Abstract
The enamel matrix protein amelogenin is secreted by ameloblasts into the extracellular space to guide the formation of highly ordered hydroxyapatite mineral crystallites, and, subsequently, is almost completely removed during mineral maturation. Amelogenin interacts with the transmembrane proteins CD63 and LAMP (lysosome-associated membrane protein) 1, which are involved in endocytosis. Exogenously added amelogenin has been observed to move rapidly into CD63/LAMP1-positive vesicles in cultured cells. In the present study, we demonstrate the protein region defined by amino acid residues 103–205 for CD63 interacts not only with amelogenin, but also with other enamel matrix proteins (ameloblastin and enamelin). A detailed characterization of binding regions in amelogenin, CD63 and LAMP1 reveals that the amelogenin region defined by residues PLSPILPELPLEAW is responsible for the interaction with CD63 through residues 165–205, with LAMP1 through residues 226–251, and with the related LAMP2 protein through residues 227–259. We predict that the amelogenin binding region is: (i) hydrophobic; (ii) largely disordered; and (iii) accessible to the external environment. In contrast, the binding region of CD63 is likely to be organized in a ‘7’ shape within the mushroom-like structure of CD63 EC2 (extracellular domain 2). In vivo, the protein interactions between the secreted enamel matrix proteins with the membrane-bound proteins are likely to occur at the specialized secretory surfaces of ameloblast cells called Tomes' processes. Such protein–protein interactions may be required to establish short-term order of the forming matrix and/or to mediate feedback signals to the transcriptional machinery of ameloblasts and/or to remove matrix protein debris during enamel biomineralization.
Keywords: amelogenin, CD63, enamel matrix, lysosome-associated membrane protein 1 (LAMP1), protein–protein interaction, yeast two-hybrid assay
Abbreviations: AD, activation domain; EC, extracellular domain; LAMP, lysosome-associated membrane protein; LRAP, leucine-rich amelogenin peptide; Y2H, yeast two-hybrid
INTRODUCTION
With the identification of a cDNA sequence for amelogenin in 1983 [1] and the subsequent discoveries of additional organic components of the enamel extracellular matrix, including ameloblastin [2], enamelin [3] and amelotin [4], our understanding of enamel formation has advanced significantly. The expression of these structural proteins remains relatively unique to the developing tooth organ; primarily to the ectoderm-derived enamel layer during amelogenesis, but amelogenin, ameloblastin and enamelin are also transiently expressed in ectomesenchyme-derived dentin during dentinogenesis [5,6]. Certain proteases, including the serine protease kallikrein-4 [7] and the matrix metalloproteinase 20 [8], are described as relatively unique to the developing enamel. The spatiotemporal expression for each of these enamel proteins continues to be defined. Of these proteins, amelogenin contributes more than 90% of the bulk of the organic matrix, and is absolutely essential for proper enamel formation [9,10]. Amelogenin is secreted by ameloblasts into the extracellular space to guide hydroxyapatite crystal formation and is subsequently almost completely removed during the enamel maturation.
At the ameloblast Tomes' processes, protein–protein interactions occur between the secreted enamel matrix proteins and membrane-bound proteins. The enamel matrix proteins amelogenin, ameloblastin and enamelin each interact with CD63 in vitro [11]. CD63 is a member of the tetraspanin family, in which most members are cell-surface proteins that are characterized by the presence of four transmembrane segments [12]. The tetraspanin proteins mediate signal transduction events that play roles in the regulation of cell development, activation, growth and motility [13]. CD63, and other tetraspanins, are known to form complexes with integrins and act as organizers of membrane microdomains and signalling complexes [13,14]. CD63 resides not only in the cytoplasmic membranes of most cell types, but also in late endosomes, lysosomes and secretory vesicles, and traffics among these different compartments [15]. This has led to the suggestion that CD63 plays a role in the recycling of membrane components and the uptake of degraded proteins from the extracellular matrix [15].
Amelogenin interacts with LAMP (lysosome-associated membrane protein) 1 [16]. LAMP1 is a transmembrane protein that is highly expressed in late endosomes and lysosomes and is often used as a marker for these two organelles [17]. LAMP1 immunoreactivity is also observed at the plasma membrane and in early endocytic compartments [18]. The presence of LAMP1 on the plasma membrane is suggestive of LAMP1 acting as a cell-surface intermediate that can be shuttled to the lysosome through endocytosis. Thus LAMP1 may be involved in endocytosis, pinocytosis or phagocytosis [17]. Recent studies have shown that exogenously added amelogenin moves rapidly into the CD63/LAMP1-positive vesicles that subsequently localize to the perinuclear region [19]. Collectively, these observations suggest a possible mechanism by which amelogenin, or degraded amelogenin peptides, can be removed from the extracellular matrix during enamel formation and maturation through direct interaction with CD63 and LAMP1 at the ameloblast Tomes' processes, and the subsequent trafficking into the cell cytoplasm [19]. In the present study, we have identified the protein–protein-interaction regions in amelogenin, CD63 and LAMP1, and explore possible structural information of the binding regions using computational biology methods.
MATERIALS AND METHODS
Detection of protein–protein interactions by Y2H (yeast two-hybrid) assay
The Y2H assay system was used to identify and to confirm interactions between enamel matrix proteins and membrane-bound proteins. The Y2H assay system uses two vectors, pGBKT7 and pGADT7 (BD Biosciences Clontech). The pGBKT7 vector expresses proteins, or protein regions, fused to the GAL4 DNA-binding domain, whereas the pGADT7 vector expresses proteins, or protein regions, fused to the GAL4 AD (activation domain). The GAL4-AD-fusion protein is targeted to the yeast nucleus by the SV40 (simian virus 40) nuclear localization sequence. The native signal peptides of proteins studied here were excluded to ensure that the fusion proteins were transported into the nucleus.
The cDNA sequences and open reading frame information for the enamel matrix proteins and membrane-bound proteins used in the present study can be accessed from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). The protein sequences and nomenclature are illustrated in Figure 1.
Figure 1. Amino acid sequences for amelogenins M180 and LRAP, and for CD63, LAMP1 and LAMP2.
The protein accession numbers and amino acid sequences are listed for mouse amelogenin isoforms M180 (A) and LRAP (B), and human CD63 (C), LAMP1 (D) and LAMP2 (E). For amelogenin, exons 3 and 5 (amino acids 3–33) and exon 6D (amino acids 155–179) are identified. For CD63, EC1 (amino acids 33–51) and EC2 (amino acids 103–203) are identified. The protein–protein-interaction regions for all proteins are underlined.
The cloning of full-length genes of mouse amelogenin M180 (using the cDNA and protein reference numbers NM_009666 and NP_033796.1; the M180 refers to the 180-amino-acid amelogenin protein that excludes the 16-amino-acid N-terminal signal peptide), mouse enamelin (NM_017468) and rat ameloblastin (NM_012900) into pGBKT7 vectors has been described previously [11]. In the present study, various fragments of amelogenin M180 were cloned into pGBKT7. The entire open reading frames of human CD63 (NM_001780 coding for protein NP_001771.1) and human LAMP1 (NM_005561 coding for protein NP_005552.3), and cDNA fragments of CD63, LAMP1 and human LAMP2 (NM_002294 coding for protein NP_002285.1) were cloned into pGADT7 vectors. The PCR primers to coding regions of the genes were synthesized. All primers except for INV48566043, which targets to the pUC ori region of pGBKT7, were designed to contain restriction enzyme sites for the efficient and in-frame cloning (Table 1). The complete details of engineering the constructs is not given here, but are available from M.L.P. on request. All constructs were engineered such that the ‘defined’ amino acid regions immediately followed insertion at the multi-cloning site, and the C-terminal amino acid was immediately followed by a stop codon. The plasmid constructs were created following standard protocols [20]. The constructed plasmids were sequenced across their cloning sites and the entire insert to confirm correct orientation, sequence and reading frame. Each pair of candidate ‘bait’ constructs (pGBKT7-insert) and candidate ‘prey’ construct (pGADT7-insert) was co-transformed into the yeast host strain PCY2. The β-galactosidase activity was detected by the filter assay described previously [21].
Table 1. Primers used in the Y2H assays.
Protein regions encoded by cDNA PCR fragments were generated with the corresponding primers. The restriction enzyme sites are underlined.
| Primer name | Protein region | Direction | Oligonucleotide sequence (5′→3′) |
|---|---|---|---|
| PA24253 | Ameloblastin-(1–396) | Forward | CGCCATATGGTGCCGGCATTTCCTCAACAACCTGGG |
| PA24254 | Ameloblastin-(1–396) | Reverse | CGCGTCGACTCAGGGCTCTTGGAAACGCCATGC |
| PA27536 | Enamelin-(1–1236) | Forward | TTCTCGAGCCCGGGGCCATTCCAGATGCCAATGCCCCGA |
| PA27537 | Enamelin-(1–1236) | Reverse | ACGACCCCCAAAGCCGTGATATCC |
| PA24255 | Amelogenin-(1–180) | Forward | CCGGAATTCATGCCCCTACCACCTCATCCTGGA |
| PA24256 | Amelogenin-(1–180) | Reverse | CGCGTCGACTTAATCCACTTCTTCCCGCTTGGTCTTG |
| PA39335A | Amelogenin-(3–33) | Forward | GGAATTCCTACCACCTCATCCTGGAAGCCC |
| PA39335B | Amelogenin-(3–33) | Reverse | GGTCGACTCACGGCTGCCTTATCATGCTCTGG |
| PA39335C | Amelogenin-(155–179) | Forward | GGAATTCCCCCTGTCCCCCATTCTTCCTG |
| PA39335D | Amelogenin-(155–179) | Reverse | GGTCGACTCACACTTCTTCCCGCTTGGTC |
| INV48566044 | Amelogenin-(169–179) | Forward | GGAATTCCCAGCGACAGACAAGACCAAGC |
| INV48566043 | Amelogenin-(169–179) | Reverse | CCTGAGAAAGCAACCTGACCTACAGGA |
| INV47327732 | Amelogenin-(155–168) | Reverse | GGTCGACTCACCAAGCTTCCAGAGGCAGCTCAG |
| PA29499 | CD63-(1–238) | Forward | AGTGAATTCATGGCGGTGGAAGGAGGAATGAAATGTGTG |
| PA29500 | CD63-(1–238) | Reverse | CCTCGAGCTACATCACCTCGTAGCCACTTCTGATACTCTTCA |
| hCD63_103 | CD63-(103–205) | Forward | TGAATTCGCTGGCTATGTGTTTAGAGATAAGGTG |
| INV47327730 | CD63-(165–205) | Forward | TGAATTCGTCCCCGACTCCTGCTGC |
| INV47327734 | CD63-(165–205) | Reverse | GCTCGAGTCACACCAGCACATTTTTCCTCAGCC |
| PA39458A | LAMP1-(1–121) or -(1–386) | Forward | GCCCGGGTATGGCGCCCCGCTCTCCGC |
| PA39335F | LAMP1-(1–386) or -(342–386) | Reverse | GGGATCCTCAGATGGGGATCAGCGTGCTG |
| PA39458B | LAMP1-(1–121) | Reverse | GGGATCCTCAGTTATAAACAAAACTCATGAGCTGGACGC |
| PA39335H | LAMP1-(342–386) | Forward | GCCCGGGTGAGCACGTCCGTGTCACGAAG |
| PA39335I | LAMP1-(95–251) | Forward | GCCCGGGTAGAGGACATACACTCACTCTCAATTTC |
| PA39335J | LAMP1-(95–251) | Reverse | GGGATCCTCACGTCGTGTTGTCCTTCCTCTCATAG |
| PA39336A | LAMP1-(226–361) | Forward | GCCCGGGTGGCACCAACGGGACCTGC |
| PA39336B | LAMP1-(226–361) | Reverse | GGGATCCTCAAGCCTGGACCCACACTTTGAA |
| INV47327729 | LAMP1-(226–251) | Forward | TGAATTCGGCACCAACGGGACCTGC |
| INV47327731 | LAMP1-(226–251) | Reverse | GCTCGAGTCACGTCGTGTTGTCCTTCCTC |
| INV47327733 | LAMP2-(227–259) | Forward | TGAATTCAATGGCAATGATACTTGTCTGCTGGC |
| INV47327735 | LAMP2-(227–259) | Reverse | GCTCGAGTCAAGTTGTATTGGGGTTGATGTTAATAACTGAAGC |
Protein secondary structure and solvent accessibility prediction
The SABLE server [22] (http://sable.cchmc.org) was used to analyse the secondary structure and solvent accessibility of amelogenin M180. The SABLE server utilizes advanced machine learning protocols, evolutionary profiles and predicted RSA (relative solvent accessibility) to compute protein secondary structures [22]. The SABLE server has been trained with a subset of 860 protein families derived from the Pfam protein family database. The prediction accuracy was estimated to be between 77.0 and 78.4% for the three-state classification on different control sets comprising 603 proteins with no homology with proteins included in the training [22]. The prediction accuracy is continuously and independently evaluated to be 70–80% by the EVA web server (http://cubic.bioc.columbia.edu/eva/) [23].
Disorder propensity of amelogenin M180 prediction
The IUPred server [24] (http://iupred.enzim.hu) was used to predict the disorder propensity of amelogenin M180. The IUPred server recognizes disorder regions from the amino acid sequence based on the estimated pairwise energy content [24]. The underlying assumption of the IUPred server is that intrinsically unstructured proteins adopt no stable structure because their amino acid composition does not allow sufficient favourable interactions to form [24]. The IUPred server predicted experimentally determined disorder at approx. 90% accuracy at CASP6 [25].
The three-dimensional structure model of CD63 EC2 (extracellular domain 2)
The Protein Homology/analogY Recognition Engine (Phyre) is a successor of 3D-PSSM [26] (http://www.sbg.bio.ic.ac.uk/phyre), which is a web-based method for protein fold recognition using one- and three-dimensional sequence profiles coupled with secondary structure and solvation potential information. The prediction performance is continuously and independently evaluated by the EVA [23]. In the present study, Phyre was used to model three-dimensional structure of the EC2 of CD63, and the Accelrys® Discovery Studio Visualizer was used to generate presentations of the three-dimensional structure.
RESULTS
Determination of protein–protein-interaction regions with the Y2H assay
Previously, Y2H screening of a mouse embryo [E17 (embryonic day 17)] cDNA library in conjunction with mouse amelogenin and rat ameloblastin as baits identified the transmembrane protein CD63 as an interacting partner of both enamel proteins [11]. Subsequently, these protein–protein interactions were confirmed using the Y2H filter assay to assess protein interactions between human CD63 and (i) mouse amelogenin, (ii) mouse enamelin, and (iii) rat ameloblastin [11]. These previously identified protein–protein interactions were used in this study as positive controls (Table 2, rows b–d). Tompkins et al. [16] have identified the interaction between mouse amelogenin M59 and mouse LAMP1 by affinity pull-down and far-Western assays. In the present study, we used a Y2H assay to demonstrate the interaction of mouse full-length amelogenin M180 with human LAMP1 (Table 2, row e).
Table 2. Y2H assays for interactions between the enamel matrix proteins and membrane-bound proteins.
The DNA inserts in the pGBKT7 and pGADT7 vectors encode the corresponding protein fragments. In the β-galactosidase activity assay, a positive reaction evident after 6 h is recorded as ++, and a positive reaction evident only after 20 h is recorded as +.
| Row | Insert of pGBKT7 vector | Insert of pGADT7 vector | β-Galactosidase activity |
|---|---|---|---|
| a | No insert | No insert | − |
| b | Amelogenin-(1–180) | CD63-(1–238) | ++ |
| c | Ameloblastin-(1–396) | CD63-(1–238) | ++ |
| d | Enamelin-(1–1236) | CD63-(1–238) | ++ |
| e | Amelogenin-(1–180) | LAMP1-(1–386) | ++ |
| f | Amelogenin-(1–180) | CD63-(103–205) | ++ |
| g | Ameloblastin-(1–396) | CD63-(103–205) | ++ |
| h | Enamelin-(1–1236) | CD63-(103–205) | ++ |
| i | Amelogenin-(1–180) | CD63-(165–205) | ++ |
| j | Amelogenin-(3–33) | LAMP1-(1–121) | − |
| k | Amelogenin-(3–33) | LAMP1-(95–251) | − |
| l | Amelogenin-(3–33) | LAMP1-(226–361) | − |
| m | Amelogenin-(3–33) | LAMP1-(342–386) | − |
| n | Amelogenin-(3–33) | LAMP1-(1–386) | − |
| o | Amelogenin-(155–179) | LAMP1-(1–121) | (+)* |
| p | Amelogenin-(155–179) | LAMP1-(95–251) | ++† |
| q | Amelogenin-(155–179) | LAMP1-(226–361) | +† |
| r | Amelogenin-(155–179) | LAMP1-(342–386) | − |
| s | Amelogenin-(155–179) | LAMP1-(1–386) | ++ |
| t | Amelogenin-(155–168) | LAMP1-(226–251) | ++ |
| u | Amelogenin-(155–168) | LAMP2-(227–259) | ++ |
| v | Amelogenin-(155–168) | CD63-(165–205) | ++ |
| w | Amelogenin-(169–179) | LAMP1-(226–251) | − |
| x | Amelogenin-(169–179) | LAMP2-(227–259) | − |
| y | Amelogenin-(169–179) | CD63-(165–205) | − |
| z | p53 | SV40 large T-antigen | + |
* The reaction is noticeably weaker than the reaction at the row q.
† The LAMP1–amelogenin-interacting region was concluded to be the overlapping region of LAMP1 identified from the reactions seen in rows p and q.
Tetraspanin CD63 has two extracellular domains and three intracellular domains, with both the N- and C-termini located within the cytoplasm. The extracellular domains are named EC1 and EC2 and are defined as amino acids 33–51 and 103–203 respectively, based on the 238 amino acids in both mouse and human full-length CD63 protein (using the amino acid sequences and numbering identified by GenBank® accession numbers NP_031679.1 and NP_001771.1 respectively). The binding of CD63-(166–205) to amelogenin has been reported previously [11]. In the present study, amino acids 103–205 from the human CD63 protein were used to establish that amelogenin, ameloblastin and enamelin proteins each interacted with CD63-(103–205) (Table 2, rows f–h, and compared with negative and positive controls at rows a and z). Subsequently amino acids 165–205 from human CD63 were subcloned into pGADT7 and used to confirm a strong interaction of CD63-(165–205) with full-length amelogenin (Table 2, row i).
The amelogenin isoform M59, also known as LRAP (leucine-rich amelogenin peptide) [16], and M180 interact with LAMP1 (Table 2, row e). Since M59 is a partial subset of the amino acid residues contained within the larger M180 protein [27], we sought to identify the possible regions responsible for the amelogenin–LAMP1 interaction. We cloned the amelogenin cDNA fragments corresponding to exons 3 and 5 (amino acids 3–33) or the amino acids corresponding to exon 6D (residues 155–179) into the ‘bait’ vector (pGBKT7). We cloned the human LAMP1 cDNA fragments corresponding to amino acids 1–121, 95–251, 226–361 or 342–386 into the ‘prey’ vector (pGADT7). The C-terminal region of LAMP1 beyond amino acid 386 was not investigated because this region is not exposed to the extracellular environment. The β-galactosidase activity from the Y2H assay suggests that amelogenin-(155–179), as defined by exon 6D, is entirely responsible for the interaction of amelogenin with LAMP1 (Table 2, rows o–s compared with rows j–n). The two regions of LAMP1 showing the strongest interaction with amelogenin-(155–179) were LAMP1-(95–251) and -(226–361) (Table 2, rows p and q).
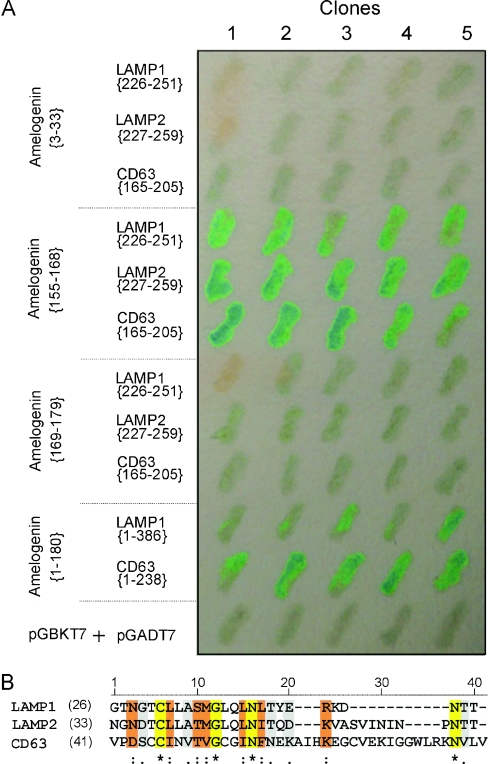
Fine mapping of amelogenin-(155–179) was performed by dividing further the domain between Trp168 and Pro169. This site was chosen since it corresponds to the primary proteolytic cleavage site responsible for the removal of the C-terminal teleopeptide from the full-length amelogenin [28]. Amelogenin-(155–168) and -(169–179) were tested against the following peptide regions: (i) the overlapping region of LAMP1 (amino acids 226–251) that showed the strongest interactions with amelogenin (Table 2, rows p and q); (ii) the highly homologous region of human LAMP2 defined by amino acids 227–259; and (iii) human CD63-(165–205). LAMP2 is predicted to exhibit a functional redundancy in vivo with LAMP1, as suggested by the mouse gene knockout study [29,30]. The results from the Y2H assay indicate that amelogenin-(155–168) was entirely responsible for the interaction with LAMP1-(226–251), LAMP2-(227–259) and CD63-(165–205) (Table 2, rows t–v, when compared with rows w–y). The assays in Table 2 were performed at various times, but always with the same positive and negative controls. A single Y2H filter assay highlighting these regions is shown (Figure 2A).
Figure 2. Identified interaction regions.
(A) Interaction confirmation by the filter assay. Five individual co-transformed clones from various Y2H experiments were assessed on one filter. (B) Alignment of the amelogenin-binding regions within LAMP1 (residues 226–251), LAMP2 (residues 227–259) and CD63 (residues 165–205). The alignment was produced with ClustalW using the Gonnet matrix. The identical, conserved substitution and semi-conserved substitution residues among those sequences are indicated with *, : and . respectively.
Regions of LAMP1 (amino acids 226–251), LAMP2 (amino acids 227–259) and CD63 (amino acids 165–205) all interact with the same small motif of amelogenin (P155LSPILPELPLEAW168). To assess whether similarities exist among the binding regions of LAMP1, LAMP2 and CD63, we aligned these peptide regions with ClustalW using the Gonnet matrix. The overall alignment shows moderate similarity (∼27%) within the three regions (Figure 2B). However, limiting the comparison to residues 3–17 shows a ∼60% amino acid similarity.
Bioinformatic analysis of the amelogenin-binding region
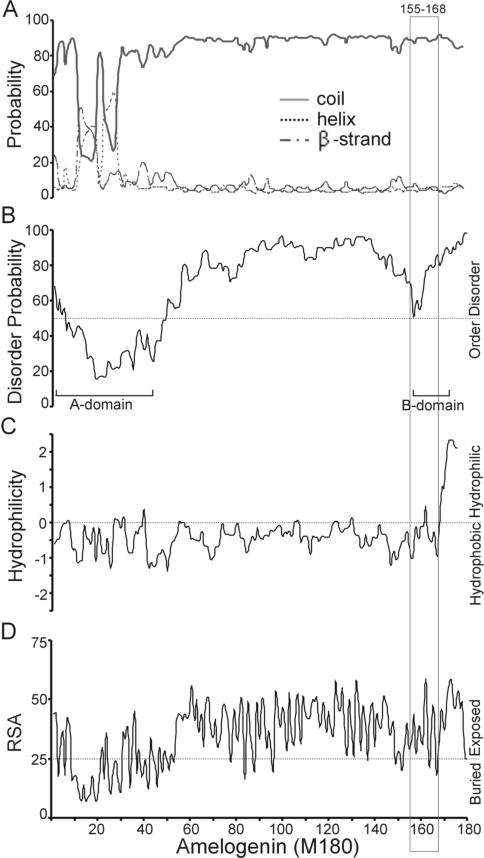
The three-dimensional molecular structure of amelogenin has not been determined. Our attempt to compute the three-dimensional molecular structure of amelogenin M180 failed to produce a reliable model. To obtain limited structural information for the amelogenin binding region (amino acids 155–168), we analysed the amelogenin protein sequence with various bioinformatics tools. The secondary structures of amelogenin M180 were predicted using SABLE (Figure 3A). We predict that there are two helices and one β-strand within the previously defined ‘A-domain’ (residues 1–42) [31]. The amelogenin region from amino acids 42 to 180 is predicted to be in a coil structure with a probability of 80–92%.
Figure 3. Structural analysis of amelogenin-(155–168) in the context of full-length M180.
(A) Probability of secondary-structure elements. The region comprising amino acids 155–168 is in the coil structure with greater than 80% probability. (B) Disorder probability. The region comprising amino acids 155–168 is largely disordered. (C) Hydrophilicity. The region comprising amino acids 155–168 is largely hydrophobic. (D) Relative solvent accessibility (RSA). The region comprising amino acids 155–168 is accessible. Also identified (in B) are the A-domain (residues 1–42) and B-domain (residues 157–173) of amelogenin.
The disorder probability for amelogenin M180 was predicted with the IUPred server (Figure 3B). The profiles for either the A-domain or the B-domain responsible for amelogenin self-assembly [31] are distinguishable from the remaining regions of amelogenin. The A-domain (residues 1–42) is predicted to be structured, whereas the remainder of the protein (amino acids 43–180) is predicted to be largely disordered. The amelogenin region (amino acids 155–168) was identified by the Y2H assay to be responsible for amelogenin interactions with members of the LAMP family and CD63, and corresponds largely to the previously identified self-assembly B-domain (residues 157–173) and is predicted to be disordered.
The hydrophilicity of amelogenin M180 was re-calculated with ProScale at the window size of 5 using Hopp and Woods values (Figure 3C) [1]. Almost the entire M180 protein, except for the short C-terminus (11 amino acids), is largely hydrophobic.
The relative solvent accessibility of amelogenin M180 was calculated with the SABLE server (Figure 3D). The majority of the buried regions of M180 are located within the A-domain, whereas, beyond the A-domain, the M180 protein is largely exposed. In short, the amelogenin M180-binding region (amino acids 155–168) is largely disordered, hydrophobic and accessible.
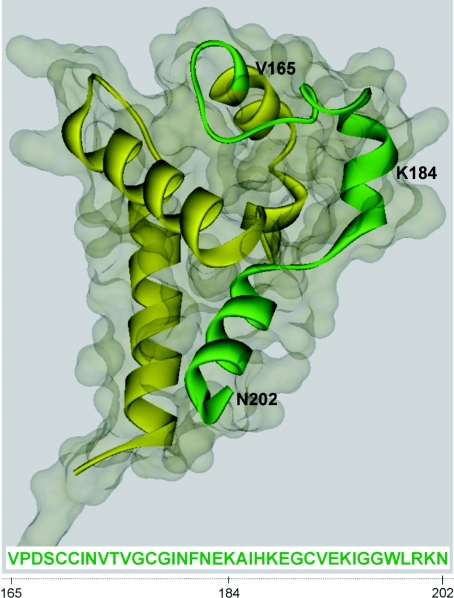
Three-dimensional structure modelling of the binding region in CD63
The molecular structure of the EC2 of tetraspanin CD81 has been determined using X-ray crystallography [32,33]. The atomic structure of CD81 EC2 was used as the template to build a three-dimensional model of CD63 EC2. The predicted three-dimensional structure of CD63 EC2 is presented in schematic backbone and solvent-accessible surface (Figure 4). As expected, the EC2 of CD63 is in a mushroom-like structure. The backbone of the amelogenin-binding region of CD63 coloured green is in a ‘7’ shape, and forms part of the ‘mushroom head’, as well as part of the ‘mushroom stalk’. The three residues V203LV205 are lacking in this model. These residues are very likely to be part of the transmembrane helix, as indicated by the computational analysis of the transmembrane region (results not shown).
Figure 4. Predicted three-dimensional structure for the EC2 of CD63.
The backbone structure is represented in the ribbon style within the solvent-accessible surface. The amelogenin-binding region is shown in green.
We were unable to build a reliable three-dimensional molecular structural model for LAMP1 and LAMP2 because the suitable structure template, which is normally derived from the experimentally determined structures, is currently unavailable. Both LAMP1 and LAMP2 belong to LAMP family and share similar domain structures. LAMP1-(226–251) and LAMP2-(227–259) regions are located within ‘Domain 2’ and are right next to the hinge region between Domain 1 and Domain 2.
DISCUSSION
The identification that amelogenin binds to CD63 [11] and LAMP1 [16] led us to hypothesize that amelogenin may be rapidly taken up by the ameloblast cell through receptor-mediated endocytosis [19]. The detailed characterization of the binding regions in amelogenin, CD63 and LAMP1 by the Y2H assay reveals that the region (P155LSPILPELPLEAW168) of amelogenin M180 is responsible for the interaction with CD63 through the domain specified by amino acids 165–205, LAMP1 through the domain specified by amino acids 226–251 and LAMP2 through the domain specified by amino acids 227–259. The binding region (residues 155–168) of amelogenin is hydrophobic, and is predicted to be largely disordered and accessible to the external environment. The region defined by amino acids 165–205 of CD63 is likely to be in a ‘7’ shape within the mushroom-like structure of CD63 EC2.
All of the data presented in Table 2 were derived from the Y2H assays. No significant ambiguity clouded our interpretation, as smaller regions of amelogenin, CD63 and LAMP1 were tested in multiple ways. However, we found that, for amelogenin, residues 155–179 interacted weakly with LAMP1-(1–121) (Table 2, row o). In this instance, there is a moderate similarity (∼38%) between LAMP1 amino acids 226–251 and 36–64 based on the ClustalW alignment. This weak homology is likely to explain the weak interaction noted. Fine dissection of regions of ameloblastin and enamelin responsible for binding to CD63, as well as the interaction of ameloblastin and enamelin with LAMP1/LAMP2, were not examined in the present study, and a separate investigation is needed to explore those interaction domains.
Attempts to characterize the structure of enamel matrix by X-ray crystallography have been made since the 1960s [10,34–39]. To date, the X-ray crystallographic structure of full-length amelogenin has yet to be solved [10]. The secondary structures of amelogenin have been characterized experimentally since the 1980s by CD and FTIR (Fourier-transform infrared) spectroscopy methods [10,40,41]. However, the composition and location for each of the selected structural elements remain vague. Secondary-structure predictions of M180 suggested that there are two helix and one β-strand element within the A-domain at the N-terminal region, whereas the remaining regions of amelogenin are in a random coil. The scarcity of highly ordered secondary structures (helix and β-strand), and the large random-coil region, imply that monomeric amelogenin M180 may not have a stable tertiary structure. Amelogenin M180 is rich in proline (24.4%) and glutamine (13.9%), and contains 13 Gln-Pro-Xaa repeats. This amino acid compositional bias and short repeats are characteristic of ‘intrinsically unstructured proteins’ [42], supporting our prediction for the propensity of mouse amelogenin M180 to exhibit a disordered structure. Our conclusion of ‘largely disordered’ is consistent with two previous reports on the structure of the enamel matrix: (i) “The organic material of dental enamel seems to be completely disordered at the molecular level” [35]; and (ii) “70% of the fetal enamel protein chains exhibit rapid (≤10−6 s), nearly isotropic molecular motion” [43]. Such disordered regions are quite often lacking from the electron-density maps obtained by X-ray crystallography [44], and are flexible and dynamic in solution [45]. A popular hypothesis is that the natively disordered proteins are malleable, leading to advantages with respect to functions such as regulation and binding of diverse ligands [39,45].
Amelogenin is a largely hydrophobic molecule with a hydrophilic C-terminus [1], that has low solubility under physiological conditions and tends to form nanosphere assemblies [46]. The amelogenin nanospheres assemble further to form microribbons [38]. Nanospheres of purified bacterially produced recombinant amelogenin have been demonstrated in vitro [46], whereas similar nanostructures are also observed in vivo [47]. Several models of amelogenin assemblies have been proposed [37,38,48]. The common feature among these models is the presence of a hydrophobic core surrounded by a hydrophilic shell. The hydrophilic shell is very likely to be composed of the highly charged C-terminal region (P169ATDKTKREEVD180), which has been referred to as the C-terminal teleopeptide of amelogenin [1,28]. This region defined by residues 169–180 is specifically cleaved shortly after its secretion [28,49], and this proteolytic processing is essential for proper enamel formation [10]. After this cleavage, the stability of the amelogenin assemblies is predicted to decrease as a result of a significant decrease for molecular charge and with a concurrent sharp increase of hydrophobicity at the outer shells. This physical change may also initiate the disassembly of amelogenin nanosphere. Also after the removal of teleopeptide region, this binding region of amelogenin (residues 155–168) would be exposed at the surface of amelogenin nanospheres, and could be made available to the plasma membrane at the Tomes' processes.
The only available experimental structure for tetraspanin family is the crystal structure of CD81 EC2, which has been resolved by Kitadokoro et al. [32,33]. Sequence analyses suggest that all tetraspanin EC2s share a mushroom-like structure, with a highly variable region embedded in the ‘mushroom head’ [50]. The crystal structures of CD81 EC2 have been used to model structures of other tetraspanins, CD53 [50], Q9V3R4 [50] and CD82 [51]. The three-dimensional structure of the CD63 EC2 presented here is built with Phyre using the atomic structure of CD81 EC2 (PDB code 1G8Q) as the template. The ‘mushroom stalk’ of the EC2 is suggested to interact with other tetraspanins in the tetraspanin web [52]. Based on our three-dimensional modelling data, amino acids 165–184 of the CD63 EC2 would be more available and accessible than the EC2 region defined by amino acids 185–205 for intermolecular interactions (e.g. interactions with amelogenin). As supported by the homology analysis (Figure 2B), we postulate that the tetraspanin region (V165PDSCCINVTVGCGINFNEK184) plays a central role in supporting a protein–protein interaction with amelogenin.
CD63, LAMP1 and LAMP2 are ubiquitously expressed and localize to the plasma membrane. They are also present in the membranes of the endosome/late endosome and lysosome [15,17]. A lysosomal targeting motif is present at the C-termini of CD63, LAMP1 and LAMP2 [53], suggesting that they all may employ a similar mechanism in their trafficking from the plasma membrane to the lysosome. Although CD63 and LAMP1 are expressed by ameloblasts at all stages of amelogenesis, CD63 is more highly expressed in late-secretory and post-secretory stages, whereas LAMP1 is more highly expressed in the early secretory stage [19]. Protein interactions between the secreted enamel matrix proteins (such as amelogenin, ameloblastin and enamelin) and membrane-bound proteins (such as CD63, LAMP1 and LAMP2) probably occur at the ameloblast Tomes' processes. Such interactions may be required to establish short-term order of the forming matrix, and/or to mediate feedback signals to the transcriptional machinery of ameloblasts, and/or to quickly remove matrix protein debris during enamel biomineralization.
Acknowledgments
This work was supported by Grants DE006988, DE013045, DE013404 and DE014867 from the National Institutes of Health, National Institute of Dental and Craniofacial Research.
References
- 1.Snead M. L., Zeichner-David M., Chandra T., Robson K. J. H., Woo S. L. C., Slavkin H. C. Construction and identification of mouse amelogenin cDNA clones. Proc. Natl. Acad. Sci. U.S.A. 1983;80:7254–7258. doi: 10.1073/pnas.80.23.7254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fong C. D., Slaby I., Hammarstrom L. Amelin: an enamel-related protein, transcribed in the cells of epithelial root sheath. J. Bone Miner. Res. 1996;11:892–898. doi: 10.1002/jbmr.5650110704. [DOI] [PubMed] [Google Scholar]
- 3.Hu C. C., Fukae M., Uchida T., Qian Q., Zhang C. H., Ryu O. H., Tanabe T., Yamakoshi Y., Murakami C., Dohi N., et al. Cloning and characterization of porcine enamelin mRNAs. J. Dent. Res. 1997;76:1720–1729. doi: 10.1177/00220345970760110201. [DOI] [PubMed] [Google Scholar]
- 4.Iwasaki K., Bajenova E., Somogyi-Ganss E., Miller M., Nguyen V., Nourkeyhani H., Gao Y., Wendel M., Ganss B. Amelotin: a novel secreted, ameloblast-specific protein. J. Dent. Res. 2005;84:1127–1132. doi: 10.1177/154405910508401207. [DOI] [PubMed] [Google Scholar]
- 5.Begue-Kirn C., Krebsbach P. H., Bartlett J. D., Butler W. T. Dentin sialoprotein, dentin phosphoprotein, enamelysin and ameloblastin: tooth-specific molecules that are distinctively expressed during murine dental differentiation. Eur. J. Oral Sci. 1998;106:963–970. doi: 10.1046/j.0909-8836.1998.eos106510.x. [DOI] [PubMed] [Google Scholar]
- 6.Nanci A., Zalzal S., Lavoie P., Kunikata M., Chen W. Y., Krebsbach P. H., Yamada Y., Hammarstrom L., Simmer J. P., Fincham A. G., et al. Comparative immunochemical analyses of the developmental expression and distribution of ameloblastin and amelogenin in rat incisors. J. Histochem. Cytochem. 1998;46:911–934. doi: 10.1177/002215549804600806. [DOI] [PubMed] [Google Scholar]
- 7.Simmer J. P., Fukae M., Tanabe T., Yamakoshi Y., Uchida T., Xue J., Margolis H. C., Shimizu M., DeHart B. C., Hu C. C., Bartlett J. D. Purification, characterization, and cloning of enamel matrix serine proteinase 1. J. Dent. Res. 1998;77:377–386. doi: 10.1177/00220345980770020601. [DOI] [PubMed] [Google Scholar]
- 8.Bartlett J. D., Simmer J. P., Xue J., Margolis H. C., Moreno E. C. Molecular cloning and mRNA tissue distribution of a novel matrix metalloproteinase isolated from porcine enamel organ. Gene. 1996;183:123–128. doi: 10.1016/s0378-1119(96)00525-2. [DOI] [PubMed] [Google Scholar]
- 9.Gibson C. W., Yuan Z. A., Hall B., Longenecker G., Chen E. H., Thyagarajan T., Sreenath T., Wright J. T., Decker S., Piddington R., et al. Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J. Biol. Chem. 2001;276:31871–31875. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- 10.Margolis H. C., Beniash E., Fowler C. E. Role of macromolecular assembly of enamel matrix proteins in enamel formation. J. Dent. Res. 2006;85:775–793. doi: 10.1177/154405910608500902. [DOI] [PubMed] [Google Scholar]
- 11.Wang H. J., Tannukit S., Zhu D. H., Snead M. L., Paine M. L. Enamel matrix protein interactions. J. Bone Miner. Res. 2005;20:1032–1040. doi: 10.1359/JBMR.050111. [DOI] [PubMed] [Google Scholar]
- 12.Stipp C. S., Kolesnikova T. V., Hemler M. E. Functional domains in tetraspanin proteins. Trends Biochem. Sci. 2003;28:106–112. doi: 10.1016/S0968-0004(02)00014-2. [DOI] [PubMed] [Google Scholar]
- 13.Yunta M., Lazo P. A. Tetraspanin proteins as organisers of membrane microdomains and signalling complexes. Cell. Signalling. 2003;15:559–564. doi: 10.1016/s0898-6568(02)00147-x. [DOI] [PubMed] [Google Scholar]
- 14.Berditchevski F. Complexes of tetraspanins with integrins: more than meets the eye. J. Cell Sci. 2001;114:4143–4151. doi: 10.1242/jcs.114.23.4143. [DOI] [PubMed] [Google Scholar]
- 15.Duffield A., Kamsteeg E. J., Brown A. N., Pagel P., Caplan M. J. The tetraspanin CD63 enhances the internalization of the H,K-ATPase β-subunit. Proc. Natl. Acad. Sci. U.S.A. 2003;100:15560–15565. doi: 10.1073/pnas.2536699100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tompkins K., George A., Veis A. Characterization of a mouse amelogenin [A-4]/M59 cell surface receptor. Bone. 2006;38:172–180. doi: 10.1016/j.bone.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 17.Cook N. R., Row P. E., Davidson H. W. Lysosome associated membrane protein 1 (Lamp1) traffics directly from the TGN to early endosomes. Traffic. 2004;5:685–699. doi: 10.1111/j.1600-0854.2004.00212.x. [DOI] [PubMed] [Google Scholar]
- 18.Kannan K., Stewart R. M., Bounds W., Carlsson S. R., Fukuda M., Betzing K. W., Holcombe R. F. Lysosome-associated membrane proteins h-LAMP1 (CD107a) and h-LAMP2 (CD107b) are activation-dependent cell surface glycoproteins in human peripheral blood mononuclear cells which mediate cell adhesion to vascular endothelium. Cell. Immunol. 1996;171:10–19. doi: 10.1006/cimm.1996.0167. [DOI] [PubMed] [Google Scholar]
- 19.Shapiro J. L., Wen X., Okamoto C. T., Wang H. J., Lyngstadaas S. P., Goldberg M., Snead M. L., Paine M. L. Cellular uptake of amelogenin, and its localization to CD63, and Lamp1-positive vesicles. Cell. Mol. Life Sci. 2007;64:244–256. doi: 10.1007/s00018-006-6429-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook J., Russell D. W. New York: Cold Spring Harbor Laboratory Press; 2001. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 21.Paine M. L., Krebsbach P. H., Chen L. S., Paine C. T., Yamada Y., Deutsch D., Snead M. L. Protein-to-protein interactions: criteria defining the assembly of the enamel organic matrix. J. Dent. Res. 1998;77:496–502. doi: 10.1177/00220345980770030901. [DOI] [PubMed] [Google Scholar]
- 22.Adamczak R., Porollo A., Meller J. Combining prediction of secondary structure and solvent accessibility in proteins. Proteins. 2005;59:467–475. doi: 10.1002/prot.20441. [DOI] [PubMed] [Google Scholar]
- 23.Koh I. Y. Y., Eyrich V. A., Marti-Renom M. A., Przybylski D., Madhusudhan M. S., Eswar N., Grana O., Pazos F., Valencia A., Sali A., Rost B. EVA: evaluation of protein structure prediction servers. Nucleic Acids Res. 2003;31:3311–3315. doi: 10.1093/nar/gkg619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dosztanyi Z., Csizmok V., Tompa P., Simon I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J. Mol. Biol. 2005;347:827–839. doi: 10.1016/j.jmb.2005.01.071. [DOI] [PubMed] [Google Scholar]
- 25.Jin Y. M., Dunbrack R. L., Jr Assessment of disorder predictions in CASP6. Proteins. 2005;61:167–175. doi: 10.1002/prot.20734. [DOI] [PubMed] [Google Scholar]
- 26.Kelley L. A., MacCallum R. M., Sternberg M. J. E. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 2000;299:499–520. doi: 10.1006/jmbi.2000.3741. [DOI] [PubMed] [Google Scholar]
- 27.Simmer J. P., Hu C. C., Lau E. C., Sarte P., Slavkin H. C., Fincham A. G. Alternative splicing of the mouse amelogenin primary RNA transcript. Calcif. Tissue Int. 1994;55:302–310. doi: 10.1007/BF00310410. [DOI] [PubMed] [Google Scholar]
- 28.Paine M. L., Wang H. J., Snead M. L. Amelogenin self-assembly and the role of the proline located within the carboxyl-teleopeptide. Connect. Tissue Res. 2003;44:52–57. [PubMed] [Google Scholar]
- 29.Andrejewski N., Punnonen E. L., Guhde G., Tanaka Y., Lullmann-Rauch R., Hartmann D., von Figura K., Saftig P. Normal lysosomal morphology and function in LAMP1-deficient mice. J. Biol. Chem. 1999;274:12692–12701. doi: 10.1074/jbc.274.18.12692. [DOI] [PubMed] [Google Scholar]
- 30.Eskelinen E. L., Schmidt C. K., Neu S., Willenborg M., Fuertes G., Salvador N., Tanaka Y., Lullmann-Rauch R., Hartmann D., Heeren J., et al. Disturbed cholesterol traffic but normal proteolytic function in LAMP-1/LAMP-2 double-deficient fibroblasts. Mol. Biol. Cell. 2004;15:3132–3145. doi: 10.1091/mbc.E04-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paine M. L., Snead M. L. Protein interactions during assembly of the enamel organic extracellular matrix. J. Bone Miner. Res. 1997;12:221–227. doi: 10.1359/jbmr.1997.12.2.221. [DOI] [PubMed] [Google Scholar]
- 32.Kitadokoro K., Bordo D., Galli G., Petracca R., Falugi F., Abrignani S., Grandi G., Bolognesi M. CD81 extracellular domain 3D structure: insight into the tetraspanin superfamily structural motifs. EMBO J. 2001;20:12–18. doi: 10.1093/emboj/20.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kitadokoro K., Ponassi M., Galli G., Petracca R., Falugi F., Grandi G., Bolognesi M. Subunit association and conformational flexibility in the head subdomain of human CD81 large extracellular loop. Biol. Chem. 2002;383:1447–1452. doi: 10.1515/BC.2002.164. [DOI] [PubMed] [Google Scholar]
- 34.Pautard F. G. An X-ray diffraction pattern from human enamel matrix. Arch. Oral Biol. 1961;3:217–220. doi: 10.1016/0003-9969(61)90139-x. [DOI] [PubMed] [Google Scholar]
- 35.Angmar-Mansson B. A polarization microscopic and micro X-ray diffraction study on organic matrix of developing human enamel. Arch. Oral Biol. 1971;16:147–156. doi: 10.1016/0003-9969(71)90102-6. [DOI] [PubMed] [Google Scholar]
- 36.Jodaikin A., Weiner S., Perltreves D., Traub W., Termine J. D. Developing enamel matrix proteins: a conformation study of enamelins and amelogenins. Int. J. Biol. Macromol. 1987;9:166–168. [Google Scholar]
- 37.Aichmayer B., Margolis H. C., Sigel R., Yamakoshi Y., Simmer J. P., Fratzl P. The onset of amelogenin nanosphere aggregation studied by small-angle X-ray scattering and dynamic light scattering. J. Struct. Biol. 2005;151:239–249. doi: 10.1016/j.jsb.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 38.Du C., Falini G., Fermani S., Abbott C., Moradian-Oldak J. Supramolecular assembly of amelogenin nanospheres into birefringent microribbons. Science. 2005;307:1450–1454. doi: 10.1126/science.1105675. [DOI] [PubMed] [Google Scholar]
- 39.Shaw W. J., Campbell A. A., Paine M. L., Snead M. L. The COOH terminus of the amelogenin, LRAP, is oriented next to the hydroxyapatite surface. J. Biol. Chem. 2004;279:40263–40266. doi: 10.1074/jbc.C400322200. [DOI] [PubMed] [Google Scholar]
- 40.Goto Y., Kogure E., Takagi T., Aimoto S., Aoba T. Molecular conformation of porcine amelogenin in solution: 3 folding units at the N-terminal, central, and C-terminal regions. J. Biochem. (Tokyo) 1993;113:55–60. doi: 10.1093/oxfordjournals.jbchem.a124003. [DOI] [PubMed] [Google Scholar]
- 41.Zheng S. D., Tu A. T., Renugopalakrishnan V., Strawich E., Glimcher M. J. A mixed β-turn and β-sheet structure for bovine tooth enamel amelogenin: Raman-spectroscopic evidence. Biopolymers. 1987;26:1809–1813. doi: 10.1002/bip.360261012. [DOI] [PubMed] [Google Scholar]
- 42.Dyson H. J., Wright P. E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 43.Termine J. D., Torchia D. A. 13C-1H magnetic double-resonance study of fetal enamel matrix proteins. Biopolymers. 1980;19:741–750. doi: 10.1002/bip.1980.360190311. [DOI] [PubMed] [Google Scholar]
- 44.Ward J. J., Sodhi J. S., McGuffin L. J., Buxton B. F., Jones D. T. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 45.Fink A. L. Natively unfolded proteins. Curr. Opin. Struct. Biol. 2005;15:35–41. doi: 10.1016/j.sbi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 46.Moradian-Oldak J., Simmer J. P., Lau E. C., Sarte P. E., Slavkin H. C., Fincham A. G. Detection of monodisperse aggregates of a recombinant amelogenin by dynamic light-scattering. Biopolymers. 1994;34:1339–1347. doi: 10.1002/bip.360341006. [DOI] [PubMed] [Google Scholar]
- 47.Robinson C., Fuchs P., Weatherell J. A. The appearance of developing rat incisor enamel using a freeze fracturing technique. J. Cryst. Growth. 1981;53:160–165. [Google Scholar]
- 48.Snead M. L. Amelogenin protein exhibits a modular design: implications for form and function. Connect. Tissue Res. 2003;44:47–51. [PubMed] [Google Scholar]
- 49.Bartlett J. D., Simmer J. P. Proteinases in developing dental enamel. Crit. Rev. Oral Biol. Med. 1999;10:425–441. doi: 10.1177/10454411990100040101. [DOI] [PubMed] [Google Scholar]
- 50.Seigneuret M., Delaguillaumie A., Lagaudriere-Gesbert C., Conjeaud H. Structure of the tetraspanin main extracellular domain: a partially conserved fold with a structurally variable domain insertion. J. Biol. Chem. 2001;276:40055–40064. doi: 10.1074/jbc.M105557200. [DOI] [PubMed] [Google Scholar]
- 51.Bienstock R. J., Barrett J. C. KAI1, a prostate metastasis suppressor: prediction of solvated structure and interactions with binding partners: integrins, cadherins, and cell-surface receptor proteins. Mol. Carcinog. 2001;32:139–153. doi: 10.1002/mc.1073. [DOI] [PubMed] [Google Scholar]
- 52.Levy S., Shoham T. The tetraspanin web modulates immune-signalling complexes. Nat. Rev. Immunol. 2005;5:136–148. doi: 10.1038/nri1548. [DOI] [PubMed] [Google Scholar]
- 53.Rous B. A., Reaves B. J., Ihrke G., Briggs J. A. G., Gray S. R., Stephens D. J., Banting G., Luzio J. P. Role of adaptor complex AP-3 in targeting wild-type and mutated CD63 to lysosomes. Mol. Biol. Cell. 2002;13:1071–1082. doi: 10.1091/mbc.01-08-0409. [DOI] [PMC free article] [PubMed] [Google Scholar]