Scholar]
- 10.Al-Ejeh F., Croucher D., Ranson M. Kinetic analysis of plasminogen activator inhibitor type-2: urokinase complex formation and subsequent internalisation by carcinoma cell lines. Exp. Cell Res. 2004;297:259–271. doi: 10.1016/j.yexcr.2004.03.030. [DOI] [PubMed] [Google Scholar]
- 11.Ellis V., Wun T. C., Behrendt N., Ronne E., Dano K. Inhibition of receptor-bound urokinase by plasminogen-activator inhibitors. J. Biol. Chem. 1990;265:9904–9908. [PubMed] [Google Scholar]
- 12.Duffy M. J. The urokinase plasminogen activator system: role in malignancy. Curr. Pharm. Des. 2004;10:39–49. doi: 10.2174/1381612043453559. [DOI] [PubMed] [Google Scholar]
- 13.Weigelt B., Peterse J. L., van't Veer L. J. Breast cancer metastasis: markers and models. Nat. Rev. Cancer. 2005;5:591–602. doi: 10.1038/nrc1670. [DOI] [PubMed] [Google Scholar]
- 14.Duffy M. J., Duggan C. The urokinase plasminogen activator system: a rich source of tumour markers for the individualised management of patients with cancer. Clin. Biochem. 2004;37:541–548. doi: 10.1016/j.clinbiochem.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 15.Liu G., Shuman M. A., Cohen R. L. Co-expression of urokinase, urokinase receptor and PAI-1 is necessary for optimum invasiveness of cultured lung cancer cells. Int. J. Cancer. 1995;60:501–506. doi: 10.1002/ijc.2910600413. [DOI] [PubMed] [Google Scholar]
- 16.Bajou K., Noel A., Gerard R. D., Masson V., Brunner N., Holst-Hansen C., Skobe M., Fusenig N. E., Carmeliet P., Collen D., Foidart J. M. Absence of host plasminogen activator inhibitor 1 prevents cancer invasion and vascularization. Nat. Med. 1998;4:923–928. doi: 10.1038/nm0898-923. [DOI] [PubMed] [Google Scholar]
- 17.Foekens J. A., Peters H. A., Look M. P., Portengen H., Schmitt M., Kramer M. D., Brunner N., Janicke F., Meijer-van Gelder M. E., Henzen-Logmans S. C., et al. The urokinase system of plasminogen activation and prognosis in 2780 breast cancer patients. Cancer Res. 2000;60:636–643. [PubMed] [Google Scholar]
- 18.Argraves K. M., Battey F. D., MacCalman C. D., McCrae K. R., Gafvels M., Kozarsky K. F., Chappell D. A., Strauss J. F., 3rd, Strickland D. K. The very low density lipoprotein receptor mediates the cellular catabolism of lipoprotein lipase and urokinase-plasminogen activator inhibitor type I complexes. J. Biol. Chem. 1995;270:26550–26557. doi: 10.1074/jbc.270.44.26550. [DOI] [PubMed] [Google Scholar]
- 19.Croucher D., Saunders D. N., Ranson M. The urokinase–PAI-2 complex: a new high affinity ligand for the endocytosis receptor low density lipoprotein receptor-related protein. J. Biol. Chem. 2006;281:10206–10213. doi: 10.1074/jbc.M513645200. [DOI] [PubMed] [Google Scholar]
- 20.Herz J., Clouthier D. E., Hammer R. E. LDL receptor-related protein internalizes and degrades uPA–PAI-1 complexes and is essential for embryo implantation. Cell. 1992;71:411–421. doi: 10.1016/0092-8674(92)90511-a. [DOI] [PubMed] [Google Scholar]
- 21.Kounnas M. Z., Henkin J., Argraves W. S., Strickland D. K. Low density lipoprotein receptor-related protein/α2-macroglobulin receptor mediates cellular uptake of pro-urokinase. J. Biol. Chem. 1993;268:21862–21867. [PubMed] [Google Scholar]
- 22.Nykjaer A., Kjoller L., Cohen R. L., Lawrence D. A., Garni-Wagner B. A., Todd R. F., 3rd, van Zonneveld A. J., Gliemann J., Andreasen P. A. Regions involved in binding of urokinase-type-1 inhibitor complex and pro-urokinase to the endocytic α2-macroglobulin receptor/low density lipoprotein receptor-related protein. Evidence that the urokinase receptor protects pro-urokinase against binding to the endocytic receptor. J. Biol. Chem. 1994;269:25668–25676. [PubMed] [Google Scholar]
- 23.Rodenburg K. W., Kjoller L., Petersen H. H., Andreasen P. A. Binding of urokinase-type plasminogen activator-plasminogen activator inhibitor-1 complex to the endocytosis receptors α2-macroglobulin receptor/low-density lipoprotein receptor-related protein and very-low-density lipoprotein receptor involves basic residues in the inhibitor. Biochem. J. 1998;329:55–63. doi: 10.1042/bj3290055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stefansson S., Muhammad S., Cheng X. F., Battey F. D., Strickland D. K., Lawrence D. A. Plasminogen activator inhibitor-1 contains a cryptic high affinity binding site for the low density lipoprotein receptor-related protein. J. Biol. Chem. 1998;273:6358–6366. doi: 10.1074/jbc.273.11.6358. [DOI] [PubMed] [Google Scholar]
- 25.Webb D. J., Nguyen D. H., Sankovic M., Gonias S. L. The very low density lipoprotein receptor regulates urokinase receptor catabolism and breast cancer cell motility in vitro. J. Biol. Chem. 1999;274:7412–7420. doi: 10.1074/jbc.274.11.7412. [DOI] [PubMed] [Google Scholar]
- 26.Herz J., Strickland D. K. LRP: a multifunctional scavenger and signaling receptor. J. Clin. Invest. 2001;108:779–784. doi: 10.1172/JCI13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen D. H., Catling A. D., Webb D. J., Sankovic M., Walker L. A., Somlyo A. V., Weber M. J., Gonias S. L. Myosin light chain kinase functions downstream of Ras/ERK to promote migration of urokinase-type plasminogen activator-stimulated cells in an integrin-selective manner. J. Cell Biol. 1999;146:149–164. doi: 10.1083/jcb.146.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Webb D. J., Thomas K. S., Gonias S. L. Plasminogen activator inhibitor 1 functions as a urokinase response modifier at the level of cell signaling and thereby promotes MCF-7 cell growth. J. Cell Biol. 2001;152:741–752. doi: 10.1083/jcb.152.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Degryse B., Neels J. G., Czekay R. P., Aertgeerts K., Kamikubo Y., Loskutoff D. J. The low density lipoprotein receptor-related protein is a motogenic receptor for plasminogen activator inhibitor-1. J. Biol. Chem. 2004;279:22595–22604. doi: 10.1074/jbc.M313004200. [DOI] [PubMed] [Google Scholar]
- 30.Ranson M., Andronicos N. M., O'Mullane M. J., Baker M. S. Increased plasminogen binding is associated with metastatic breast cancer cells: differential expression of plasminogen binding proteins. Br. J. Cancer. 1998;77:1586–1597. doi: 10.1038/bjc.1998.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruiz J., Kouiavskaia D., Migliorini M., Robinson S., Saenko E. L., Gorlatova N., Li D., Lawrence D., Hyman B. T., Weisgraber K. H., Strickland D. K. The apoE isoform binding properties of the VLDL receptor reveal marked differences from LRP and the LDL receptor. J. Lipid Res. 2005;46:1721–1731. doi: 10.1194/jlr.M500114-JLR200. [DOI] [PubMed] [Google Scholar]
- 32.Aertgeerts K., De Bondt H. L., De Ranter C. J., Declerck P. Mechanisms contributing to the conformational and functional flexibility of plasminogen activator inhibitor-1. Nat. Struc. Biol. 1995;2:891–897. doi: 10.1038/nsb1095-891. [DOI] [PubMed] [Google Scholar]
- 33.Jankova L., Harrop S. J., Saunders D. N., Andrews J. L., Bertram K. C., Gould A. R., Baker M. S., Curmi P. M. Crystal structure of the complex of plasminogen activator inhibitor 2 with a peptide mimicking the reactive center loop. J. Biol. Chem. 2001;276:43374–43382. doi: 10.1074/jbc.M103021200. [DOI] [PubMed] [Google Scholar]
- 34.Huber R., Carrell R. W. Implications of the three-dimensional structure of α1-antitrypsin for structure and function of serpins. Biochemistry. 1989;28:8951–8966. doi: 10.1021/bi00449a001. [DOI] [PubMed] [Google Scholar]
- 35.Fisher C., Beglova N., Blacklow S. C. Structure of an LDLR–RAP complex reveals a general mode for ligand recognition by lipoprotein receptors. Mol. Cell. 2006;22:277–283. doi: 10.1016/j.molcel.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 36.Hussain M. M., Strickland D. K., Bakillah A. The mammalian low-density lipoprotein receptor family. Annu. Rev. Nutr. 1999;19:141–172. doi: 10.1146/annurev.nutr.19.1.141. [DOI] [PubMed] [Google Scholar]
- 37.Jensen G. A., Andersen O. M., Bonvin A. M., Bjerrum-Bohr I., Etzerodt M., Thogersen H. C., O'shea C., Poulsen F. M., Kragelund B. B. Binding site structure of one LRP–RAP complex: implications for a common ligand-receptor binding motif. J. Mol. Biol. 2006;362:700–716. doi: 10.1016/j.jmb.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 38.Stefansson S., Lawrence D. A. The serpin PAI-1 inhibits cell migration by blocking integrin αVβ3 binding to vitronectin. Nature. 1996;383:441–443. doi: 10.1038/383441a0. [DOI] [PubMed] [Google Scholar]
- 39.Mikhailenko I., Considine W., Argraves K. M., Loukinov D., Hyman B. T., Strickland D. K. Functional domains of the very low density lipoprotein receptor: molecular analysis of ligand binding and acid-dependent ligand dissociation mechanisms. J. Cell Sci. 1999;112:3269–3281. doi: 10.1242/jcs.112.19.3269. [DOI] [PubMed] [Google Scholar]
- 40.Heegaard C. W., Simonsen A. C., Oka K., Kjoller L., Christensen A., Madsen B., Ellgaard L., Chan L., Andreasen P. A. Very low density lipoprotein receptor binds and mediates endocytosis of urokinase-type plasminogen activator-type-1 plasminogen activator inhibitor complex. J. Biol. Chem. 1995;270:20855–20861. doi: 10.1074/jbc.270.35.20855. [DOI] [PubMed] [Google Scholar]
- 41.Kasza A., Petersen H. H., Heegaard C. W., Oka K., Christensen A., Dubin A., Chan L., Andreasen P. A. Specificity of serine proteinase/serpin complex binding to very-low-density lipoprotein receptor and α2-macroglobulin receptor/low-density-lipoprotein-receptor-related protein. Eur. J. Biochem. 1997;248:270–281. doi: 10.1111/j.1432-1033.1997.00270.x. [DOI] [PubMed] [Google Scholar]
- 42.Skeldal S., Larsen J. V., Pedersen K. E., Petersen H. H., Egelund R., Christensen A., Jensen J. K., Gliemann J., Andreasen P. A. Binding areas of urokinase-type plasminogen activator-plasminogen activator inhibitor-1 complex for endocytosis receptors of the low-density lipoprotein receptor family, determined by site-directed mutagenesis. FEBS J. 2006;273:5143–5159. doi: 10.1111/j.1742-4658.2006.05511.x. [DOI] [PubMed] [Google Scholar]
- 43.Horn I. R., van den Berg B. M., Moestrup S. K., Pannekoek H., van Zonneveld A. J. Plasminogen activator inhibitor 1 contains a cryptic high affinity receptor binding site that is exposed upon complex formation with tissue-type plasminogen activator. Thromb. Haemostasis. 1998;80:822–828. [PubMed] [Google Scholar]