Abstract
Sialidase NEU3 is also known as the plasma-membrane-associated form of mammalian sialidases, exhibiting a high substrate specificity towards gangliosides. In this respect, sialidase NEU3 modulates cell-surface biological events and plays a pivotal role in different cellular processes, including cell adhesion, recognition and differentiation. At the moment, no detailed studies concerning the subcellular localization of NEU3 are available, and the mechanism of its association with cellular membranes is still unknown. In the present study, we have demonstrated that sialidase NEU3, besides its localization at the plasma membrane, is present in intracellular structures at least partially represented by a subset of the endosomal compartment. Moreover, we have shown that NEU3 present at the plasma membrane is internalized and locates then to the recycling endosomal compartment. The enzyme is associated with the outer leaflet of the plasma membrane, as shown by selective cell-surface protein biotinylation. This evidence is in agreement with the ability of NEU3 to degrade gangliosides inserted into the plasma membrane of adjacent cells. Moreover, the mechanism of the protein association with the lipid bilayer was elucidated by carbonate extraction. Under these experimental conditions, we have succeeded in solubilizing NEU3, thus demonstrating that the enzyme is a peripheral membrane protein. In addition, Triton X-114 phase separation demonstrates further the hydrophilic nature of the protein. Overall, these results provide important information about the biology of NEU3, the most studied member of the mammalian sialidase family.
Keywords: endosome, ganglioside, peripheral membrane protein, plasma membrane, sialidase NEU3
Abbreviations: Cav-1, caveolin-1; DMEM, Dulbecco's modified Eagle's medium; EEA1, early endosomal antigen 1; GPI, glycosylphosphatidylinositol; HA, haemagglutinin; HRP, horseradish peroxidase; HsNEU3, Homo sapiens sialidase NEU3; LAMP1, lysosome-associated membrane protein 1; LBPA, lysobisphosphatidic acid; MmNEU3, Mus musculus sialidase NEU3; 4MU-NeuAc, 4-methylumbelliferyl-N-acetyl-α-D-neuraminic acid; PBS/BSA, 1% BSA in PBS; PBS/Sap, 0.5% saponin in PBS; PBST, PBS containing 0.1% Tween 20; PDI, protein disulfide-isomerase; TfR, transferrin receptor
INTRODUCTION
Sialidases or neuraminidases (EC 3.2.1.18) are glycohydrolytic enzymes that remove sialic acid residues from sialoglycoconjugates, such as gangliosides, sialoglycoproteins and sialo-oligosaccharides. They are widely distributed in Nature, from viruses and bacteria to vertebrates [1]. In mammals, four different forms of sialidases have been cloned so far, and their classification is based mainly on their subcellular distribution: the lysosomal NEU1, the cytosolic NEU2, the plasma-membrane-associated NEU3 and the lysosomal/mitochondrial NEU4 [2]. Among them, the plasma-membrane-associated sialidase NEU3 has been shown to be preferentially active towards gangliosidic substrates [3–8]. Moreover, NEU3 is present within lipid rafts [9,10] that represent specialized functional areas of the membranes, highly enriched in cholesterol and sphingolipids [11,12]. Sphingolipids, and among them gangliosides, are known to affect different biological events, including tumorigenic transformation [13–18], cell differentiation [19,20] and motility [21–25]. The evidence that the overexpression of sialidase NEU3 induces dramatic changes in the cellular ganglioside pattern [6] is of particular interest because of the relevance of these amphiphilic molecules in mediating important modes of cellular behaviour [26]. Indeed, sialidase NEU3 has been reported to play a pivotal role in different normal cellular processes, including axonal growth and regeneration [27], and tumorigenic transformation [28]. Importantly, the activity of sialidase NEU3 is exerted also on gangliosides exposed on the extracellular leaflet of the plasma membrane of adjacent cells by cell–cell interaction [6]. This evidence has led to the hypothesis that the enzyme is located at the external leaflet of the plasma membrane, facing the extracellular environment, thus allowing its interaction with the gangliosidic substrates exposed on the plasma membrane of neighbouring cells.
Detailed studies concerning the membrane-anchoring of sialidase NEU3 have not been reported so far. Analysis of the NEU3 amino acid sequence reveals a striking similarity with its soluble counterpart NEU2. Indeed, on the basis of the crystal structure of human NEU2 [29] and on the amino acid sequence homology, a modelling of HsNEU3 (Homo sapiens sialidase NEU3) was performed, suggesting a common β-propeller folding for both enzymes [30]. The possible presence of canonical amino acid motif(s) necessary for post-translational modification(s) potentially involved in NEU3 membrane anchoring can be explored by bioinformatic analysis [31–33]. In order to gain more information about the subcellular distribution of sialidase NEU3 and its mechanism of association with the lipid bilayer, we expressed the mouse protein MmNEU3 (Mus musculus sialidase NEU3) in HeLa and COS-7 cells. We found that expressed sialidase NEU3 is present both at the plasma membrane and in intracellular tubulovesicular structures that represent a subset of the endosomal compartment. In addition, experiments of cell-surface protein biotinylation and indirect immunofluorescence gave the first direct evidence that sialidase NEU3 is associated with the external leaflet of the plasma membrane. Finally, using different extraction and solubilization methods, we provide evidence that sialidase NEU3 has hydrophilic characteristics and behaves as a peripheral membrane protein.
EXPERIMENTAL
Cell culture and transfection
COS-7 and HeLa cells were cultured in DMEM (Dulbecco's modified Eagle's medium) containing 4 mM L-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin and 10% (v/v) fetal bovine serum, and were maintained at 37 °C and 5% CO2 in a humidified incubator. Cells were transiently transfected with a C-terminal HA (haemagglutinin)-tagged form of mouse NEU3 (MmNEU3–HA), subcloned into the pcDNA1/Amp (Invitrogen) vector. Transfection was performed in serum-free medium (OptiMEM; Gibco-BRL) employing FuGENE 6 (Roche) and incubating the cells at 37 °C for 6 h. After transfection, cells were grown in DMEM for 36 h and then processed.
Antibodies
For indirect immunofluorescence and immunoblotting experiments, the following primary antibodies were used: rabbit anti-HA (Sigma), mouse anti-EEA1 (early endosomal antigen 1) (Transduction Laboratories), mouse anti-TfR (transferrin receptor) (Zymed Laboratories), mouse anti-PDI (protein disulfide-isomerase) (Stressgene), mouse anti-LBPA (lysobisphosphatidic acid) (from Dr J. Grünberg, Department of Biochemistry, University of Geneva, Geneva, Switzerland) and rabbit anti-Cav-1 (caveolin-1) (Santa Cruz Biotechnology). Detection of HsNEU3 in immunoblotting experiments was achieved using a rabbit anti-HsNEU3 antibody (from Dr N. Stamatos, Institute of Human Virology, University of Maryland, Baltimore, MD, U.S.A.). For immunofluorescence experiments, donkey Alexa Fluor® 488-conjugated anti-rabbit and donkey Alexa Fluor® 555-conjugated anti-mouse (Molecular Probes) secondary antibodies were used. For immunoblotting experiments, HRP (horseradish peroxidase)-conjugated donkey anti-rabbit and sheep anti-mouse secondary antibodies (GE Healthcare) were used.
Indirect immunofluorescence and confocal microscopy analysis
Indirect immunofluorescence experiments were performed as described previously [6], with minor modifications. COS-7 or HeLa cells were seeded on to glass coverslips and transfected with MmNEU3–HA. At 36 h post-transfection, cells were fixed with 3% (w/v) paraformaldehyde in PBS for 15 min at room temperature (25 °C). Paraformaldehyde was then quenched by incubating the samples with 50 mM NH4Cl in PBS for 15 min. After three washes with PBS, cells were permeabilized with 0.5% saponin in PBS (PBS/Sap) for 30 min and incubated with primary antibodies diluted in PBS/Sap for 1 h. Subsequently, cells were washed three times with PBS/Sap and incubated with secondary antibodies diluted in PBS/Sap for the same period. Finally, after three washes with PBS/Sap followed by washes with PBS, specimens were mounted using DakoCytomation Fluorescent Mounting Medium and analysed using the confocal system LSM-510 META (Carl Zeiss). For indirect immunofluorescence of non-permeabilized cells, the same procedure was used, substituting 1% BSA for 0.5% saponin (PBS/BSA). Images were processed with LSM Image Browser (Carl Zeiss) and Adobe Photoshop software.
Antibody uptake
COS-7 cells seeded on to glass coverslips were transfected with MmNEU3–HA as described above. After 36 h of transfection, cells were incubated for 3 h at 37 °C in growth medium supplemented with rabbit anti-HA antibody at 2.5 μg/ml final concentration. Cells were then chilled on ice, washed three times with ice-cold PBS and fixed with methanol. After saturation with PBS/BSA, specimens were incubated with mouse anti-TfR antibody diluted in PBS/BSA for 1 h. After extensive washes with PBS/BSA, Alexa Fluor® 488-conjugated donkey anti-rabbit and Alexa Fluor® 555-conjugated donkey anti-mouse secondary antibodies were used. Specimens were then mounted and analysed as described above.
Immunoblotting
Proteins were separated by SDS/12% PAGE and transferred on to a Hybond™-P PVDF membrane (GE Healthcare). Membranes were then blocked with 5% (w/v) non-fat dried skimmed milk in PBS, washed three times with PBST (PBS containing 0.1% Tween 20) and incubated with primary antibody diluted in PBST containing 1% (w/v) non-fat dried skimmed milk for 2 h at room temperature. After three washes with PBST, membranes were incubated with HRP-conjugated secondary antibody diluted in PBST for 1 h at room temperature. Detection of the immunocomplexes was performed using an enhanced chemiluminescence-based system (SuperSignal West Pico Chemiluminescent Substrate; Pierce), followed by densitometric analysis using GelPro 3.1 software (Media Cybernetics).
Cell-surface protein biotinylation and purification
For biotinylation experiments, all steps were carried out at 4 °C. Surface proteins of subconfluent cultured COS-7 and HeLa cells expressing MmNEU3–HA for 36 h were labelled with 0.5 mg/ml membrane-impermeable sulfo-NHS-SS-biotin [sulfosuccinimidyl-2-(biotinamido)ethyl-1,3-dithiopropionate] (Pierce) in PBS for 30 min with gentle agitation, according to the manufacturer's instructions. Cells were then washed three times with PBS, and free biotin was quenched by one wash with 25 mM Tris/HCl (pH 8.0), followed by three washes with PBS. Cells were then scraped into PBS and lysed in RIPA buffer (25 mM Tris/HCl pH 7.4, 0.15 M NaCl, 0.1% SDS, 1% Triton X-100 and 1% sodium deoxycholate) by sonication. After lysis, total cell extracts were clarified by centrifugation at 800 g for 10 min, and biotinylated proteins were separated from non-biotinylated proteins using Immobilized Monomeric Avidin resin (Pierce). Fractions were adjusted to the same final volume and were analysed by immunoblotting.
Bioinformatic analysis
Analysis of the protein motifs involved in post-translational modifications was performed using the amino acid sequence of MmNEU3 (GenBank® accession number Q9JMH7) [34] and HsNEU3 (GenBank® accession number Q9UQ49) [5] with public domain software. Prediction of GPI (glycosylphosphatidylinositol)-anchor motifs was performed using the following software: big-PI Predictor GPI modification site prediction (http://mendel.imp.ac.at/sat/gpi/gpi_server.html), DGPI prediction of GPI-anchor and cleavage sites (http://129.194.185.165/dgpi/index_en.html), and GPI-SOM identification of GPI-anchor signals by a Kohonen self-organizing map (http://gpi.unibe.ch/). Prediction of fatty acyl- or prenyl-anchor motifs was performed with the following software: Myristoylator prediction of N-terminal myristoylation by neural networks (http://www.expasy.org/tools/myristoylator/), NMT prediction of N-terminal N-myristoylation (http://mendel.imp.ac.at/myristate/SUPLpredictor.htm), and PrePS Prenylation Prediction Suite (http://mendel.imp.ac.at/sat/PrePS/index.html). For the prediction of transmembrane sequences the following software were used: HMMTOP prediction of transmembrane helices and topology of proteins (http://www.enzim.hu/hmmtop/), SOSUI prediction of transmembrane regions (http://bp.nuap.nagoya-u.ac.jp/sosui/), TMHMM prediction of transmembrane helices in proteins (http://www.cbs.dtu.dk/services/TMHMM-2.0/), and TMpred prediction of transmembrane regions and protein orientation (http://www.ch.embnet.org/software/TMPRED_form.html). Finally, we analysed the amino acid sequences of MmNEU3 with AmphipaSeeK (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_amphipaseek.html), a monotopic membrane protein prediction tool.
Membrane protein extraction
HeLa or COS-7 cells 36 h after transfection with MmNEU3–HA were broken by bundle sonication in ice-cold lysis buffer (10 mM Tris/HCl, pH 7.5, containing 0.032 μg/ml aprotinin, 0.032 μg/ml leupeptin, 0.008 μg/ml pepstatin A, 0.06 μg/ml chymostatin and 0.2 mM PMSF as protease inhibitors). After cell lysis, samples were clarified by centrifugation at 800 g for 10 min at 4 °C, and the resulting supernatant (total cell extract) was centrifuged at 100000 g for 1 h at 4 °C in order to obtain a total cell membrane fraction. The resulting pellet was washed once with ice-cold distilled water, resuspended in the lysis buffer and split into aliquots of 200 μl. Extraction of peripheral proteins was performed by the exposure of total cell membranes either to pH 11.5 or 1 M KCl or 0.025 M EGTA and incubated on ice for 30 min. The above extraction conditions were achieved by the addition to the samples of an equal volume of 0.2 M Na2CO3 (pH 12) or 2 M KCl or 0.05 M EGTA respectively. As a control sample, membranes were incubated in presence of lysis buffer alone and processed as described above. Finally, solubilized and non-extractable proteins were separated by centrifugation at 100000 g, and carbonate-containing samples were immediately neutralized to pH 7.5 by the addition of ethanoic (acetic) acid. Soluble and membrane fractions were then adjusted to the same final volume before protein repartition analysis by sialidase activity and/or immunoblotting assay.
Triton X-114 phase separation
Triton X-114 phase separation was performed as described in [35]. Briefly, at 36 h post-transfection with MmNEU3–HA, HeLa cells were chilled at 4 °C. After three washes with ice-cold PBS, cells were scraped off and pelleted at 800 g for 10 min. Cells were then resuspended in 10 mM Tris/HCl (pH 7.4) and lysed by sonication, and the resulting total cell extracts were diluted to a protein concentration corresponding to 1.0 mg/ml in 0.1 ml of the same buffer. Proteins were extracted by adding 0.1 ml of 2% (v/v) pre-condensed Triton X-114 (Sigma) and incubating the sample for 1 h on ice. Detergent-extracted samples (200 μl) were then layered on to a 6% (w/v) sucrose cushion (300 μl), incubated at 30 °C for 3 min and finally centrifuged at 300 g for 3 min. After centrifugation, the upper aqueous phase was removed, re-extracted with 1% Triton X-114 and subjected to a second separation through the same sucrose cushion. The detergent and aqueous phases were adjusted to the same final volume and MmNEU3–HA repartition, together with the endogenous protein markers, was analysed by immunoblotting.
Sialidase activity assay
The enzymatic activity of MmNEU3–HA was determined as described previously [6] using [3H]GD1a ganglioside (radiolabelled at position 3 of the sphingosine moiety), prepared according to Ghidoni et al. [36] (specific radioactivity, 1.2 Ci/mmol; homogeneity >99%), and 4MU-NeuAc (4-methylumbelliferyl-N-acetyl-α-D-neuraminic acid) (Sigma) as substrates, in 100 mM sodium citrate/phosphate buffer at pH 3.8. Reaction mixtures were adjusted to the appropriate final concentrations of buffered carbonate or Triton X-114 before the addition of the enzymatic source. Samples were incubated at 37 °C for 30 min. To examine NEU3 enzymatic stability at alkaline pH, total membranes were incubated at pH 11.5 for different time periods (up to 30 min), neutralized to pH 7.5 and assayed as described above without ultracentrifugation. NEU3 activity related to Triton X-114 fractions was measured in both the single aqueous and detergent phases, as well as in the pooled phases.
RESULTS
Subcellular localization of sialidase NEU3
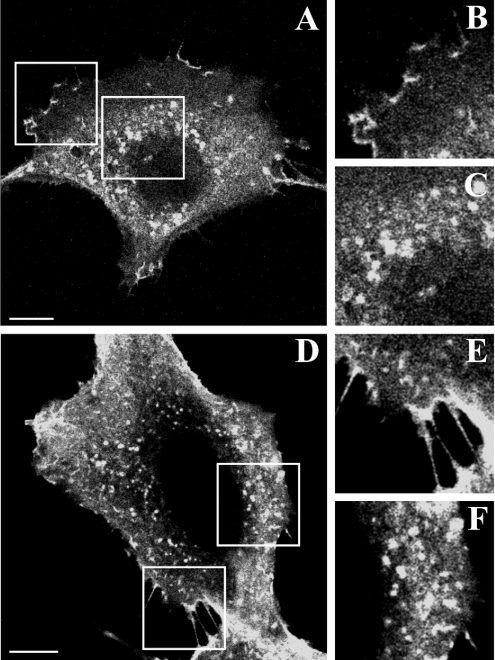
In order to study the subcellular distribution and localization of sialidase NEU3, MmNEU3–HA was transiently expressed in both HeLa and COS-7 cells. The expression of this construct gives rise to a fusion protein that is catalytically active and has already been used extensively in biochemical and functional studies [6–8,10]. After 36 h of expression, cells were fixed and permeabilized with saponin, and MmNEU3–HA was subsequently detected using anti-HA antibodies. Laser confocal microscopy analysis clearly shows that, irrespective to the cell type considered (Figures 1A and 1D), MmNEU3–HA localizes both at the plasma membrane (Figures 1B and 1E) and in intracellular vesicular structures that concentrate in the juxtanuclear region of the cells (Figures 1C and 1F). Although the presence of NEU3 at the plasma membrane has been demonstrated previously [3,5–7], intracellular localization of the enzyme has only been reported once, but without giving any information about the precise nature of this intracellular labelling [8].
Figure 1. Sialidase NEU3 localizes both at the plasma membrane and intracellularly.
COS-7 (A) and HeLa (D) cells transfected with MmNEU3–HA were fixed and, after permeabilization, subjected to indirect immunofluorescence using anti-HA antibody. Transfectants were then analysed by laser confocal microscopy. Close-ups of the plasma membrane (B and E) and intracellular labelling (C and F) are given for both cell types. Single confocal planes are shown. Scale bars, 10 μm.
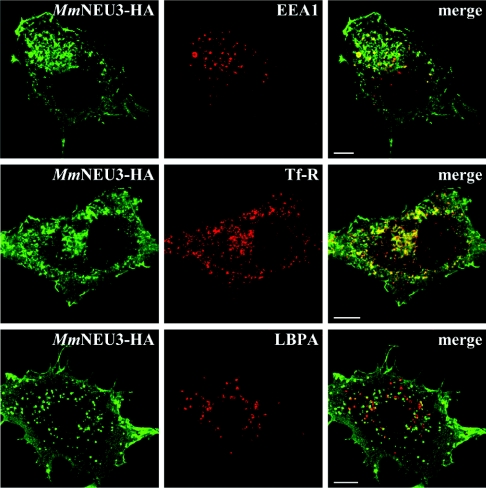
In order to better characterize the intracellular distribution of NEU3, co-localization experiments were carried out using markers of different cellular compartments. When MmNEU3–HA distribution was related either to the endoplasmic reticulum marker PDI or to the Golgi complex marker GM-130 (Golgi matrix protein of 130 kDa), no co-localization was observed (results not shown). Instead, co-localization between MmNEU3–HA and markers of the endosomal compartment was observed. As shown in Figure 2, COS-7 cells showed intracellular vesicular structures labelled for both MmNEU3–HA and EEA1, a marker of the early endosomes (Figure 2, top row). Co-localization of MmNEU3–HA is even more striking with the juxtanuclear TfR-positive structures, representing the recycling endosomal compartment (Figure 2, middle row). Interestingly, MmNEU3–HA did not co-localize with the late endosome marker LBPA (Figure 2, bottom row), as well as with the LAMP1 (lysosome-associated membrane protein 1) (results not shown). Taken together, these results indicate a restricted localization of sialidase NEU3 to a subset of the endosomal compartment. Moreover, the protein does not follow the degradative lysosomal pathway, as demonstrated by the lack of co-localization with both LBPA and LAMP1. A similar intracellular distribution and co-localization pattern was also observed in HeLa cells (results not shown). Overall, we can conclude that MmNEU3–HA, besides its localization at the plasma membrane, is present also in intracellular membranous structures that are mainly represented by the early and recycling endosomes.
Figure 2. Sialidase NEU3 co-localizes with endosomal markers.
COS-7 cells were transfected with MmNEU3–HA and subjected to immunofluorescence for co-localization experiments. Subcellular distribution of MmNEU3–HA was related to EEA1 (top row), TfR (middle row) and LBPA (bottom row), markers of the endocytic/recycling pathway. Single confocal planes are shown. Scale bars, 10 μm.
Sialidase NEU3 is associated with the external leaflet of the plasma membrane
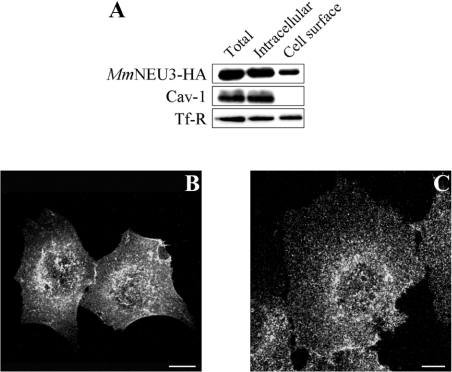
In order to confirm the presence of sialidase NEU3 both at the plasma membrane and in intracellular compartments, COS-7 cells expressing MmNEU3–HA were subjected to cell-surface protein biotinylation at 4 °C, and labelled proteins were isolated by affinity chromatography using an avidin resin. Distribution of MmNEU3–HA between avidin-unbound and avidin-bound fractions was then analysed by Western blotting. As shown in Figure 3(A), MmNEU3–HA was found in both the non-biotinylated and the biotinylated fractions, confirming that the protein is present both in intracellular compartments and at the cell surface. Densitometric analysis of the immunoblotting indicates that 23.8±5.4% (n=3) of MmNEU3–HA localizes to the cell surface, while the remaining portion of the protein is localized intracellularly. We also analysed the distribution of two other proteins, i.e. Cav-1, a protein known to be associated with the internal leaflet of the plasma membrane [37,38], and TfR, a transmembrane protein that cycles between the plasma membrane and intracellular compartments [39]. As shown in Figure 3(A), in the same cell extracts used for the detection and distribution of MmNEU3–HA, Cav-1 was detected only in the non-biotinylated fraction, as expected, confirming that, under these experimental conditions, intracellularly localized proteins are not accessible to biotin. Conversely, TfR was detected in both the biotinylated and the non-biotinylated fractions, as expected. Superimposable results were obtained also using HeLa cells (results not shown). On the basis of this evidence, we can reasonably assume that the portion of MmNEU3–HA that is bound to the plasma membrane is accessible to biotinylation, hence exposed to the extracellular environment. In other words, the enzyme is associated with the external leaflet of the plasma membrane. In order to confirm this finding, non-permeabilized COS-7 and HeLa cells expressing MmNEU3–HA were processed for indirect immunofluorescence using, at the same time, anti-HA antibodies and, as a control, anti-PDI antibodies. Under these experimental conditions, no labelling of the intracellular endoplasmic reticulum marker PDI was observed at all (results not shown), demonstrating the integrity of the plasma membrane, while a specific labelling for MmNEU3–HA was detected (Figures 3B and 3C). This result strengthens further the notion that sialidase NEU3 is associated with the external leaflet of the plasma membrane.
Figure 3. Sialidase NEU3 is associated with the extracellular leaflet of the plasma membrane.
(A) Cell-surface proteins of COS-7 cells transfected with MmNEU3–HA were selectively biotinylated and isolated by avidin–biotin-affinity chromatography. Equal volumes of the total cell extract (Total), avidin-unbound (Intracellular) and avidin-bound (Cell surface) fractions were analysed by immunoblotting in order to reveal MmNEU3–HA and the endogenous markers Cav-1 and TfR. Non-permeabilized HeLa (B) and COS-7 (C) cells transfected with MmNEU3–HA were subjected to indirect immunofluorescence using anti-HA antibody. Projections of sequential confocal planes (0.37 μm distance for each plane) acquired through the cell thickness are shown. Scale bars, 10 μm.
NEU3 is internalized from the cell surface to the recycling compartment
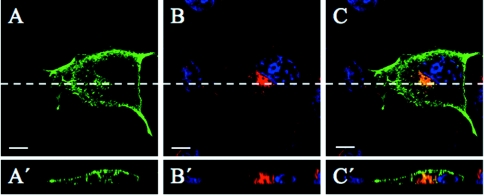
As described above, we have demonstrated that sialidase NEU3 localizes to the external leaflet of the plasma membrane and in the endosomal compartment. We then tried to assess directly the propensity of the protein to move from the surface to the cell interior. For this purpose, cultured COS-7 cells transfected with MmNEU3–HA were incubated for 3 h in presence of anti-HA antibody. Cells were then fixed and permeabilized, and the distribution of the MmNEU3–HA–anti-HA antibody immunocomplexes was analysed and compared with TfR distribution. As shown in Figure 4(A), in transfected cells, a significant labelling for NEU3 could be detected at the plasma membrane, but also in vesicular structures that are supposed to be located intracellularly. Co-localization between the anti-HA antibody signal and TfR (Figure 4B) is observed in the juxtanuclear region of the cell (Figure 4C), suggesting that the MmNEU3–HA–anti-HA antibody immunocomplexes were internalized from the cell surface and localize to the recycling endosomal compartment. The evidence that the anti-HA antibody is internalized and localizes intracellularly is given by the direct vertical confocal sections shown in Figures 4(A′)–4(C′). Indeed, a significant labelling for MmNEU3–HA both at the surface and in intracellular structures that concentrate at the juxtanuclear region of the cell is evident. Notably, these structures co-localize with the intracellular labelling for TfR. Taken together, these results clearly indicate that NEU3 internalizes from the plasma membrane and that internalized molecules reach the recycling endosomal compartment.
Figure 4. Cell-surface sialidase NEU3 internalizes to the recycling endosomal compartment.
MmNEU3–HA transfected COS-7 cells were subjected to anti-HA antibody uptake for 3 h, processed for indirect immunofluorescence and analysed by laser confocal microscopy as described in the Experimental section. Subcellular distribution of MmNEU3–HA–anti-HA antibody immunocomplexes (A, A′) was related to TfR staining (B, B′). Nuclei are shown by DAPI (4′,6-diamidino-2-phenylindole) staining. The merged image is given in (C, C′). The broken white lines in (A)–(C) represent the plane through which the corresponding direct vertical confocal sections (A′)–(C′) were acquired. Scale bars, 10 μm.
Sialidase NEU3 is a peripheral membrane-associated protein
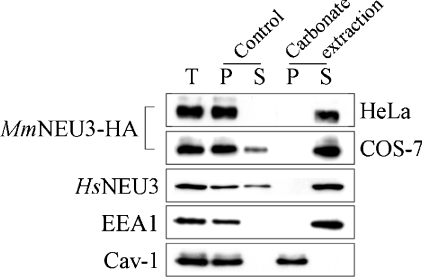
The amino acid sequence of MmNEU3 was analysed for the prediction of trasmembrane domain(s) that would anchor the protein to the lipid bilayer. Only the TMpred program (see the Experimental section) predicted the presence of a single hydrophobic amino acid stretch, spanning between residues Ala178 and Ser203. If we assume this stretch to be a transmembrane portion of the enzyme, the protein would additionally feature an intracellular and an extracellular domain. This structural model is unlikely, because, in this configuration, putative amino acid residues essential for the catalytic activity would be located at opposite sides of the membrane, which would be clearly inconsistent with catalytic activity [2]. Moreover, on the basis of the crystal structure of NEU2 [29] and considering the high homology between the two proteins, a common β-propeller fold for HsNEU3 was suggested recently [30]. Bioinformatic analysis of MmNEU3 amino acid sequence also excludes the possibility that the protein may associate with membranes as a monotopic protein [40]. Finally, sequence analysis does not reveal the presence of any amino acid motifs for post-translational modifications that would anchor the protein to the lipid bilayer, such as GPI, fatty acyl or prenyl linkages. On this basis, we took into consideration the possibility that sialidase NEU3 would associate with cellular membranes as a peripheral protein. For this purpose, total cell membranes obtained from HeLa and COS-7 cells expressing MmNEU3–HA were incubated for 30 min in presence of 0.1 M sodium carbonate at pH 11.5 and then soluble and membrane fractions were obtained by ultracentrifugation [41–43]. The resulting fractions were then adjusted to pH 7.5 and analysed for their enzyme content by Western blotting and sialidase activity. As control, total cell membranes were incubated for the same time period in 0.01 M Tris/HCl (pH 7.5) and processed as described above. As expected, incubation of membranes under control conditions had no significant effect on the association of MmNEU3–HA with the membranes (Figure 5) and the sialidase activity was almost completely recovered in the particulate fraction (82.5–87.3%, depending on the substrate used) (see Table 1). The small amount of MmNEU3–HA activity in the soluble fraction may derive from a spontaneous release of the protein, as a consequence of membrane manipulation. In contrast, a remarkable, almost complete, solubilization of MmNEU3–HA was achieved when membranes were incubated under alkaline conditions, irrespective of the cell line taken into consideration (Figure 5). Under these experimental conditions, the peripheral membrane protein EEA1 was completely recovered in the soluble fraction, as expected [44]. Noteworthily, carbonate treatment preserved the lipid structure without affecting the association of integral proteins with the lipid bilayer, as confirmed by the analysis of the repartition of Cav-1 in the same cell extracts. In fact, Cav-1 always remained associated with the membranous fractions.
Figure 5. Sialidase NEU3 is a peripheral membrane-associated protein.
A crude preparation of cell membranes from COS-7 and HeLa cells transfected with MmNEU3–HA were extracted with Tris buffer (Control) or with sodium carbonate (Carbonate extraction). After treatment, membranes were collected by ultracentrifugation, and equal volumes corresponding to the starting crude cell membranes (T), membranes (P) and soluble (S) fractions were analysed by Western blotting using anti-HA, anti-HsNEU3, anti-EEA1 and anti-Cav-1 antibodies.
Table 1. Repartition of sialidase activity.
Total NEU3 enzymatic activity in particulate (P) and soluble (S) fractions obtained from total cell membranes (T) of transfected COS-7 cells (MmNEU3–HA) was assayed under control and alkaline (carbonate extraction) conditions. Values, representative of three independent experiments, are expressed as total activity recovered in each fraction. Mock, COS-7 cells transfected with vector alone.
| Mock | MmNEU3–HA | Control | Carbonate extraction | |||
|---|---|---|---|---|---|---|
| T | T | P | S | P | S | |
| 4MU-NeuAc (nmol/h) | 15.7±1.8 | 125.7±20.8 | 110.7±14.1 | 8.1±2.8 | 0.4±0.2 | 0.3±0.2 |
| [3H]GD1a (μmol/h) | 73.5±7.8 | 1090.0±185.3 | 900.0±117.0 | 160.0±56.0 | 30.0±14.4 | 10.0±6.7 |
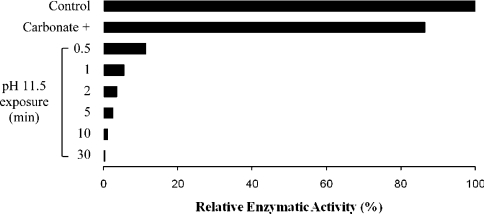
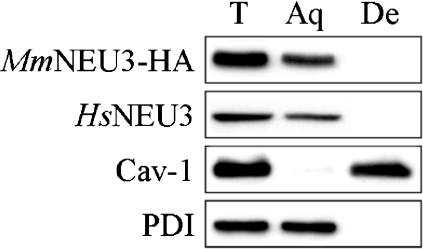
In order to investigate whether alkaline extraction of MmNEU3–HA could be somehow ascribed to the overexpression of the heterologous protein, solubilization of the human endogenous protein HsNEU3 was analysed in untransfected (Mock) HeLa cells. For this purpose, we took advantage of a rabbit HsNEU3 antiserum that specifically recognizes the human protein in immunoblotting experiments [45]. The endogenous enzyme showed a roughly superimposable behaviour compared with MmNEU3–HA, with a complete extraction of the protein only after alkaline treatment (Figure 5). However, using both the artificial 4MU-NeuAc and the [3H]GD1a ganglioside as substrates, almost no sialidase activity could be detected in the particulate and in the soluble fractions after 30 min of carbonate treatment, although samples were adjusted to pH 7.5 immediately after ultracentrifugation (Table 1). Indeed, the enzymatic activity measured after carbonate extraction was markedly lower compared with that measured under control experimental conditions (3–7%, when assayed with [3H]GD1a). Loss of activity might be ascribed either to the presence of carbonate in the enzyme assay mixture or to exposure of the protein to the alkaline pH. We therefore determined the sialidase activity of total membranes by adding buffered carbonate (see the Experimental section), at the appropriate final concentration, to the enzymatic assay mixture before the addition of the enzyme source. A small reduction in sialidase activity was observed as compared with that observed under control conditions (Figure 6), indicating that the presence of carbonate in the reaction mixture is compatible with the catalytic activity. We then subjected total membranes to alkaline pH treatment for different time intervals and, after adjustment to pH 7.5, tested the sialidase activity. As shown in Figure 6, already after 1 min exposure to alkaline pH, more than 90% of the enzymatic activity was lost. By considering that the enzymatic activity was determined without separation of the soluble and membranous fractions, we can conclude that loss of catalytic activity can be ascribed to exposure to alkaline pH. To gain further information about the mechanism of NEU3 membrane anchoring, HeLa cells expressing MmNEU3–HA were subjected to Triton X-114 extraction followed by aqueous/detergent phase separation, and the distribution of the protein was analysed by Western blotting. These experimental conditions allow the separation of hydrophilic peripheral membrane proteins from transmembrane, GPI-anchored and fatty acyl- or prenyl-anchored membrane proteins [35,46]. After phase separation, we observed a significant enrichment or a complete segregation of MmNEU3–HA in the aqueous phase (Figure 7). The same results were obtained for the endogenous HsNEU3 in HeLa cells. On the other hand, Cav-1 that is associated with membranes by a hydrophobic domain and palmitoylations [38], segregates exclusively in the detergent phase. As in the case of carbonate extraction, we were not able to recover any activity in either the single or the reconstituted fractions after Triton X-114 phase separation, regardless of the substrate used in the assay. In addition to these findings, treatment of membranes derived from transfected cells with high-ionic-strength solution (1 M KCl) or with bivalent cation chelator (25 mM EGTA) did not result in any solubilization of sialidase NEU3 (results not shown).
Figure 6. Sialidase NEU3 enzymatic activity is highly sensitive to alkaline pH.
NEU3 enzymatic activity toward the artificial substrate 4MU-NeuAc was assayed using total cell membranes in presence of buffered carbonate in the reaction mixture (Carbonate +) or following exposure to alkaline pH for different time periods (0.5–30 min). Values are given as the percentage of recovered activity related to control conditions. Results are representative of two independent experiments.
Figure 7. Sialidase NEU3 is a hydrophilic protein.
Total cell extracts derived from HeLa cells transfected with MmNEU3–HA were extracted and subjected to phase separation with Triton X-114. Aliquots of the aqueous (Aq) and detergent (De) phases were then analysed by Western blotting using anti-HA, anti-HsNEU3, anti-Cav-1 and anti-PDI antibodies.
Overall, these findings demonstrate that: (i) NEU3 behaves as a hydrophilic peripheral protein; (ii) its biological activity is highly sensitive to alkaline pH; and (iii) the interaction of the protein with the membrane does not seem to depend either on direct or calcium-mediated electrostatic interactions with phospholipids.
DISCUSSION
The plasma-membrane-associated sialidase NEU3 is known to be highly active towards gangliosidic substrates [3,5,6,34] and to be involved in several biological processes, possibly through the modulation of sialic acid content at the cell surface. Sialidase NEU3 is present in lipid rafts and co-fractionates with the lipid raft marker Cav-1 [9,10]. These organized subsets of the membrane represent specialized areas where cell–cell interactions and signal transduction events take place [13,26]. Up-regulation of sialidase NEU3 has been correlated with apoptosis suppression in human colon carcinoma [47], in renal carcinoma [25], as well as in other tumour cell lines [28]. In addition, it has been recently demonstrated that silencing of NEU3 results in the activation of apoptosis mechanisms in tumour-derived cells [48]. Also, sialidase NEU3 plays an important role in normal cellular processes such as axonal growth and regeneration [27], as well as axonal fate in unpolarized neurons [49]. Transfection and overexpression of sialidase NEU3 induces dramatic changes in the ganglioside composition of transfected cells and in neighbouring non-transfected cells [6], supporting the hypothesis that the protein is associated with the external leaflet of the plasma membrane.
In the present study, we provide the first direct demonstration that sialidase NEU3 is topologically associated with the outer leaflet of the plasma membrane, where its natural gangliosidic substrates are present. In fact, after cell-surface protein biotinylation, sialidase NEU3 was recovered in the biotinylated fraction. However, we also found that most sialidase NEU3 was not accessible to biotin, suggesting that the protein is present in intracellular compartments too. We confirmed this hypothesis by indirect immunofluorescence experiments coupled to laser confocal microscopic analysis where we observed a diffused cytoplasmic labelling for sialidase NEU3, with a specific localization in vesicular structures. The presence of sialidase NEU3 in intracellular structures had been observed previously by Yamaguchi et al. [8], but no precise topological identification was given. We could demonstrate that a subset of the intracellular labelling corresponds to the endosomal compartment, since intracellular NEU3-positive structures were found to co-localize with EEA1 and TfR. These results were independent of the cell type used (HeLa and COS-7 cells) and of the expression level of the protein, indicating that the presence of sialidase NEU3 in the endosomal compartment is not a consequence of any possible impairment of intracellular trafficking and distribution of the protein, due to transfection. Possibly, sialidase NEU3 is present in different cellular pools in a dynamic equilibrium with each other, and such a possibility is strongly suggested by the specific internalization of exogenously administrated antibody recognizing the HA epitope of the transfected protein. Interestingly, internalized antibody co-localized with TfR at the recycling endosomal compartment, indicating further a possible dynamic equilibrium between these two pools of protein. It has been demonstrated recently that sialidase NEU3 dynamically accumulates at peripheral areas of the plasma membrane upon stimulation of starved cells with EGF (epidermal growth factor) [8]. Under these experimental conditions, sialidase NEU3 co-localizes with Rac-1, a small GTPase protein that is involved in cell motility events. This indicates that sialidase NEU3 undergoes a specific recruitment to membrane ruffles upon external stimuli. Nothing is known concerning the possibility that sialidase NEU3 may traffic between the plasma membrane and intracellular compartments and whether this event is dependent on the presence of extracellular factors. Our findings that, at steady-state, the protein is present both at the plasma membrane and in the recycling endosomal compartment and that NEU3 internalizes from the plasma membrane, strongly supports such a possibility. The molecular mechanisms by which sialidase NEU3 is internalized and factors that may regulate this phenomenon are under investigation.
Moreover, the mechanism of sialidase NEU3 anchorage to the lipid bilayer was studied. The bioinformatic analysis of the MmNEU3 amino acid sequence excludes the possibility that the protein is a member of the GPI-anchored protein family or is a fatty acylated or prenylated protein because none of the consensus sequences necessary for these post-translational modifications have been found. This evidence adds to the already established lack of transmembrane domains in sialidase NEU3 [2]. Therefore we considered the possibility that sialidase NEU3 would associate with the lipid bilayer as a peripheral membrane protein. These are hydrophilic proteins that associate with membranes usually by ionic and/or hydrogen interactions between the protein itself and either phospholipids (protein–lipid interactions) or with integral membrane proteins (protein–protein interactions). In any case, peripheral membrane proteins can be released by different treatments such as extreme pH, high ionic strength and chaotropic agents, without disruption of the lipid bilayer organization [50]. Extracted peripheral proteins can be then isolated from intrinsic membrane proteins by ultracentrifugation. Under control conditions, sialidase NEU3 remained associated with the particulate fraction and fully maintained its catalytic activity. Alkaline treatment [41] resulted in the extraction of the peripheral membrane protein EEA1 [44], as well as of both MmNEU3 and HsNEU3, as assessed by Western blot analysis. The same treatment did not affect at all the association to membranes of Cav-1, an integral membrane protein. We were not able to recover appreciable activity in the soluble and particulate fractions obtained after carbonate extraction, although the incubating conditions for the enzyme assay were optimal. Loss of activity can not be ascribed either to carbonate itself or to the separation between the soluble and the particulate fractions. Presumably, the enzyme is highly sensitive to exposure to alkaline pH, then becoming inactive.
In order to support further the hydrophilic nature of NEU3 we analysed the partitioning of the protein between the aqueous and detergent phases obtained after protein extraction in the presence of Triton X-114, followed by temperature-induced phase separation. By this method, hydrophilic proteins partition in the aqueous phase, whereas proteins that associate with biological membranes via fatty acyl or prenyl chains, GPI anchors or transmembrane domains distribute into the detergent phase [35,46]. Triton X-114 treatment allows the enrichment of both the murine and the HsNEU3 in the aqueous phase, thus demonstrating the hydrophilic nature of the two proteins. However, also in this case, phase separation results in a complete lack of NEU3 enzymatic activity. A possible explanation could reside in the segregation of the enzyme in the aqueous phase and a putative cofactor, important for catalytic activity, in the detergent phase, although upon pooling the two phases the enzymatic activity could not be recovered. Possibly, the lack of activity in the reconstituted system can be ascribed to the Triton X-114 insolubility at temperatures higher than 0 °C. In addition, when membranes were incubated in presence of high salt concentrations or treated with a bivalent cation chelator (results not shown), sialidase NEU3 was not released from the lipid bilayer. As a whole, these observations clearly demonstrate that NEU3 is a hydrophilic peripheral membrane protein.
In summary, in the present study, we give the first direct demonstration that sialidase NEU3 is a peripheral membrane protein that resides on the external leaflet of the plasma membrane. Moreover, the protein is localized also in intracellular compartments that are in relation with the endocytic/recycling pathway. Presence of the protein in these two different cellular compartments suggests a possible dynamic equilibrium between the different pools of NEU3. This hypothesis is suggested by the evidence that NEU3 can internalize from the cell surface and then localize to the recycling compartment. Finally, given the hydrophilic nature of the protein and the phospholipid-independent mechanism of the association with membranes, it is possible that NEU3 may interact with an integral protein partner. The latter may also represent the driving force for the recycling of NEU3 in relation to extracellular stimuli, thus regulating the presence of sialidase NEU3 on the cell surface. The latter aspect may be critical in relation to the role of NEU3 and gangliosides in cell growth and malignant transformation.
Acknowledgments
We thank Dr Nicholas Stamatos (Institute of Human Virology, University of Maryland, Baltimore, MD 21201, U.S.A.) for providing the HsNEU3 antiserum and Dr Jean Grünberg (Biochemistry Department, University of Geneva, Geneva, Switzerland) for the anti-LBPA antibody. We also thank Mario P. Franguelli for technical support. This work was supported by a grant from FIRB (Fondo Investimenti Ricerca di Base) 2001 to A. P. and from MIUR (Ministero dell'Istruzione, dell'Università e della Ricerca Scientifica)-PRIN (Progetti di Ricerca di Interesse Nazionale) 2004 to E. M. and G. T.
References
- 1.Saito M., Yu R. K. Biochemistry and function of sialidases. In: Rosenberg A., editor. Biology of the Sialic Acids. New York: Plenum Press; 1995. pp. 261–313. [Google Scholar]
- 2.Monti E., Preti A., Venerando B., Borsani G. Recent development in mammalian sialidase molecular biology. Neurochem. Res. 2002;27:649–663. doi: 10.1023/a:1020276000901. [DOI] [PubMed] [Google Scholar]
- 3.Miyagi T., Wada T., Iwamatsu A., Hata K., Yoshikawa Y., Tokuyama S., Sawada M. Molecular cloning and characterization of a plasma membrane-associated sialidase specific for gangliosides. J. Biol. Chem. 1999;274:5004–5011. doi: 10.1074/jbc.274.8.5004. [DOI] [PubMed] [Google Scholar]
- 4.Wada T., Yoshikawa Y., Tokuyama S., Kuwabara M., Akita H., Miyagi T. Cloning, expression, and chromosomal mapping of a human ganglioside sialidase. Biochem. Biophys. Res. Commun. 1999;261:21–27. doi: 10.1006/bbrc.1999.0973. [DOI] [PubMed] [Google Scholar]
- 5.Monti E., Bassi M. T., Papini N., Riboni M., Manzoni M., Venerando B., Croci G., Preti A., Ballabio A., Tettamanti G., Borsani G. Identification and expression of NEU3, a novel human sialidase associated to the plasma membrane. Biochem. J. 2000;349:343–351. doi: 10.1042/0264-6021:3490343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papini N., Anastasia L., Tringali C., Croci G., Bresciani R., Yamaguchi K., Miyagi T., Preti A., Prinetti A., Prioni S., et al. The plasma membrane-associated sialidase MmNEU3 modifies the ganglioside pattern of adjacent cells supporting its involvement in cell-to-cell interactions. J. Biol. Chem. 2004;279:16989–16995. doi: 10.1074/jbc.M400881200. [DOI] [PubMed] [Google Scholar]
- 7.Valaperta R., Chigorno V., Basso L., Prinetti A., Bresciani R., Preti A., Miyagi T., Sonnino S. Plasma membrane production of ceramide from ganglioside GM3 in human fibroblasts. FASEB J. 2006;20:1227–1229. doi: 10.1096/fj.05-5077fje. [DOI] [PubMed] [Google Scholar]
- 8.Yamaguchi K., Hata K., Wada T., Moriya S., Miyagi T. Epidermal growth factor-induced mobilization of a ganglioside-specific sialidase (NEU3) to membrane ruffles. Biochem. Biophys. Res. Commun. 2006;346:484–490. doi: 10.1016/j.bbrc.2006.05.136. [DOI] [PubMed] [Google Scholar]
- 9.Kalka D., von Reitzenstein C., Kopitz J., Cantz M. The plasma membrane ganglioside sialidase cofractionates with markers of lipid rafts. Biochem. Biophys. Res. Commun. 2001;283:989–993. doi: 10.1006/bbrc.2001.4864. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y., Yamaguchi K., Wada T., Hata K., Zhao X., Fujimoto T., Miyagi T. A close association of the ganglioside-specific sialidase Neu3 with caveolin in membrane microdomains. J. Biol. Chem. 2002;277:26252–26259. doi: 10.1074/jbc.M110515200. [DOI] [PubMed] [Google Scholar]
- 11.Simons K., Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 12.Munro S. Lipid rafts: elusive or illusive? Cell. 2003;115:377–388. doi: 10.1016/s0092-8674(03)00882-1. [DOI] [PubMed] [Google Scholar]
- 13.Hakomori S., Handa K. Glycosphingolipid-dependent cross-talk between glycosynapses interfacing tumor cells with their host cells: essential basis to define tumor malignancy. FEBS Lett. 2002;531:88–92.. doi: 10.1016/s0014-5793(02)03479-8. [DOI] [PubMed] [Google Scholar]
- 14.Toledo M. S., Suzuki E., Handa K., Hakomori S. Cell growth regulation through GM3-enriched microdomain (glycosynapse) in human lung embryonal fibroblast WI38 and its oncogenic transformant VA13. J. Biol. Chem. 2004;279:34655–34664. doi: 10.1074/jbc.M403857200. [DOI] [PubMed] [Google Scholar]
- 15.Bektas M., Spiegel S. Glycosphingolipids and cell death. Glycoconjugate J. 2004;20:39–47. doi: 10.1023/B:GLYC.0000016741.88476.8b. [DOI] [PubMed] [Google Scholar]
- 16.Bieberich E. Integration of glycosphingolipid metabolism and cell-fate decisions in cancer and stem cells: review and hypothesis. Glycoconjugate J. 2004;21:315–327. doi: 10.1023/B:GLYC.0000046274.35732.47. [DOI] [PubMed] [Google Scholar]
- 17.Mitsuzuka K., Handa K., Satoh M., Arai Y., Hakomori S. A specific microdomain (“glycosynapse 3”) controls phenotypic conversion and reversion of bladder cancer cells through GM3-mediated interaction of α3β1 integrin with CD9. J. Biol. Chem. 2005;280:35545–35553. doi: 10.1074/jbc.M505630200. [DOI] [PubMed] [Google Scholar]
- 18.d'Azzo A., Tessitore A., Sano R. Gangliosides as apoptotic signals in ER stress response. Cell Death Differ. 2006;13:404–414. doi: 10.1038/sj.cdd.4401834. [DOI] [PubMed] [Google Scholar]
- 19.Sonnino S., Chigorno V. Ganglioside molecular species containing C18 and C20-sphingosine in mammalian nervous tissues and neuronal cell cultures. Biochim. Biophys. Acta. 2000;1469:63–77. doi: 10.1016/s0005-2736(00)00210-8. [DOI] [PubMed] [Google Scholar]
- 20.Malisan F., Testi R. GD3 ganglioside and apoptosis. Biochim. Biophys. Acta. 2002;1585:179–187. doi: 10.1016/s1388-1981(02)00339-6. [DOI] [PubMed] [Google Scholar]
- 21.Ono M., Handa K., Sonnino S., Withers D. A., Nagai H., Hakomori S. GM3 ganglioside inhibits CD9-facilitated haptotactic cell motility: coexpression of GM3 and CD9 is essential in the downregulation of tumor cell motility and malignancy. Biochemistry. 2001;40:6414–6421. doi: 10.1021/bi0101998. [DOI] [PubMed] [Google Scholar]
- 22.Lang Z., Guerrera M., Li R., Ladisch S. Ganglioside GD1a enhances VEGF-induced endothelial cell proliferation and migration. Biochem. Biophys. Res. Commun. 2001;282:1031–1037. doi: 10.1006/bbrc.2001.4630. [DOI] [PubMed] [Google Scholar]
- 23.Kawakami Y., Kawakami K., Steelant W. F., Ono M., Baek R. C., Handa K., Withers D. A., Hakomori S. Tetraspanin CD9 is a “proteolipid,” and its interaction with α3 integrin in microdomain is promoted by GM3 ganglioside, leading to inhibition of laminin-5-dependent cell motility. J. Biol. Chem. 2002;277:34349–34358. doi: 10.1074/jbc.M200771200. [DOI] [PubMed] [Google Scholar]
- 24.Wang X. Q., Sun P., Paller A. S. Gangliosides inhibit urokinase-type plasminogen activator (uPA)-dependent squamous carcinoma cell migration by preventing uPA receptor/αβ integrin/epidermal growth factor receptor interactions. J. Invest. Dermatol. 2005;124:839–848. doi: 10.1111/j.0022-202X.2005.23669.x. [DOI] [PubMed] [Google Scholar]
- 25.Ueno S., Saito S., Wada T., Yamaguchi K., Satoh M., Arai Y., Miyagi T. Plasma membrane-associated sialidase is up-regulated in renal cell carcinoma and promotes interleukin-6-induced apoptosis suppression and cell motility. J. Biol. Chem. 2006;281:7756–7764. doi: 10.1074/jbc.M509668200. [DOI] [PubMed] [Google Scholar]
- 26.Hakomori S. The glycosynapse. Proc. Natl. Acad. Sci. U.S.A. 2002;99:225–232. doi: 10.1073/pnas.012540899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodriguez J. A., Piddini E., Hasegawa T., Miyagi T., Dotti C. G. Plasma membrane ganglioside sialidase regulates axonal growth and regeneration in hippocampal neurons in culture. J. Neurosci. 2001;21:8387–8395. doi: 10.1523/JNEUROSCI.21-21-08387.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyagi T., Wada T., Yamaguchi K., Hata K. Sialidase and malignancy. Glycoconjugate J. 2004;20:189–198. doi: 10.1023/B:GLYC.0000024250.48506.bf. [DOI] [PubMed] [Google Scholar]
- 29.Chavas L. M., Tringali C., Fusi P., Venerando B., Tettamanti G., Kato R., Monti E., Wakatsuki S. Crystal structure of the human cytosolic sialidase Neu2: evidence for the dynamic nature of substrate recognition. J. Biol. Chem. 2005;280:469–475. doi: 10.1074/jbc.M411506200. [DOI] [PubMed] [Google Scholar]
- 30.Magesh S., Suzuki T., Miyagi T., Ishida H., Kiso M. Homology modeling of human sialidase enzymes NEU1, NEU3 and NEU4 based on the crystal structure of NEU2: hints for the design of selective NEU3 inhibitors. J. Mol. Graphics Modell. 2006;25:196–207. doi: 10.1016/j.jmgm.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 31.Tusnady G. E., Simon I. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J. Mol. Biol. 1998;283:489–506. doi: 10.1006/jmbi.1998.2107. [DOI] [PubMed] [Google Scholar]
- 32.Eisenhaber B., Bork P., Eisenhaber F. Prediction of potential GPI-modification sites in proprotein sequences. J. Mol. Biol. 1999;292:741–758. doi: 10.1006/jmbi.1999.3069. [DOI] [PubMed] [Google Scholar]
- 33.Resh M. D. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta. 1999;1451:1–16. doi: 10.1016/s0167-4889(99)00075-0. [DOI] [PubMed] [Google Scholar]
- 34.Hasegawa T., Yamaguchi K., Wada T., Takeda A., Itoyama Y., Miyagi T. Molecular cloning of mouse ganglioside sialidase and its increased expression in neuro2a cell differentiation. J. Biol. Chem. 2000;275:14778. doi: 10.1074/jbc.275.11.8007. [DOI] [PubMed] [Google Scholar]
- 35.Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- 36.Ghidoni R., Sonnino S., Masserini M., Orlando P., Tettamanti G. Specific tritium labeling of gangliosides at the 3-position of sphingosine. J. Lipid Res. 1981;22:1286–1295. [PubMed] [Google Scholar]
- 37.Schlegel A., Lisanti M. P. A molecular dissection of caveolin-1 membrane attachment and oligomerization: two separate regions of the caveolin-1 C-terminal domain mediate membrane binding and oligomer/oligomer interactions in vivo. J. Biol. Chem. 2000;275:21605–21617. doi: 10.1074/jbc.M002558200. [DOI] [PubMed] [Google Scholar]
- 38.Parton R. G., Hanzal-Bayer M., Hancock J. F. Biogenesis of caveolae: a structural model for caveolin-induced domain formation. J. Cell Sci. 2006;119:787–796. doi: 10.1242/jcs.02853. [DOI] [PubMed] [Google Scholar]
- 39.Trowbridge I. S. Endocytosis and signals for internalization. Curr. Opin. Cell Biol. 1991;3:634–641. doi: 10.1016/0955-0674(91)90034-v. [DOI] [PubMed] [Google Scholar]
- 40.Sapay N., Guermeur Y., Deleage G. Prediction of amphipathic in-plane membrane anchors in monotopic proteins using a SVM classifier. BMC Bioinformatics. 2006;7:255. doi: 10.1186/1471-2105-7-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fujiki Y., Hubbard A. L., Fowler S., Lazarow P. B. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Funaki T., Fujiwara T., Hong H. S., Misumi Y., Nishioka M., Ikehara Y. Identification and characterization of a 230-kDa Golgi-associated protein recognized by autoantibodies from a patient with HBV hepatitis. Cell Struct. Funct. 1996;21:63–72. doi: 10.1247/csf.21.63. [DOI] [PubMed] [Google Scholar]
- 43.Morel-Huaux V. M., Pypaert M., Wouters S., Tartakoff A. M., Jurgan U., Gevaert K., Courtoy P. J. The calcium-binding protein p54/NEFA is a novel luminal resident of medial Golgi cisternae that traffics independently of mannosidase II. Eur. J. Cell Biol. 2002;81:87–100. doi: 10.1078/0171-9335-00224. [DOI] [PubMed] [Google Scholar]
- 44.Mu F. T., Callaghan J. M., Steele-Mortimer O., Stenmark H., Parton R. G., Campbell P. L., McCluskey J., Yeo J. P., Tock E. P., Toh B. H. EEA1, an early endosome-associated protein. EEA1 is a conserved α-helical peripheral membrane protein flanked by cysteine “fingers” and contains a calmodulin-binding IQ motif. J. Biol. Chem. 1995;270:13503–13511. doi: 10.1074/jbc.270.22.13503. [DOI] [PubMed] [Google Scholar]
- 45.Stamatos N. M., Liang F., Nan X., Landry K., Cross A. S., Wang L. X., Pshezhetsky A. V. Differential expression of endogenous sialidases of human monocytes during cellular differentiation into macrophages. FEBS J. 2005;272:2545–2556. doi: 10.1111/j.1742-4658.2005.04679.x. [DOI] [PubMed] [Google Scholar]
- 46.Hooper N. M., Bashir A. Glycosyl-phosphatidylinositol-anchored membrane proteins can be distinguished from transmembrane polypeptide-anchored proteins by differential solubilization and temperature-induced phase separation in Triton X-114. Biochem. J. 1991;280:745–751. doi: 10.1042/bj2800745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kakugawa Y., Wada T., Yamaguchi K., Yamanami H., Ouchi K., Sato I., Miyagi T. Up-regulation of plasma membrane-associated ganglioside sialidase (Neu3) in human colon cancer and its involvement in apoptosis suppression. Proc. Natl. Acad. Sci. U.S.A. 2002;99:10718–10723. doi: 10.1073/pnas.152597199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wada T., Hata K., Yamaguchi K., Shiozaki K., Koseki K., Moriya S., Miyagi T. A crucial role of plasma membrane-associated sialidase in the survival of human cancer cells. Oncogene. 2007;26:2483–2490. doi: 10.1038/sj.onc.1210341. [DOI] [PubMed] [Google Scholar]
- 49.Da Silva J. S., Hasegawa T., Miyagi T., Dotti C. G., Abad-Rodriguez J. Asymmetric membrane ganglioside sialidase activity specifies axonal fate. Nat. Neurosci. 2005;8:606–615. doi: 10.1038/nn1442. [DOI] [PubMed] [Google Scholar]
- 50.Jennings M. L. Topography of membrane proteins. Annu. Rev. Biochem. 1989;58:999–1027. doi: 10.1146/annurev.bi.58.070189.005031. [DOI] [PubMed] [Google Scholar]